Abstract
In tomato, carotenoids are important with regard to major breeding traits such as fruit colour and human health. The enzyme phytoene synthase (PSY1) directs metabolic flux towards carotenoid synthesis. Through TILLING (Targeting Induced Local Lesions IN Genomes), we have identified two point mutations in the Psy1 gene. The first mutation is a knockout allele (W180*) and the second mutation leads to an amino acid substitution (P192L). Plants carrying the Psy1 knockout allele show fruit with a yellow flesh colour similar to the r, r mutant, with no further change in colour during ripening. In the line with P192L substitution, fruit remain yellow until 3 days post-breaker and eventually turn red. Metabolite profiling verified the absence of carotenoids in the W180* line and thereby confirms that PSY1 is the only enzyme introducing substrate into the carotenoid pathway in ripening fruit. More subtle effects on carotenoid accumulation were observed in the P192L line with a delay in lycopene and β-carotene accumulation clearly linked to a very slow synthesis of phytoene. The observation of lutein degradation with ripening in both lines showed that lutein and its precursors are still synthesised in ripening fruit. Gene expression analysis of key genes involved in carotenoid biosynthesis revealed that expression levels of genes in the pathway are not feedback-regulated by low levels or absence of carotenoid compounds. Furthermore, protein secondary structure modelling indicated that the P192L mutation affects PSY1 activity through misfolding, leading to the low phytoene accumulation.
Keywords: Carotenoids, Point mutation, Psy1, Tomato, TILLING
Introduction
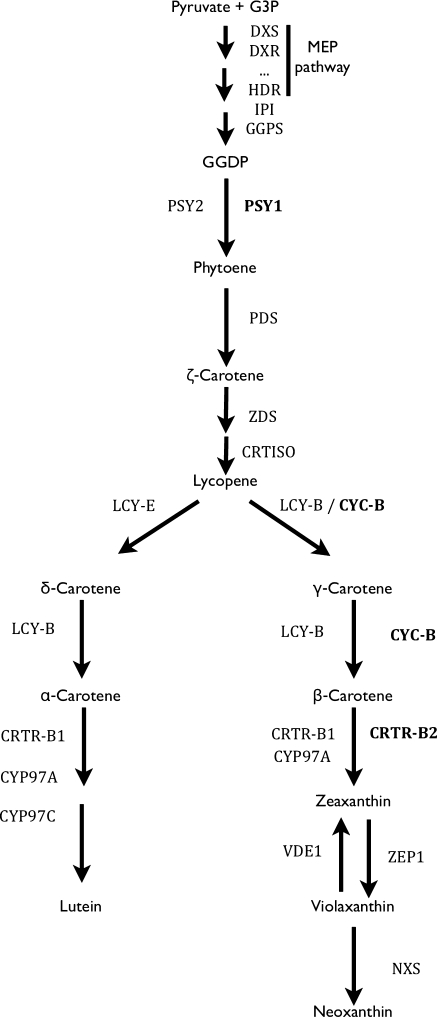
Tomato fruit are characterised by their red colour. Customer awareness that the red colour is associated with promoting human health is increasing (Fraser et al. 2009). Recent studies have shown the health-promoting role of tomato consumption in preventing various cancers, cardio-vascular diseases and blindness. These properties are attributable to a set of compounds such as flavonoids, phenylpropanoids, vitamin C, vitamin E and carotenoids. (Friishansen and Mccullough 1962; Fraser et al. 2005; Rao and Rao 2007; Ried and Fakler 2010). Carotenoids are part of the isoprenoid family and the carotenoid pathway starts from the transformation of GGDP to phytoene through the action of the enzyme phytoene synthase (PSY). The pathway (Fig. 1) consists of a cascade of enzymatic reactions taking place in plastids. Most of the reactions occur in chloroplasts where they play a key role in the photosynthetic machinery (Niyogi 1999). Therefore, most carotenoids are present in leaves and green fruit. Development of tomato fruit is characterised by the transition from green to coloured tissues. This change in colour is coupled to the development of chromoplasts during fruit ripening. Chloroplast to chromoplast transition is characterised by the activation of a different set of genes in the carotenoid biosynthesis pathway. PSY1, which is chromoplast-specific, takes over the enzymatic role of PSY2 in the green tissue.
Fig. 1.
Simplified overview of the carotenoid pathway. Highlighted in bold are the genes up-regulated in chromoplast-containing tissues. DXS 1-deoxy-d-xylulose-5-phosphate synthase, DXR deoxyxylulose 5-phosphate reductoisomerase, HDR hydroxymethylbutenyl diphosphate reductase, IPI IPP isomerase, GGPS geranylgeranyl diphosphatesynthase, PSY phytoene synthase, PDS phytoene desaturase, ZDS z-carotene desaturase, CRTISO carotenoid cis-trans-isomerase, LCY-B lycopene β-cyclase, CYC-B β-lycopene cyclase, CRTR-B β-carotene hydroxylase, CYP97A and C P450 carotenoid β- and ε-hydroxylases, VDE violaxanthin desaturase, ZEP zeaxanthin de-epoxidase, NXS neoxantin synthase, LCY-E ε-lycopene cyclase
Studying natural colour variation has allowed the discovery of genes involved in the carotenoid pathway through map-based cloning. This strategy led to the identification of a knock-out allele of the Psy1 gene called the r, r mutant that displays a characteristic yellow flesh phenotype, first reported by Ray et al. (1992) and Fray and Grierson (1993). In the r, r mutant, the suppression of PSY1 activity is sufficient to stop the accumulation and synthesis of phytoene and also of the downstream compounds of the carotenoid pathway.
To increase diversity in their available germplasm collection, plant breeders and researchers augment their material using mutant populations. Among the mutagen treatments available, the most commonly used is ethyl methanesulfonate (EMS). EMS primarily introduces single G–A or C–T point mutations randomly throughout genomes. After mutagen treatment, the population is phenotyped to identify lines with changed characteristics for the trait of interest. In this way, only lines with visible phenotypes are picked from the screening. McCallum et al. (2000) introduced TILLING (Targeting Induced Local Lesions IN Genomes) as a high-throughput method for the identification of point mutations in candidate genes. TILLING makes use of mutant populations without requiring phenotypic data. This allows the user to identify single mutant families carrying mutations in a gene of interest. Allelic series ranging from silent mutations to knockout alleles can be identified for each candidate gene.
Here we present two EMS-induced mutations in the Psy1 gene that were isolated using TILLING combined with high resolution melting (HRM) screening in an EMS-mutated M2 TPAADASU tomato population (Gady et al. 2009). These point mutations introduced a stop codon and an amino acid substitution in the Psy1 coding sequence. We describe the molecular characterisation of these two mutant lines.
Materials and methods
TILLING
Mutations were identified by screening the TPAADASU EMS M2 mutant population for three PCR fragments with the TILLING protocol adapted for a HRM LightScanner® platform as described previously (Gady et al. 2009). The two mutant lines presented here were both identified in one of the three fragments amplified using the following primers: 5′-TCCTCCCTTTTTCTCCACTTCAAGC-3′ (forward) and 5′-AAGCCCTCAGCAAAAGTGACATCA-3′ (reverse). PCRs were performed as follows on fourfold genomic DNA pools and after positive pool selection on single family genomic DNA: 94°C for 2 min; 40 cycles of 94°C for 5 s, 68°C for 10 s, and 72°C for 10 s; a denaturation step of 30 s at 94°C; and renaturation by cooling to 30°C.
Plant materials
For each identified mutant line, 15 M3 seeds were grown under greenhouse conditions. Plantlets were genotyped for the previously identified mutations. All plants were self-pollinated for seed production. Homozygous M4 plants were then grown in the greenhouse and for each line two variants of homozygous plants were grown: homozygous mutants and homozygous plants not carrying the mutation identified by TILLING. For each mutant line, five plants with the mutation and five plants without the mutation were grown as biological replicates. For each plant, three fruit were harvested at three different ripening stages: mature green (MG), breaker stage +3 days (Breaker + 3) and breaker stage +7 days (Breaker + 7).
For both carotenoid profiling and gene expression analysis experiments, samples were analysed from five biological repeats from the two mutant lines and their non-mutant siblings at the three ripening stages.
Carotenoid extraction and HPLC measurements
The flesh of all fruit samples described in the previous section was frozen and ground to powder. Extraction and analysis of carotenoids were performed as described by Bino et al. (2005).
RNA extraction and gene expression measurements
Total RNA was extracted from the same fruit samples as for the carotenoid extractions. The extraction procedure was performed using a standard TRIzol extraction method. Prior to DNase 1 (Invitrogen) treatment, all RNA samples were normalised to 100 ng/μl in a 10 μl reaction volume. cDNA synthesis was then performed using Bio-Rad iScript™ cDNA Synthesis kit in a 20 μl reaction volume. Following cDNA synthesis, samples were diluted 20-fold. Quantitative real-time PCR (QRT-PCR) was performed on a Bio-Rad CFX96™ Real-Time System according to the manufacturer’s recommendations. In addition to the reference gene, Actin, 11 genes involved in the carotenoid pathway were targeted with the primers listed in Table 1.
Table 1.
Gene-specific primers for QRT-PCR expression analysis
| Gene name | Forward primer (5′ → 3′) | Reverse primer (5′ → 3′) | Source |
|---|---|---|---|
| Actin | GAAATAGCATAAGATGGC | ATACCCACCATCACACCA | |
| Psy1 | TGACGTCTCAAATGGGACAAGT | CCTCGATGAATCAAAAAAACGG | Giorio et al. (2007) |
| Psy2 | AGGCAAGGCTGGAAGATATTTTT | GAAACAGTGTCGGATAAAGCTGC | Giorio et al. (2007) |
| Pds | TGGGTGGTTTGTCTACAGCAAA | ATCCCTTGCCTCCAGCAGTA | Giorio et al. (2007) |
| Zds | CAATGGCTGGGTTACAGAGTTG | CAATCCTGCAGCGCGC | Giorio et al. (2007) |
| Lcy-b | TGCTTATGGCATTTTGGCTG | CGCCAATCCATGAAAACCA | Giorio et al. (2007) |
| CrtR-b1 | TGTTGGTGCTGCTGTAGGAATG | AGTGAAGCATGCCACAGTGC | Giorio et al. (2007) |
| Zep | AAGGTTCCACAGAAGAAGTTGAAAG | TGCCAAAGCAAACACTAACCC | Giorio et al. (2007) |
| Cyc-B | GGGTAATGAGCCATATTTAAGGG | TAGGATCAAGATCAAAGAAAGCG | Ronen et al. (2000) |
| Lcy-e | GGCAGCCTCGGGGAAATTC | CACACGGAAGAATGCGCGC | Ronen et al. (1999) |
| Dxs | GCGGAGCTATTTCACATGGT | CTGCTGAGCATCCCAAT | Fraser et al. (2007) |
For each target gene, relative expression was calculated as ΔCt values as follows: (Ct value of target gene) − (Ct value of Actin), to correct for differential total cDNA concentration between samples.
SIFT, PARSESNP and protein secondary structure modelling analysis
To predict whether a point mutation would have an effect at the protein level, the PSY1 amino acid sequence was analysed with SIFT (http://www.sift.jcvi.org/). Amino acid substitutions identified with TILLING were compared with SIFT output.
To position the mutations on the genomic sequence, Psy1 sequence and mutation position were analysed with PARSESNP (http://www.proweb.org/parsesnp/).
Amino acid sequences of PSY1 proteins were submitted to the GOR4 secondary structure prediction tool from the Network Protein Sequence Analysis of the Pole Bioinformatique Lyonnais (http://www.npsa-pbil.ibcp.fr).
Results
Identification of Psy1 mutants
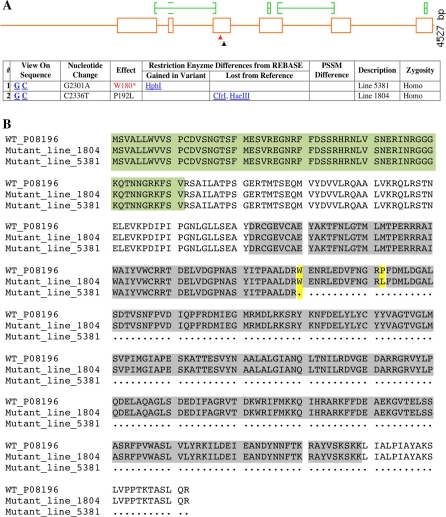
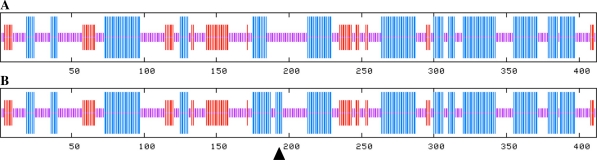
The TPAADASU M2 population was screened for point mutations in the phytoene synthase 1 (Psy1) gene. Using HRM-based screening, eight point mutations were identified within the target gene: T68I, R73Q, W180*, P192L and four mutations in introns (Gady et al. 2009). In the present study we focus on two of the identified mutant lines. These two mutations were identified in exon 3 of the Psy1 genomic sequence (Fig. 2A). The two lines were selected for further phenotyping experiments, as they were predicted to contain non-tolerated mutations using the SIFT online software, which is designed for prediction of the influence of amino acid substitutions on protein activity (Ng and Henikoff 2003). Line 1804 was shown to carry a C–T mutation leading to a proline (Pro) to leucine (Leu) amino acid substitution in position 192 of the PSY1 protein sequence (P192L). In line 5381 the EMS mutagenesis treatment induced a G to A transition at position 180 of the protein sequence leading to a premature stop codon in the first half of the coding sequence (W180*). The PSY1 protein produced in line W180* is truncated to a 179-amino-acid chain instead of the 412-amino-acid wild-type protein in tomato (Fig. 2B).
Fig. 2.
Graphical representation of mutation positions in the Psy1 gene and wild-type and mutant proteins sequence alignment. A PARSESNP representation (Taylor and Greene 2003) of the Psy1 gene based on genomic sequence. Mutations are positioned in the third exon. Mutation inducing the W180* is represented with a red arrowhead and mutation P192L by a black arrowhead. B ClustalW protein alignment of the wild-type and the two mutant proteins. Mutation positions are highlighted in yellow. Highlighted in green is the TARGETP-predicted chloroplastic transit peptide (Giorio et al. 2008). Highlighted in grey is the Trans_IPPS_HH conserved domain (trans-isoprenyl diphosphate synthases) identified with NCBI CDD tool
In addition to the identified mutation, EMS-mutated plants carry a large number of background mutations. To make sure that the observed phenotype is due to the targeted mutation, plants carrying the mutation are analysed in comparison with sibling plants of the same line which therefore have most of the same background mutations but lack the mutation of interest. In the present study, five plants homozygous for the mutation were grown along with five plants not carrying the mutation in Psy1 but belonging to the same EMS-mutated line. Consistent observation of the same phenotype in the five repetitions was taken as a confirmation that the phenotype is due to the identified mutation and not to the background mutations. In addition, to eliminate ambiguities it is preferable to have at least one other mutant line for the same target gene in order to check for a similar phenotype in a different mutant background, as was also suggested by Ostergaard and Yanofsky (2004). In the case of the Psy1 mutants, a natural null mutant allele has been identified and described as the yellow flesh mutant r, r, giving a clear indication of the expected phenotype (Fray and Grierson 1993).
The PSY1 protein sequence contains two important functional domains: firstly, the chloroplastic transit peptide, starting from the first amino acid to valine61, that is essential for protein targeting to the plastid compartment (Giorio et al. 2008), and secondly, the region from aspartate122 to lysine389, that was suggested to be a trans-isoprenyl diphosphate synthase (trans–IPP–HH) domain (Marchler-Bauer et al. 2009). Both mutations identified in this study are localised in this second domain (Fig. 2B).
Mutant line phenotypes
Homozygous plants for each Psy1 mutation were grown in the greenhouse together with their siblings without the Psy1 mutations as controls. During fruit development, all non-mutant lines had fruit turning from green to orange to red, following a normal developmental and maturation process corresponding to the MG to Breaker + 3 (turning) and Breaker + 7 (ripe) stages, respectively.
In the mutant line carrying the W180* mutation, fruit turned yellow and reached a deeper yellow colour at the ripe stage (Fig. 3). This phenotype is similar to the one described for the r, r mutant, confirming the sequence-based hypothesis of loss of function of Psy1 in this line.
Fig. 3.
Control and mutant fruit phenotypes at Mature Green, Breaker + 3 and Breaker + 7 ripening stages (left–right). Control fruit come from the same line as the mutant ones and thus, share background mutations but do not carry a mutation in Psy1. A Whole and open control fruit from line 1804. B Whole and open control fruit from line 5381. C Whole and open fruit from line 1804 carrying the PSY1 P192L amino acid substitution. D Whole and open fruit from line 5381 carrying the PSY1 W180* point mutation resulting in a Psy1 knockout allele
In the line carrying the P192L mutation, the fruit colour at Breaker + 3 stage remained dark yellow before eventually turning red at the ripe stage (Fig. 3). At breaker stage, where the fruit of the control plants turn orange, the mutant fruit remains yellow in colour, indicating an effect on the enzyme activity as predicted by SIFT. This mutated allele does not cause a loss of function, as fruit turns red at the ripe stage showing that lycopene is produced and therefore also its precursors.
Carotenoid profiling
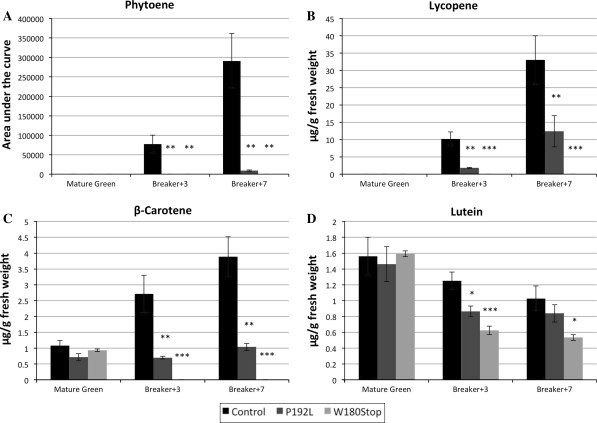
To further investigate the colour phenotypes, flesh samples from all fruit were analysed for carotenoid content using HPLC, as described in Materials and Methods (Fig. 4). Four metabolites were measured: phytoene, the direct PSY product; lycopene, the main compound responsible for the red colour of tomato fruit; β-carotene; and lutein.
Fig. 4.
HPLC carotenoid profiling of control lines, line 1804 carrying the P192L substitution and line 5381 carrying the W180* mutation in Psy1. Carotenoids were measured from fruit at three developmental and ripening stages: Mature Green, Breaker + 3 and Breaker + 7 stages. A Fruit phytoene content expressed as area under the phytoene curve. B Fruit lycopene content expressed in μg of lycopene/mg of fruit fresh weight. C Fruit β-carotene content expressed in μg of lycopene/mg of fruit fresh weight. D Fruit lutein content expressed in μg of lycopene per mg of fruit fresh weight. Significant difference (Student’s t test) in compound content in mutant lines compared to controls are represented with one or more asterisks with the following levels of significance: *P < 0.05; **P < 0.01; ***P < 0.001
In the Psy1 non-mutated lines, all measured compounds accumulated upon ripening, except for lutein, the level of which was highest in mature green fruit and decreased as ripening progressed. In line W180*, none of the measured carotenoids were detectable, with the exception of lutein. This data supports the hypothesis that the EMS point mutation completely inactivated the PSY1 protein.
In line P192L, phytoene, lycopene and β-carotene accumulated at much lower levels than in the controls both at Breaker + 3 and Breaker + 7 stages, while their starting levels in MG fruit were comparable. Phytoene was not detectable in mutant fruit until the ripe stage and at that time point the concentration was over seven-fold lower than in Breaker + 7 control fruit. Lycopene levels were over 5-fold lower in the mutant to than control fruit 3 days after breaker stage; however, they increased as ripening progressed, reaching final levels two times lower than control fruit at ripe stage. Similarly, β-carotene accumulated in the mutant fruit at a lesser rate than in controls. Mutant ripe fruit (Breaker + 7) contained approximately 50% of the β-carotene levels detected in the controls.
At MG stage, lutein levels were comparable in all lines. At later stages of ripening, lutein degradation occurred more rapidly in mutant lines than in controls. The most dramatic decrease in lutein content was observed in the line carrying the W180* mutation, as it showed a significantly lower concentration during the whole ripening process (Student’s t test, P < 0.05). Interestingly, in line P192L the decrease in lutein content was only significantly different from control 3 days after breaker stage while in ripe fruit lutein levels were comparable. This suggests that lutein is still produced through the carotenoid pathway in ripening fruit as it is in green tissues.
Carotenoid pathway-related gene expression analysis
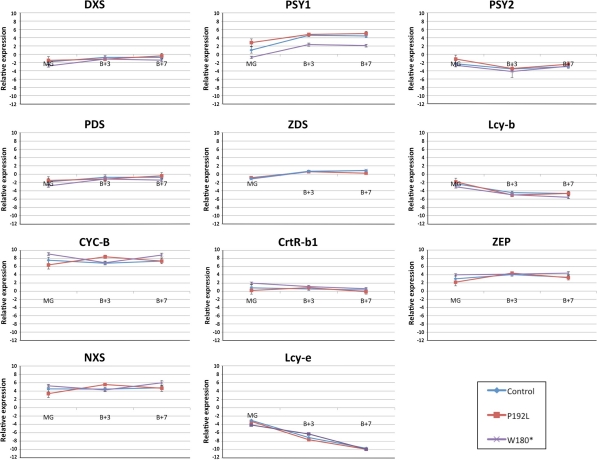
Genes involved in the carotenoid pathway were analysed for their expression levels to assess if one or both mutations lead to a modulation of the biosynthesis pathway. Apart for the first enzyme of the plastidial methylerythritol phosphate (MEP) pathway (DXS), the expression levels of genes involved in the biosynthesis of isoprenoids precursors were not assessed, as the focus of this study is on carotenoid biosynthesis. Eleven genes involved in both chloroplast and chromoplast specific carotenoid metabolism were analysed for their expression profiles at the three ripening stages: MG, Breaker + 3 and Breaker + 7. Although, we detected a large variation between mutant and control lines in the accumulation of carotenoids, we did not observe any difference in expression levels for the genes involved in the turnover of these compounds apart for Psy1 (Fig. 5). In line W180*, Psy1 is significantly reduced in expression in fruit at Breaker + 3 and Breaker + 7 stages (Student’s t test, P < 0.05).
Fig. 5.
Quantitative PCR-based expression analysis of genes involved in carotenoid biosynthesis during ripening in control and mutant lines. Eleven genes selected for their role in the tomato carotenoid pathway (Fig. 1) were studied at three ripening stages, Mature Green (MG), Breaker + 3 (B + 3) and Breaker + 7 (B + 7), to follow their expression levels in the two mutant lines (W180* and P192L) and in their respective controls. For each ripening and for each mutant line and non-mutant sibling, five biological repeats were sampled and analysed for gene expression. Relative expression is plotted as ΔCt values, obtained as follows: (Ct value of target gene) − (Ct value of Actin), to correct for differential cDNA concentration between samples. ΔCt values of the two control lines were averaged and shown on the graph. Statistical calculations were performed with both individual and averaged control values. Errors bars represent the variation between biological replicas. For all genes, significant difference (Student’s t test) in relative expression in mutant lines compared to controls was calculated for all lines and ripening stages. Only in line W180* was Psy1 expression significantly (P < 0.05) lower than control at B + 3 and B + 7 ripening stages
PSY1 wild-type and P192L mutant alleles secondary structure modelling
In order to visualise the effect of the P192L mutation on PSY1 structure and in the absence of crystallographic data, we analysed wild-type and mutant P192L alleles with the GOR4 secondary structure prediction tool from the PBL Network Protein Sequence Analysis. A short additional α-helix is predicted to form at the mutation position around the centre of the amino acid chain (Fig. 6). Even though the modification does not have a major influence on the protein structure, its position may result in differential enzyme activity occurring in the PSY1 P192L mutant protein. Additionally as a phenotype is observed due to the proline-to-leucine substitution that is linked to protein secondary structure effect, it is possible that the active site of PSY1 is located in the proximity of this mutation.
Fig. 6.
PSY wild-type and P192L mutant proteins secondary structure prediction using PBIL GOR4 tool. A Secondary structure prediction of the PSY1 wild-type protein. B Secondary structure prediction of the PSY1 P192L mutant protein. An additional alpha helix is predicted to form at the mutation position (black arrowhead)
Discussion
Carotenoid regulation in the mutant lines
In the knockout line of the Psy1 gene there is no carotenoid accumulation in ripening fruit, confirming the absence of an alternative route for the formation of carotenoids during ripening. The lack of phytoene results in an absence of lycopene and β-carotene. However, in the line carrying the mutation at residue 192 resulting in a proline-to-leucine substitution (P192L), all phytoene produced in the first day after breaker is converted into lycopene and downstream products. The low but detectable increase in carotenoid formation in P192L strongly suggests that PSY1 enzymatic activity is affected by the amino acid substitution. Indeed, phytoene is barely detectable during the whole ripening process. However, lycopene is still produced at very low quantities at Breaker + 3 stage, resulting in a yellow- instead of orange-coloured fruit and indicating that phytoene is produced through the action of the mutated enzyme. Four days later, lycopene content increases to reach half the level of control fruit. At Breaker + 7, lycopene accumulation is sufficient to give a red colour to the fruit; however, the intensity is weaker than in the control fruit. The yellow fruit colour observed in P192L Breaker + 3 fruit and in all ripening fruit from line W180* is due to the accumulation of the flavonoid naringenin chalcone. In control fruit the orange colour of Breaker + 3 fruit is due to the accumulation of both naringenin chalcone and lycopene and, in later ripening stages, the accumulation of lycopene that turns the fruit deep red.
As P192L fruit ripen, enzymes LCY-B, LCY-E and CYC-B controlling the branched pathways to the downstream carotenoids are the rate-limiting enzymatic steps, and thus, lycopene does accumulate in this line giving the red colour to the fruit, albeit at a substantially slower rate. These observations together with the absence of any carotenoid in line W180* confirm PSY1 as the enzyme responsible for directing products into the carotenoid pathway, thus, being the most influential step in this pathway as was suggested by Fraser et al. (2002).
Lutein content decreased during ripening in both mutant and control lines, and therefore the lutein pool is synthesised prior to ripening, at green stage, through the carotenoid chloroplast-specific pathway starting with PSY2 activity (Fig. 1). Measurement of lutein and its steady-state presence in ripening fruit is additional evidence for the existence of two overlapping pathways. In both mutant lines, the decrease in lutein content was more pronounced. In the line carrying the P192L substitution a significantly lower lutein content was only observed at Breaker + 3 stage. In the W180* line, lutein concentration was significantly lower at all ripening stages. These observations suggest that lutein was synthesised in ripening fruit. From the expression dataset we observe in all lines that the gene specific to branching from lycopene towards the xanthophyll pathway, Lcy-e, is strongly down-regulated during the ripening process. Nevertheless, observation of a slower degradation of the lutein pool in the ripening control fruit compared to the mutant lines suggests that Lcy-e and additional genes necessary for lutein synthesis are still expressed in ripening chromoplast-containing tissues or that residual enzymes coded by these genes are still active at those stages. The notion that these genes are expressed or that their corresponding translated proteins are still active is demonstrated by the difference observed between control and Psy1 knockout lines for lutein content in ripening fruit.
As lutein, β-carotene is already present in MG fruit. In line W180*, directly after the beginning of the ripening process, β-carotene is no longer detectable because no precursors for its synthesis are available in the Psy1 knockout line, and it may also be that it is metabolised by the downstream enzymatic reactions of the carotenoid pathway. In line P192L, β-carotene accumulates at a much later stage: Breaker + 7 instead of Breaker + 3 in control plants. In line with this, the production of precursors such as phytoene and lycopene is barely sufficient to maintain β-carotene levels at Breaker + 3 stage, resulting in a slow accumulation.
During ripening, a specific carotenoid pathway is activated through ripening-specific genes (Ronen et al. 1999; Ronen et al. 2000; Alba et al. 2005). At breaker stage, Psy2, Lcy-e, Lcy-b and CrtR-b1 genes are shut down and another set of genes with overlapping roles, with the exception of genes allowing the branching to xanthophyll synthesis, are switched on, including Psy1, Cyc-b and CrtR-b2. In addition, the burst of ethylene synthesis initiating ripening events plays a signalling role in activating several fruit-ripening genes specific to carotenoid synthesis, such as Dxs, Pds and Cyc-b (Alba et al. 2005). These regulatory events modify the carotenoid accumulation profile in ripe fruit compared to green tissues, as different compounds are then accumulated, including phytoene, lycopene and β-carotene. In addition, fruit do not accumulate xanthophylls such as lutein, even though they are still produced, because Lcy-e is down-regulated.
Decreased carotenoid content does not affect gene expression
The complete inhibition of the carotenoid pathway does not appear to lead to increased expression of genes encoding for enzymes involved in carotenoid synthesis. We conclude that there is not likely to be a feedback regulation from either the quantity or quality of the synthesised metabolite products on the genes involved in this pathway. It is interesting to note that the regulatory system appears to fail to sense the absence of carotenoids, whereas it can detect an over-accumulation (Fraser et al. 2007; Giorio et al. 2007). In line with this, no differences in relative expression were observed for the genes involved in carotenoid biosynthesis in mutant 1804 as well as in the knockout line 5381.
PSY1 protein structure
In Fig. 2B the trans-IPP-HH active domain identified with the NCBI CDD tool is highlighted in grey. This domain covers 268 amino acids. The P192L mutation and its linked decreased enzymatic activity might pinpoint a specific domain of importance such as the substrate binding site. The substitution was predicted as deleterious for enzyme activity using SIFT prediction software. SIFT does not account for active or conserved domains but bases the predictions on amino acid properties and on the level of conservation of each amino acid in the sequence after an alignment search in protein sequence databases. The secondary structure of the PSY1 wild-type and P192L mutant proteins was predicted using the GOR4 (Garnier et al. 1996) secondary structure prediction tool from the Pole Bioinformatique Lyonnais (PBIL, Lyons University, France). It is predicted that the protein secondary structure is affected by the mutation, as a short α-helix is formed instead of a long coil joining two α-helixes (Fig. 6). This structural modification could be the cause of a reduced enzyme activity as it may disturb the spatial conformation of the protein. To understand further catalytic domains of the PSY1 enzyme, a 3-D structure of the protein would be required. With such a model amino acid substitutions and their influence on the protein spatial conformation could be visualised. In the case of the present P192L mutation, such a visualisation tool together with experimental enzymatic activity assays would deepen our understanding of the PSY1 enzyme.
Conclusion
TILLING was proposed as a tool for discovering new alleles for selected candidate genes (Comai and Henikoff 2006; Barkley and Wang 2008) with the claim that the discovered mutation would provide a range of effects on the gene function. This range could span from no effect, when a synonymous mutation occurs, to a knockout of the gene when a premature stop codon is introduced. Between these extreme phenotypes, mild effects on enzyme activity can potentially be found depending on the quality of the induced amino acid substitutions.
In this study we have characterised two TILLING mutants carrying an amino acid substitution and an early stop codon, respectively. This work confirms that a range of effects on enzyme activity and consequently on plant biology can be obtained following a TILLING strategy. Indeed, looking at fruit phenotypes and carotenoid profiling data, the Psy1 W180* mutation proved to be a knockout allele of the Psy1 gene. Based on similar observations, the P192L mutation is strongly suspected to affect PSY1 activity. Analysis of the mutants provides important insights into the regulatory processes involved in the tomato fruit carotenoid pathway.
The study of mutants carrying amino acid substitutions affecting enzyme activity provides an opportunity to modify the phenotype more subtly and thus, could target genes important for development without killing the plant.
In the context of plant breeding, a TILLING approach allows the creation of novel alleles that can be used in breeding programs, bringing additional variation to traits of interest. The TILLING method is applicable to any trait as long as sequence information for the candidate genes is available. In the last decade tremendous efforts have been made to sequence plant genomes, providing a wealth of sequence information in most of the major crop species. These data, together with the rapidly growing knowledge in plant biology and plant defence mechanisms against diseases, makes a TILLING approach even more sensible for breeders.
Acknowledgments
The authors thank the greenhouse employees of Nunhems B. V. for their help with the propagation of the mutant lines. We thank Harry H. Jonker (Plant Research International, Bioscience Unit, Wageningen) for his guidance through the HPLC protocol and data analysis software. A. G. was supported by a joint project between Nunhems B. V. and the Laboratory of Plant Breeding, Wageningen-UR.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Contributor Information
Antoine L. F. Gady, Email: antoine.gady@wur.nl
Wim H. Vriezen, Email: wim.vriezen@bayer.com
Marion H. B. J. Van de Wal, Email: marion.vandewal@bayer.com
Pingping Huang, Email: pingping.huang@wur.nl.
Arnaud G. Bovy, Email: arnaud.bovy@wur.nl
Richard G. F. Visser, Email: richard.visser@wur.nl
Christian W. B. Bachem, Phone: +31-317-482854, Email: christian.bachem@wur.nl
References
- Alba R, Payton P, Fei Z, McQuinn R, Debbie P, Martin GB, Tanksley SD, Giovannoni JJ. Transcriptome and selected metabolite analyses reveal multiple points of ethylene control during tomato fruit development. Plant Cell. 2005;17:2954–2965. doi: 10.1105/tpc.105.036053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkley NA, Wang ML. Application of TILLING and EcoTILLING as reverse genetic approaches to elucidate the function of genes in plants and animals. Curr Genomics. 2008;9:212–226. doi: 10.2174/138920208784533656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bino RJ, De Vos CHR, Lieberman M, Hall RD, Bovy A, Jonker HH, Tikunov Y, Lommen A, Moco S, Levin I. The light-hyperresponsive high pigment-2<sup>dg</sup> mutation of tomato: alterations in the fruit metabolome. New Phytol. 2005;166:427–438. doi: 10.1111/j.1469-8137.2005.01362.x. [DOI] [PubMed] [Google Scholar]
- Comai L, Henikoff S. TILLING: practical single-nucleotide mutation discovery. Plant J. 2006;45:684–694. doi: 10.1111/j.1365-313X.2006.02670.x. [DOI] [PubMed] [Google Scholar]
- Fraser PD, Romer S, Shipton CA, Mills PB, Kiano JW, Misawa N, Drake RG, Schuch W, Bramley PM. Evaluation of transgenic tomato plants expressing an additional phytoene synthase in a fruit-specific manner. Proc Natl Acad Sci USA. 2002;99:1092–1097. doi: 10.1073/pnas.241374598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser ML, Lee AH, Binns CW. Lycopene and prostate cancer: emerging evidence. Expert Rev Anticancer Ther. 2005;5:847–854. doi: 10.1586/14737140.5.5.847. [DOI] [PubMed] [Google Scholar]
- Fraser PD, Enfissi EMA, Halket JM, Truesdale MR, Yu D, Gerrish C, Bramley PM. Manipulation of phytoene levels in tomato fruit: effects on isoprenoids, plastids, and intermediary metabolism. Plant Cell. 2007;19:3194–3211. doi: 10.1105/tpc.106.049817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser PD, Enfissi EM, Bramley PM. Genetic engineering of carotenoid formation in tomato fruit and the potential application of systems and synthetic biology approaches. Arch Biochem Biophys. 2009;483:196–204. doi: 10.1016/j.abb.2008.10.009. [DOI] [PubMed] [Google Scholar]
- Fray RG, Grierson D. Identification and genetic analysis of normal and mutant phytoene synthase genes of tomato by sequencing, complementation and co-suppression. Plant Mol Biol. 1993;22:589–602. doi: 10.1007/BF00047400. [DOI] [PubMed] [Google Scholar]
- Friishansen B, Mccullough FS. Vitamin A deficiency in African children in Northern Rhodesia. J Pediatr. 1962;60:114. doi: 10.1016/S0022-3476(62)80017-1. [DOI] [PubMed] [Google Scholar]
- Gady A, Hermans F, Van de Wal M, van Loo E, Visser R, Bachem C. Implementation of two high through-put techniques in a novel application: detecting point mutations in large EMS mutated plant populations. Plant Methods. 2009;5:13. doi: 10.1186/1746-4811-5-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garnier J, Gibrat JF, Robson B. GOR method for predicting protein secondary structure from amino acid sequence. Methods Enzymol. 1996;266:540–553. doi: 10.1016/S0076-6879(96)66034-0. [DOI] [PubMed] [Google Scholar]
- Giorio G, Stigliani AL, D’Ambrosio C. Agronomic performance and transcriptional analysis of carotenoid biosynthesis in fruit of transgenic HighCaro and control tomato lines under field conditions. Transgenic Res. 2007;16:15–28. doi: 10.1007/s11248-006-9025-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giorio G, Stigliani AL, D’Ambrosio C. Phytoene synthase genes in tomato (S. lycopersicum L.): new data on the structures, the deduced amino acid sequences and the expression patterns. FEBS J. 2008;275:527–535. doi: 10.1111/j.1742-4658.2007.06219.x. [DOI] [PubMed] [Google Scholar]
- Marchler-Bauer A, Anderson JB, Chitsaz F, Derbyshire MK, DeWeese-Scott C, Fong JH, Geer LY, Geer RC, Gonzales NR, Gwadz M, He S, Hurwitz DI, Jackson JD, Ke Z, Lanczycki CJ, Liebert CA, Liu C, Lu F, Lu S, Marchler GH, Mullokandov M, Song JS, Tasneem A, Thanki N, Yamashita RA, Zhang D, Zhang N, Bryant SH. CDD: specific functional annotation with the conserved domain database. Nucleic Acids Res. 2009;37:D205–D210. doi: 10.1093/nar/gkn845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCallum CM, Comai L, Greene EA, Henikoff S. Targeting induced local lesions IN genomes (TILLING) for plant functional genomics. Plant Physiol. 2000;123:439–442. doi: 10.1104/pp.123.2.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng P, Henikoff S. SIFT: predicting amino acid changes that affect protein function. Nucleic Acids Res. 2003;31:3812–3814. doi: 10.1093/nar/gkg509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niyogi KK. Photoprotection revisited: genetic and molecular approaches. Annu Rev Plant Physiol Plant Mol Biol. 1999;50:333–359. doi: 10.1146/annurev.arplant.50.1.333. [DOI] [PubMed] [Google Scholar]
- Ostergaard L, Yanofsky MF. Establishing gene function by mutagenesis in A. thaliana. Plant J. 2004;39:682–696. doi: 10.1111/j.1365-313X.2004.02149.x. [DOI] [PubMed] [Google Scholar]
- Rao AV, Rao LG. Carotenoids and human health. Pharmacol Res. 2007;55:207–216. doi: 10.1016/j.phrs.2007.01.012. [DOI] [PubMed] [Google Scholar]
- Ray J, Moureau P, Bird C, Bird A, Grierson D, Maunders M, Truesdale M, Bramley P, Schuch W. Cloning and characterization of a gene involved in phytoene synthesis from tomato. Plant Mol Biol. 1992;19:401–404. doi: 10.1007/BF00023387. [DOI] [PubMed] [Google Scholar]
- Ried K, Fakler P. Protective effect of lycopene on serum cholesterol and blood pressure: meta-analyses of intervention trials. Maturitas. 2010;68:299–310. doi: 10.1016/j.maturitas.2010.11.018. [DOI] [PubMed] [Google Scholar]
- Ronen G, Cohen M, Zamir D, Hirschberg J. Regulation of carotenoid biosynthesis during tomato fruit development: expression of the gene for lycopene epsilon-cyclase is down-regulated during ripening and is elevated in the mutant delta. Plant J. 1999;17:341–351. doi: 10.1046/j.1365-313X.1999.00381.x. [DOI] [PubMed] [Google Scholar]
- Ronen G, Carmel-Goren L, Zamir D, Hirschberg J. An alternative pathway to beta-carotene formation in plant chromoplasts discovered by map-based cloning of beta and old-gold color mutations in tomato. Proc Natl Acad Sci USA. 2000;97:11102–11107. doi: 10.1073/pnas.190177497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor NE, Greene EA. PARSESNP: a tool for the analysis of nucleotide polymorphisms. Nucleic Acids Res. 2003;31:3808–3811. doi: 10.1093/nar/gkg574. [DOI] [PMC free article] [PubMed] [Google Scholar]