Abstract
We report the variation with temperature of the ensemble distribution of conformations spanned by the tau protein in its dynamical states measured by small-angle X-ray scattering (SAXS) using synchrotron radiation. The SAXS data show a clear temperature variation of the distribution of occupied protein conformations from 293 to 318 K. More conformations with a smaller radius of gyration are occupied at higher temperature. The protein–solvent interactions are shown by computer simulation to be essential for controlling the dynamics of protein conformations, providing evidence for the key role of water solvent in the protein dynamics, as proposed by Giorgio Careri.
Keywords: Protein conformations, Intrinsic disordered proteins, Water effect in conformational landscape, Temperature effects in protein fluctuations
Introduction
From the “lock-and- key” model to the dynamics-function relation
The dogma that the function of a protein depends critically on its fixed three-dimensional structure was rapidly accepted after the discovery of the DNA and protein structures by X-ray diffraction in the 1950s. Enzymes have been described as being able to bind to specific substrates because their shapes match perfectly, with the ‘lock-and-key’ mechanical model first proposed by chemist Emil Fischer in 1894. The genome encodes a molecular primary structure that, once expressed, folds in a secondary and tertiary structure. The expressed structure, following the lock-and-key paradigm, allows protein–protein or protein–ligand recognition, and therefore the protein function in the living cell. In the 1970s, the general validity of the ‘lock-and-key’ paradigm was questioned by Giorgio Careri with Paolo Fasella and Emilio Gratton [1, 2], who proposed a key role for fluctuations of protein conformations in biochemical reactions, described as statistical events. The main point was the scenario of proteins as macromolecules fluctuating between quasi-stationary states [3], their dynamics being a key feature for understanding living matter. As a matter of fact, it was observed that in some proteins the binding site was not uniquely well defined. Paolo Fasella, a close collaborator of Giorgio Careri and inspired by Giorgio’s discussions, focused on the brain protein calmodulin (CaM: CALcium MODULated proteIN). Calmodulin is expressed in all eukaryotic cells that bind to and regulate a number of different protein targets, thereby affecting many different cellular functions such as short-term and long-term memory, nerve growth and the immune response, inflammation, metabolism, apoptosis, smooth muscle contraction, and intracellular movement. The instantaneous structure of the conformations of the active site of calmodulin and other Ca-binding proteins in solution [4, 5] was measured by a new fast structural method “X-ray absorption near-edge structure” (XANES), probing the higher-order correlation functions of the local structure with a time scale of 10 − 13 s [6, 7]. The instantaneous structure of these proteins in solution was found to deviate from the averaged structure of the crystallized protein detected by X-ray diffraction [8], showing the existence of fast protein fluctuations between different conformations. Calmodulin and parvalbumin [9–11] were later identified as flexible biological macromolecules that show an intrinsic dynamical state spanning a wide set of protein conformations determined by NMR [12]. The set of time-resolved protein conformations and the percent of occupation time of the multiple protein conformational states has been confirmed by other physical methods probing the protein dynamics such as inelastic neutron scattering and dielectric methods [13], a method that was used by G. Careri to investigate the dynamics of proteins controlled by the solvent [14].
In the first 10 years of this century, the key role of protein dynamics and flexibility has attracted a wide scientific audience due to the identification of a large number of proteins called intrinsic disordered proteins (IDPs) [15, 16]. The number IDPs that perform their biological function having no fixed structural shape is rapidly growing [17]. Their functionality is related to their dynamics, as proposed by Careri and collaborators [1], rather than to the fixed structure needed for the lock-and-key model.
In the same first 10 years of the XXI century, the description of the cell as an ensemble of networks has become widely accepted [18]. Protein function in cell organization has been associated with protein–protein interaction networks in the cell where most of the IDPs play the key role of “hub” proteins. The fluctuations of IDPs between multiple conformations as shown in Fig. 1 allow these proteins to bind to a wide range of different target proteins. IDPs constantly probe and sense signals from many partners in the complex cellular environment, i.e., by fluctuating between multiple conformations, they express the capability of many links. In these networks, the protein conformations can be described as nodes, and the potential barriers as links in “link-weighted” networks [19]. The network’s dynamical state is determined by the landscape of the potential barriers that separate the states of quasi-stationary protein conformations. Therefore, the network topology with weighted links based on the data of the complex energy landscape can be used to determine the fluctuations and correlations in the network dynamics [20].
Fig. 1.
Pictorial view of the intrinsic fluctuations of the first flexible protein: the tau protein (some of the multiple conformers of the tau protein are shown in the lower part of the figure connected by the dashed line) in its dynamical functional state. The biological function of the protein in the cell is determined by coherent biochemical reactions as proposed by G. Careri in the seventies. The picture shows a second fluctuating protein that fluctuates between different landscapes of multiple protein conformations (pictorially indicated by the different geometrical multiform shapes inside the solid line in the upper part of the figure) with complex association and dissociation molecular processes controlling the biochemical reactions between flexible proteins
Between intrinsic disordered proteins, much attention has been addressed to the cellular transcription factor “cAMP response element-binding” (CREB), a gene-regulatory protein involved in learning and memory, and CREB-binding protein (CBP). Several bonds form cooperatively within CREB and with CBP [21]. A well-known IDP protein is the tumor suppressor p53 [22], a hub in the multiple signaling networks implicated in human cancer. The key intrinsic dynamics of IDPs relating to their functionality has been shown in the case of the Sic1 protein [23], the cyclin-dependent kinase (CDK) inhibitor that interacts with its receptor Cdc4 [24]. The complex is a mixture of different conformations shifting around dynamically. The observation of this unusual binding mode between Sic1 and Cdc4 extends the understanding of protein interactions from predominantly static complexes to a new scenario of dynamic ensembles of intrinsically fluctuating quasi-stationary states.
Protein conformational fluctuations are relevant also for folded proteins [25–27] and today are considered an essential key feature for understanding protein function as proposed by Careri [2]. The investigation of the dynamics of rebinding of ligands and relaxation in the myoglobin [28] pocket has been a first case where it has been shown that the water solvent has a key role in slaving the protein fluctuations [29] in the protein energy landscape [30–32]. The protein fluctuations between conformers have been found to be slaved by fluctuations in the hydration shell measured by dielectric relaxation spectroscopy [33–35].
The case of tau protein
The dynamics of a protein is expected to depend on environmental conditions, for example on temperature and on solvent. Here, we have investigated the role of temperature and solvent on the landscape of protein conformations spanned by the tau protein, one of the largest IDPs. Because of its intrinsically disordered character, tau in solution explores a large region of the conformational space, fluctuating between a large number of conformers. Tau is a microtubule-associated protein expressed primarily in neurons. It is found in the human central nervous system (CNS) in six isoforms, ranging from 352 to 441 amino acids. In its physiological state, tau promotes the growth and assembly of microtubules [36, 37]; but under pathological conditions, the same tau aggregates in paired helical filaments (PHFs). This phenomenon prevents the tau protein from carrying out its physiological stabilization role, and, together with several other factors, is associated with the degeneration of the microtubules and the death of neurons [38]. Tau entails no permanent secondary structures, and the aggregation in PHFs is supposed to start from a segment forming a transient β-structure. As the application to the monomeric tau protein of standard structural techniques, such as macromolecular crystallography or electron microscopy, is not possible, small-angle X-ray scattering provides a unique tool to obtain structural information on tau in solution and, more generally, on the whole IDP category [39–46].
In this work, we investigate the influence of temperature and solvent on the behavior of the longest CNS tau isoform, htau40 (441 residues, and molecular mass 45.8 kD [43]). For the former, we report SAXS measurements at 293 K and 318 K; as for the latter, we have performed a Molecular Dynamics computer simulation of tau in water or methanol solvent.
Materials and methods
Protein preparation
The htau40 protein was purchased from Sigma-Aldrich (St. Louis, MO). The protein powder was reconstituted in 50 mM MES (pH = 6.8), 100 mM NaCl, and 0.5 mM EGTA, desalted with standard procedures, and concentrated to a nominal protein concentration of 2 mg/ml in 0.1 M PBS (pH = 6.8). The solution was centrifuged for 10 min at 10,000 × g and the supernatant was filtered to eliminate aggregates. Protein quality was assayed by SDS-PAGE in 12% (w/v) polyacrylamide. The gels were stained with Coomassie brilliant blue R-250. The SDS-PAGE analysis revealed the occurrence of a major protein band with the expected size (approx. 45 kDa) and a 90% purity. As described in the following section, the protein was subjected to temperature variation in situ during the SAXS measurements. The temperature variation does not induce protein aggregation, as demonstrated by SDS-PAGE analysis at varying temperatures.
Small-angle X-ray scattering (SAXS) measurements
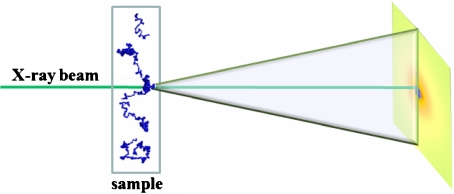
SAXS measurements [44, 45] have been acquired at the BioSAXS beamline ID 14–3 at ESRF (Grenoble, France) [46]. A pictorial view of the typical experimental geometry of a SAXS experiment is presented in Fig. 2. A volume of 50 μl of solution has been placed in a 1.8-mm-diameter quartz capillary with a few tens of microns wall thickness. Ten consecutive 2-s exposures were performed at constant temperature without observing any radiation damage. SAXS profiles have been acquired at the standard room temperature of 293 K; the temperature effect has been studied at 318 K after a temperature equilibration in the capillary sample holder. An exposure time of 3 s was used. Measurements at different temperatures have been acquired at different positions of the capillary sample holder, in order to further exclude the presence of radiation damage. Solvent scattering was measured to allow for subtraction of the background scattering.
Fig. 2.
Pictorial view of the small angle X-ray scattering experiment using high luminance synchrotron radiation X-ray beam to probe the low-resolution shape of the fluctuating tau protein conformations in the solvent
Ensemble optimization method
An explicit description of the structural ensemble of the tau protein can be obtained by the software RANCH, which produces an ensemble of conformers. The ensemble is based on a data bank of residue-specific (ψ, ϕ) values, derived from coil segments of globular proteins, and takes properly into account the coexistence of multiple conformations in solution. This ensemble is then optimized through the genetic algorithm GAJOE, which selects those conformers that, on average, best reproduce (using the program CRYSOL) the SAXS experimental curve.
Molecular dynamics simulation
A 3D structure of tau protein was produced following a procedure described in detail in [47]. First, an extended 3D structure was obtained from its primary sequence using the program VMD [48], and placed in a large box with periodic boundary conditions (box volume = 15,253 nm3). A dynamic evolution in vacuo at 300 K was performed with the package GROMACS using the ffG53a6 force field [49], which led to a rapid compaction of the molecule, as monitored through its gyration radius. This evolution was stopped when the gyration radius reached its experimental average value Rg = 6.57 nm [50]; at this point the box was filled with about 5 × 105 water molecules (space water model) or with about 2.5 × 105 methanol molecules [51]; the energy of the system was then minimized, and a short equilibration was performed, first at constant temperature (modified Berendsen thermostat) and volume, then at constant temperature and pressure (Parrinello–Rahman pressure coupling). The conformation produced by this procedure was used as the starting point to simulate the dynamic evolution in time of the protein, using GROMACS with a time step of 2 fs.
Results
SAXS results
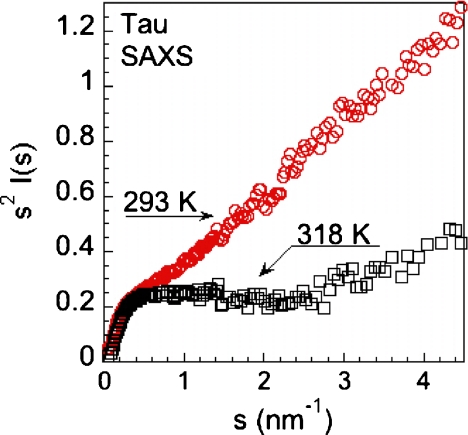
In Fig. 3, we report the Kratky plot of the tau protein measured at two different temperatures, 293 K and 318 K. In the Kratky plot, the scattered intensity profile I(s) is multiplied by the square of the scattering wave vector s2 (i.e., s2I(s) versus s). The scattering profile for an unfolded state of a molecule falls off as 1/s at high values of the exchanged momentum s. The clearly defined surface of a folded protein leads to a scattering profile which falls off as 1/s4 [46]. Therefore, the occurrence of a well-defined peak in the Kratky plot indicates that the molecule is in a compact state.
Fig. 3.
The small angle X-ray diffraction intensity I(s) multiplied by the square of the momentum transfer s, called a Kratky plot, as a function of the momentum transfer s of the tau protein at two different temperatures
The increase of s2I(s) at high value of momentum transfer s, observed in Fig. 3, points out that the tau protein has an expanded disordered state at both temperatures, in our experimental conditions. It can be also noted that the Kratky plot, in the intermediate s region, increases faster at 293 K than at 318 K.
EOM method
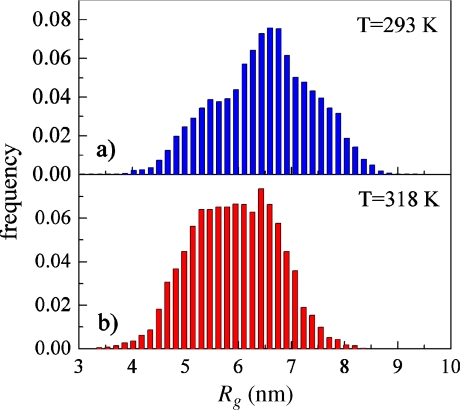
A pool of 10,000 representative backbone models of the tau protein were created using the program RANCH. The corresponding theoretical scattering intensity of the models was calculated by the software CRYSOL. The genetic algorithm GAJOE was then employed, at both experimental temperatures, to select from the set of theoretical scattering curves an ensemble that provided the best fit of the experimental SAXS data. In Fig. 4, we report the distribution of the tau conformations spanned by the tau protein characterized by their radius of gyration (Rg) at 293 K (panel a) and at 318 K (panel b). Given the multiple nature of the conformations of the molecule, the average Rg of the tau protein corresponds to the average computed over the selected ensemble of conformers. This average turns out to be 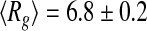 nm at T = 293 K, and
nm at T = 293 K, and 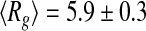 nm at T = 318 K. A close inspection of the selected distribution highlights that these values derive from an increased propensity of tau to populate more compact conformational states (
nm at T = 318 K. A close inspection of the selected distribution highlights that these values derive from an increased propensity of tau to populate more compact conformational states (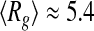 nm) at the higher temperature, rather than extended states (peak at
nm) at the higher temperature, rather than extended states (peak at 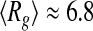 nm) at the lower one. This behavior is consistent with the information provided by the Kratky plots, which indicates an overall compaction of the tau protein with increasing temperature.
nm) at the lower one. This behavior is consistent with the information provided by the Kratky plots, which indicates an overall compaction of the tau protein with increasing temperature.
Fig. 4.
The distribution of different conformations of the tau protein characterized by their radius of gyration at 293 K (a) and at 318 K (b)
Molecular simulation results
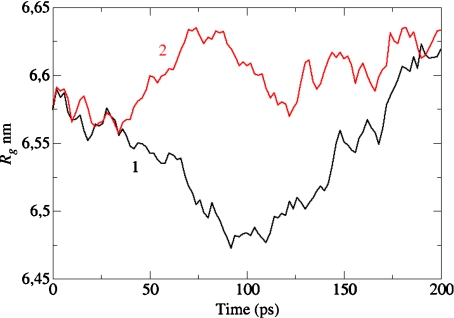
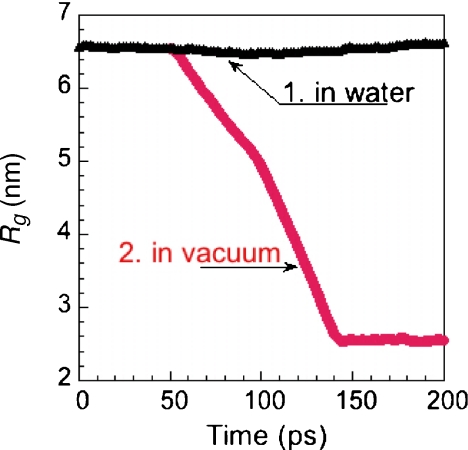
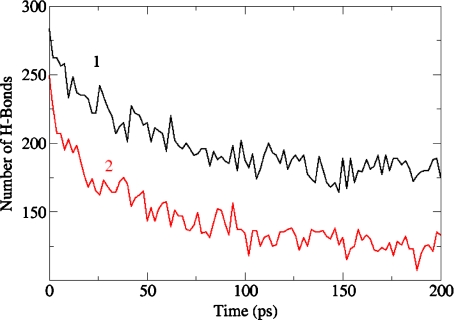
We performed a MD simulation of a tau molecule embedded in water or methanol solvent, using the GROMACS package specified before, at T = 300 K. In Fig. 5, we report the value of the gyration radius Rg in water (curve #1) and in methanol (curve #2) during a segment of 200 ps; during these 200 ps the gyration radius oscillates in both cases around its experimental average value Rg = 6.6 nm [51]. If at time t = 50 ps of this segment one takes away the water molecules and lets the tau molecule evolve in vacuo, the latter collapses to a compact form (Rg = 2.5 nm) in less than 100 ps (Fig. 6, curve #2), and keeps this form afterwards. We have also measured, in the same time interval, the number of intramolecular hydrogen bonds in tau, and in Fig. 7 we report the results in water (curve #1) and in methanol (curve #2). Both curves show an initial decrease in the number of H-bonds, due to their replacement by H-bonds between tau and solvent; after 200 ps these numbers appear to be stabilized around 175 in water, and around 125 in methanol.
Fig. 5.
Molecular dynamics calculation at T = 300 K of the conformations spanned in time by the tau protein, characterized by their gyration radius Rg in water (curve #1) and in methanol (curve #2)
Fig. 6.
Molecular dynamics calculation at T = 300 K. Gyration radius Rg of tau in water (curve #1). Removing the water and running the calculations in vacuo (curve #2), induces a rapid evolution of the protein toward collapsed conformations on a time scale of 100 ps
Fig. 7.
Molecular dynamics calculation at T = 300 K. Number of intramolecular hydrogen bonds of tau in water (curve #1) and in methanol (curve #2)
Discussion
The free energy landscape of disordered proteins is characterized by small potential barriers between conformers, and depends on solvent conditions [40]. We have shown that the tau protein dynamics in this landscape depends on the temperature, which affects the probability of the system to be found in different regions of the accessible phase space. We found at 318 K a large variation of the SAXS spectrum of tau from the spectrum at 293 K, corresponding to a compaction of the molecule as in the Sic1 protein [23]. This compaction suggests that it is related to the different time spent by the protein in different parts of the phase space region at the two temperatures. Figure 4 shows that the two distributions of the conformers, as monitored through the values of the gyration radius Rg, show an increase with temperature of the population of conformers characterized by smaller values of Rg, while at the lower temperature extended conformers are preferred. This interpretation is consistent with recent results, showing that the average radius of gyration in the intrinsically disordered protein prothymosin α decreases with increasing temperature [41].
As already mentioned, the tau protein is one of the largest totally disordered IDPs [39], and the solvent is expected to play an important role in the dynamical evolution of the conformational landscape of the protein. We have performed a computer simulation of tau in water and in methanol, which provides a hint as to the relation between the overall shape of tau and the solvent–protein interaction. It is interesting to notice that the two solvents behave quite differently in the replacement of intramolecular H-bonds, as shown in Fig. 7, but this difference does not seem to be of relevance for the overall configuration of tau, the value of Rg after 200 ps being almost the same in both cases, as shown in Fig. 5. On the other hand, we have found that the absence of water effectively induces a dramatic change in the overall shape of tau, and highlights the critical role of the solvent in determining the configuration of the molecule. The case study described in the present work is just an example of a growing body of properties that characterize the relation between a biomolecule’s function and its environment.
Conclusions
Protein dynamics is being accepted as the key feature of protein function, as proposed by Careri [2]. Both temperature and solvent can affect the dynamics of the protein, in different ways; therefore, we have separately tested their influence on the behavior of tau, an intrinsically disordered protein. Our experimental and simulation results point out that the tau protein undergoes a significant reduction in the average protein radius of gyration, both when the temperature is raised and when the solvent is removed. The larger temperature drives the molecule’s trajectory preferentially in to phase space regions characterized by conformations with more compact shapes. On the other hand, the removal of the solvent changes completely the phase space landscape, inducing an even more drastic compaction of the molecule. Unveiling the complexity of protein dynamics between multiple quasi-stationary conformational states will be a hot topic in next years as the development of new time-resolved experiments join with space-resolved nanoscale microscopy to reveal space time correlation dynamics [52, 53]. Further investigations of coherence in living matter could be unveiled if quantum phenomena such as quantum criticality and shape resonances [54, 55] are found to underlie the mysterious dynamical functionality of intrinsically disordered proteins.
References
- 1.Careri G, Fasella P, Gratton E. Statistical time events in enzymes: a physical assessment. CRC Crit. Rev. Biochem. 1975;3:141–164. doi: 10.3109/10409237509102555. [DOI] [PubMed] [Google Scholar]
- 2.Careri, G.: Order and Disorder in Matter. Benjamin/Cummings Pub. Co., Advanced Book Program. ISBN: 9780805317251 (1984)
- 3.Frauenfelder H, Parak F, Young RD. Conformational sub-states in proteins. Ann. Rev. Biophys. Biophys. Chem. 1988;17:451–479. doi: 10.1146/annurev.bb.17.060188.002315. [DOI] [PubMed] [Google Scholar]
- 4.Bianconi A, Oesh B, Alemà S, Castellani L, Davoli I, Fasella P, Mobilio S, et al. Structure of the calcium binding sites in troponin-C and calmodulin studied by EXAFS. In: Siegel FL, et al., editors. Calcium Binding Proteins: Structure and Function. Amsterdam: Elsevier North Holland; 1980. p. 297. [Google Scholar]
- 5.Bianconi A, Giovannelli A, Castellani L, Alemà S, Fasella P, Oesch B, Mobilio S. XANES determination of calcium sites of troponin-C and parvalbumin. J. Mol. Biol. 1983;165:125–138. doi: 10.1016/S0022-2836(83)80246-0. [DOI] [PubMed] [Google Scholar]
- 6.Bianconi A, Doniach S, Lublin D.X-ray Ca K edge of calcium adenosine triphosphate system and of simple Ca compounds Chem. Phys. Lett. 197859121–124.1978CPL....59..121B 10.1016/0009-2614(78)85629-2 [DOI] [Google Scholar]
- 7.Longa S, Soldatov A, Pompa M, Bianconi A. Atomic and electronic structure probed by X-ray absorption spectroscopy: Full multiple scattering analysis with the G4XANES package. Comput. Mater. Sci. 1995;4:199–210. doi: 10.1016/0927-0256(95)00027-N. [DOI] [Google Scholar]
- 8.Kretsinger RH. Structure and evolution of calcium modulated proteins. CRC Crit. Rev. Biochem. 1980;8:119–174. doi: 10.3109/10409238009105467. [DOI] [PubMed] [Google Scholar]
- 9.Chou JJ, Li S, Klee CB, Bax A. Solution structure of Ca2 + –calmodulin reveals flexible hand-like properties of its domains. Nat. Struct. Mol. Biol. 2001;8:990–997. doi: 10.1038/nsb1101-990. [DOI] [PubMed] [Google Scholar]
- 10.Permyakov SE, Bakunts AG, Denesyuk AI, Knyazeva EL, Uversky VN, Permyakov EA. Apo-parvalbumin as an intrinsically disordered protein. Proteins. 2008;72:822–836. doi: 10.1002/prot.21974. [DOI] [PubMed] [Google Scholar]
- 11.Motáčková V, Nováček J, Zawadzka-Kazimierczuk A, Kazimierczuk K, Zídek L, Sanderová H, Krásný L, Koźmiński W, Sklenář V. Strategy for complete NMR assignment of disordered proteins with highly repetitive sequences based on resolution-enhanced 5D experiments. J. Biomol. NMR. 2010;48:169–177. doi: 10.1007/s10858-010-9447-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bertini I, Kursula P, Luchinat C, Parigi G, Vahokoski J, Willmans M, Yuan J. Accurate solution structures of proteins from X-ray data and minimal set of NMR data: calmodulin peptide complexes as examples. J. Am. Chem. Soc. 2009;131:5134–5144. doi: 10.1021/ja8080764. [DOI] [PubMed] [Google Scholar]
- 13.Chen G, Fenimore PW, Frauenfelder H, Mezei F, Swenson J, Young RD.Protein fluctuations explored by inelastic neutron scattering and dielectric relaxation spectroscopy Phil. Mag. 2008883877–3883.2008PMag...88.3877C 10.1080/14786430802585117 [DOI] [Google Scholar]
- 14.Careri G, Giansanti A, Rupley JA.Proton percolation on hydrated lysozyme powders Proc. Nat. Acad. Sci. USA 1986836810–6814.1986PNAS...83.6810C 10.1073/pnas.83.18.6810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dyson HJ, Wright PE. Intrinsically unstructured proteins and their functions. Nat. Rev. Mol. Cell Biol. 2005;6:197–208. doi: 10.1038/nrm1589. [DOI] [PubMed] [Google Scholar]
- 16.Chouard T.Structural biology: breaking the protein rules Nature 2011471151–153.2011Natur.471..151C 10.1038/471151a [DOI] [PubMed] [Google Scholar]
- 17.Lobanov MY, Furletova EI, Bogatyreva NS, Roytberg MA, Galzitskaya OV. Library of disordered patterns in 3D protein structures. PLoS Comput. Biol. 2010;6:e1000958+. doi: 10.1371/journal.pcbi.1000958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Barabási A-L, Oltvai ZN. Network biology: understanding the cell’s functional organization. Nat. Rev. Genet. 2004;5:101–113. doi: 10.1038/nrg1272. [DOI] [PubMed] [Google Scholar]
- 19.Bianconi G.Emergence of weight-topology correlations in complex scale-free networks Europhys. Lett. 2005711029–1035.2005EL.....71.1029B 10.1209/epl/i2005-10167-2 [DOI] [Google Scholar]
- 20.Bradde S, Caccioli F, Dall’Asta L, Bianconi G.Critical fluctuations in spatial complex networks Phys. Rev. Lett. 20101042187012010PhRvL.104u8701B 10.1103/PhysRevLett.104.218701 [DOI] [PubMed] [Google Scholar]
- 21.Sugase K, Dyson HJ, Wright PE.Mechanism of coupled folding and binding of an intrinsically disordered protein Nature 20074471021–1025.2007Natur.447.1021S 10.1038/nature05858 [DOI] [PubMed] [Google Scholar]
- 22.Oldfield CJ, Meng J, Yang JY, Yang MQ, Uversky VN, Dunker AK. Flexible nets: disorder and induced fit in the associations of p53 and 14–3–3 with their partners. BMC Genomics. 2008;9(Suppl 1):S1. doi: 10.1186/1471-2164-9-S1-S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brocca S, Testa L, Sobott F, Šamalikova M, Natalello A, Papaleo E, Lotti M, Gioia L, Doglia SM, Alberghina L, Grandori R.Compaction properties of an intrinsically disordered protein: Sic1 and its kinase inhibitor domain Biophys. J. 20111002243–2252.2011BpJ...100.2243B 10.1016/j.bpj.2011.02.055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mittag T, Orlicky S, Choy W-Y, Tang X, Lin H, Sicheri F, Kay LE, Tyers M, Forman-Kay JD.Dynamic equilibrium engagement of a polyvalent ligand with a single-site receptor Proc. Nat. Acad. Sci. USA 200810517772–17777.2008PNAS..10517772M 10.1073/pnas.0809222105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang LC, Pang YX, Holder T, Brender JR, Kurochkin AV.Functional dynamics in the active site of the ribonuclease binase Proc. Natl. Acad. Sci. USA 2001987684–7689.2001PNAS...98.7684W 10.1073/pnas.121069998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Eisenmesser EZ, Bosco DA, Akke M, Kern D.Enzyme dynamics during catalysis Science 20022951520–1523.2002Sci...295.1520E 10.1126/science.1066176 [DOI] [PubMed] [Google Scholar]
- 27.Lindorff-Larsen K, Best RB, DePristo MA, Dobson CM, Vendruscolo M.Simultaneous determination of protein structure and dynamics Nature 2005433128–132.2005Natur.433..128L 10.1038/nature03199 [DOI] [PubMed] [Google Scholar]
- 28.Bianconi A, Congiu-Castellano A, Durham PJ, Hasnain SS, Phillips S.The CO bond angle of carboxymyoglobin determined by angular-resolved XANES spectroscopy Nature 1985318685–687.1985Natur.318..685B 10.1038/318685a0 [DOI] [PubMed] [Google Scholar]
- 29.Ansari A, Berendzen J, Braunstein D, Cowen BR, Frauenfelder H, Hong MK, Iben IET, Johnson, Ormos P, Sauke TB. Rebinding and relaxation in the myoglobin pocket. Biophys. Chem. 1987;26:337–355. doi: 10.1016/0301-4622(87)80034-0. [DOI] [PubMed] [Google Scholar]
- 30.Frauenfelder H, Sligar SG, Wolynes PG.The energy landscapes and motions of proteins Science 19912541598–1603.1991Sci...254.1598F 10.1126/science.1749933 [DOI] [PubMed] [Google Scholar]
- 31.Frauenfelder H. Energy landscape and dynamics of biomolecules. J. Biol. Phys. 2005;31:413–416. doi: 10.1007/s10867-005-0696-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bianconi A, Natali F, Alosi ML, Grande S, Lanzara A, Saini NL, Brunelli M, TDXAS study of the conformational landscape of MbCO. J. Synchrotron Radiat. 1999;6:389–391. doi: 10.1107/S0909049598017063. [DOI] [PubMed] [Google Scholar]
- 33.Fenimore PW, Frauenfelder H, McMahon BH, Parak FG.Slaving: Solvent fluctuations dominate protein dynamics and functions Proc. Nat. Acad. Sci. USA 20029916047–16051.2002PNAS...9916047F 10.1073/pnas.212637899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fenimore P, Frauenfelder H, McMahon B, Young R. Proteins are paradigms of stochastic complexity. Phys. A: Stat. Mech. Appl. 2005;351:1–13. doi: 10.1016/j.physa.2004.12.004. [DOI] [Google Scholar]
- 35.Frauenfelder H, Fenimore PW, Chen G, McMahon BH.Protein folding is slaved to solvent motions Proc. Nat. Acad. Sci. USA 200610315469–15472.2006PNAS..10315469F 10.1073/pnas.0607168103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rosenberg KJ, Ross JL, Feinstein HE, Feinstein SC, Israelachvili J.Complementary dimerization of microtubule-associated tau protein: implications for microtubule bundling and tau-mediated pathogenesis Proc. Natl. Acad. Sci. USA 20081057445–7450.2008PNAS..105.7445R 10.1073/pnas.0802036105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Avila J, Lucas JJ, Pérez M, Hernández F. Role of tau protein in both physiological and pathological conditions. Physiol. Rev. 2004;84:361–384. doi: 10.1152/physrev.00024.2003. [DOI] [PubMed] [Google Scholar]
- 38.Spires-Jones TL, Stoothoff WH, Calignon A, Alix J, Phillip B, Hyman BT. Tau pathophysiology in neurodegeneration: a tangled issue. Trends Neurosci. 2008;32:150–159. doi: 10.1016/j.tins.2008.11.007. [DOI] [PubMed] [Google Scholar]
- 39.Tompa, P.: Intrinsically disordered proteins. In: Sussman, J., Silman, I. (eds.) Structural Proteomics and its Impact on the Life Sciences. World Scientific (2008)
- 40.Uversky V. Intrinsically disordered proteins and their environment: effects of strong denaturants, temperature, pH, counter ions, membranes, binding partners, osmolytes, and macromolecular crowding. The Protein J. 2009;28:305–325. doi: 10.1007/s10930-009-9201-4. [DOI] [PubMed] [Google Scholar]
- 41.Nettels D, Müller-Späth S, Küster F, Hofmann H, Haenni D, Rüegger S, Reymond L, Hoffmann A, Kubelka J, Heinz B, Gast K, Best RB, Schuler B.Single-molecule spectroscopy of the temperature-induced collapse of unfolded proteins Proc. Natl. Acad. Sci. USA 200910620740–20745.2009PNAS..10620740N 10.1073/pnas.0900622106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kohn JE, Millett IS, Jacob J, Zagrovic B, Dillon TM, Cingel N, Dothager RS, Seifert S, Thiyagarajan P, Sosnick TR, Hasan MZ, Pande VS, Ruczinski I, Doniach S, Plaxco KW.Random-coil behavior and the dimensions of chemically unfolded proteins Proc. Natl. Acad. Sci. U.S.A. 200410112491–12496.2004PNAS..10112491K 10.1073/pnas.0403643101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schweers O, Schonbrunn-Hanebeck E, Marx A, Mandelkow E. Structural studies of tau protein and Alzheimer paired helical filaments show no evidence for beta-structure. J. Biol. Chem. 1994;269:24290–24297. [PubMed] [Google Scholar]
- 44.Glatter, O., Kratky, O.: Small-angle X-ray Scattering. Academic Press (1982) [DOI] [PubMed]
- 45.Beaucage G. Small-angle scattering from polymeric mass fractals of arbitrary mass-fractal dimension. J. Appl. Crystallogr. 1996;29:134–146. doi: 10.1107/S0021889895011605. [DOI] [Google Scholar]
- 46.Pernot P, Theveneau P, Giraud T, Nogueira Fernandes R, Nurizzo D, Spruce D, Surr J, McSweeney S, Round A, Felisaz F, Foedinger L, Gobbo A, Huet J, Villard C, Cipriani F.New beamline dedicated to solution scattering from biological macromolecules at the ESRF J. Phys.: Conf. Ser. 2010247012009–012028.2010JPhCS.247a2009P 10.1088/1742-6596/247/1/012009 [DOI] [Google Scholar]
- 47.Battisti A, Tenenbaum A. Molecular dynamics simulation of intrinsically disordered proteins. Mol. Simulat. 2011 [Google Scholar]
- 48.Humphrey W, Dalke A, Schulten K. VMD – Visual Molecular Dynamics. J. Mol. Graphics. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 49.GROMACS release 4.5.3. www.gromacs.org
- 50.Mylonas E, Hascher A, Bernado P, Blackledge M, Mandelkow E, Svergun DI. Domain conformation of tau protein studied by solution small-angle X-ray scattering. Biochemistry. 2008;47:10345–10353. doi: 10.1021/bi800900d. [DOI] [PubMed] [Google Scholar]
- 51.Caldwell JW, Kollman PA. Structure and properties of neat liquids using nonadditive molecular-dynamics – water, methanol, and N-methylacetamide. J. Phys. Chem. 1995;99:6208–6219. doi: 10.1021/j100016a067. [DOI] [Google Scholar]
- 52.Fratini M, Poccia N, Ricci A, Campi G, Burghammer M, Aeppli G, Bianconi A.Scale-free structural organization of oxygen interstitials in La2CuO4 + y Nature 2010466841–844.2010Natur.466..841F 10.1038/nature09260 [DOI] [PubMed] [Google Scholar]
- 53.Poccia N, Fratini M, Ricci A, Campi G, Barba L, Vittorini-Orgeas A, Bianconi G, Aeppli G, Bianconi A.Evolution and control of oxygen order in a cuprate superconductor Nat. Mater. 201110733–736.2011NatMa..10..733P 10.1038/nmat3088 [DOI] [PubMed] [Google Scholar]
- 54.Poccia N, Ricci A, Innocenti D, Bianconi A. A possible mechanism for evading temperature quantum decoherence in living matter by Feshbach resonance. Int. J. Mol. Sci. 2009;10:2084–2106. doi: 10.3390/ijms10052084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Poccia N, Bianconi A. The physics of life and quantum complex matter: a case of cross-fertilization. Life. 2011;1:3–6. doi: 10.3390/life1010003. [DOI] [PMC free article] [PubMed] [Google Scholar]