Abstract
The combination of induced pluripotent stem (iPS) cell technology and targeted gene modification by homologous recombination (HR) represents a promising new approach to generate genetically corrected, patient-derived cells that could be used for autologous transplantation therapies. This strategy has several potential advantages over conventional gene therapy including eliminating the need for immunosuppression, avoiding the risk of insertional mutagenesis by therapeutic vectors, and maintaining expression of the corrected gene by endogenous control elements rather than a constitutive promoter. However, gene targeting in human pluripotent cells has remained challenging and inefficient. Recently, engineered zinc finger nucleases (ZFNs) have been shown to substantially increase HR frequencies in human iPS cells, raising the prospect of employing this technology to correct disease-causing mutations. Here we describe the generation of iPS cell lines from sickle cell anemia patients and in situ correction of the disease-causing mutation using three ZFN pairs made by the publicly available Oligomerized Pool Engineering (OPEN) method. Gene-corrected cells retained full pluripotency and a normal karyotype following removal of reprogramming factor and drug-resistance genes. By testing various conditions, we also demonstrated that HR events in human iPS cells can occur as far as 82 bps from a ZFN-induced break. Our approach delineates a roadmap for using ZFNs made by an open-source method to achieve efficient, transgene-free correction of monogenic disease mutations in patient-derived iPS cells. Our results provide an important proof of principle that ZFNs can be used to produce gene-corrected human iPS cells that could be used for therapeutic applications.
Introduction
The development of induced pluripotent stem (iPS) cell technology has raised prospects for patient-specific therapies of various human diseases.1, 2 Human iPS cells can grow to virtually unlimited numbers, can differentiate to all somatic tissues, and can be derived from a variety of somatic cells such as fibroblasts obtained by a simple skin biopsy.3–6 For treatment of monogenic diseases, patient-derived iPS cells might be corrected, differentiated into therapeutically relevant cells, and transplanted back into the patient for restoration of function. A critical component of this envisioned therapeutic strategy is the efficient genetic correction of the disease-causing gene mutation in the iPS cells. One strategy for correction is to use homologous recombination (HR) with an exogenous DNA donor template to modify specific genomic sequences. This in situ approach would retain regulation by endogenous genomic elements, a key advantage for genes that must be expressed in a staged fashion through differentiation or development. In a previous report, we used gene targeting in murine iPS cells to correct the sickle cell anemia mutation in a mouse model of this disease.7 However, in contrast to mouse cells, efficiencies of gene targeting in human iPS and embryonic stem cells are low8–12 or require the construction of large donor templates,13, 14 thereby rendering this approach difficult to practice.
Engineered zinc finger nucleases (ZFNs) can be used to substantially increase the rate of gene targeting at specific genomic loci.15–17 ZFNs are customizable restriction enzymes that consist of an engineered zinc finger array fused to a non-specific DNA cleavage domain from the FokI endonuclease.18 Introduction of a ZFN-induced DNA double-strand break (DSB) substantially stimulates gene targeting mediated by homologous recombination (HR) with a “donor template” at the cleavage site. Various publicly available19–24 and proprietary methods25 exist to engineer the zinc finger arrays required for targeting the FokI cleavage domain to a specific genomic locus. We recently described a rapid, “open-source” ZFN engineering method known as “Oligomerized Pool Engineering” (OPEN)22 and have used ZFNs made by this approach to modify endogenous genes in zebrafish, plants, and various human cells.11, 22, 26–28 Previous work in human iPS cells has shown that ZFNs can be used to enhance the efficiency of HR-mediated reporter gene or drug-resistance marker insertion into ZFN-induced DSBs.9, 11 One recent report also described insertion of a wild-type gene into a ZFN-induced break induced at a potential “safe harbor” locus in human iPS cells derived from patients with X-linked chronic granulomatous disease, thereby complementing the gene defect.29 However, in situ gene correction by ZFNs is a more challenging problem than simple reporter gene insertion because the currently limited targeting range of publicly available ZFN engineering methods can make it difficult to introduce a DSB directly at the site of the mutation. In addition, high efficiency ZFN-mediated HR in human iPS cells requires co-integration of a drug-resistance gene into a nearby intron, thereby necessitating the introduction of transgenic sequences some distance away from the ZFN cleavage site.
Here we demonstrate facile and efficient correction of the sickle cell anemia mutation in patient-derived human iPS cells using ZFNs engineered by the publicly available Oligomerized Pool ENgineering (OPEN) method.22, 23 We accomplished this by using OPEN ZFNs to simultaneously correct the mutation and insert a drug-resistance cassette into a neighboring intron up to 82 base pairs away from the ZFN cut site. Following excision of the reprogramming factor and drug-resistance cassettes, corrected and transgene-free iPS cells retained full pluripotency and normal karyotypes. Our successful genetic correction of a disease-causing mutation in human iPS cells provides an important proof-of-principle for therapeutic applications and delineates a roadmap for using ZFNs as an efficient tool for targeted manipulation of human iPS cells.
Materials and Methods
Detailed Methods are provided in the Supplementary Materials
Derivation and Culture of iPS cells
Derivation, modification, and analysis of patient-derived iPS cells were performed under IRB protocols approved by Stanford University and Massachusetts General Hospital.
Dermal fibroblasts from two sickle cell anemia patients were infected with the polycistronic STEMCCA lentiviral reprogramming vector. 30 105 fibroblasts were seeded in MEF medium and infected 24 hours later. After 6 days, cells were transferred onto inactivated MEFs. The following day MEF medium was replaced with hES medium and the cells were grown for up to 8 weeks until hES-like colonies started to emerge. iPS colonies were manually picked and expanded on MEFs. After approximately 10 passages the clones were transferred to feeder-free culture conditions in mTeSR1 medium (Stem Cell Technologies).
Engineering ZFNs targeted to the human β-globin gene
Zinc finger arrays binding each of these target sites were selected using either the previously described OPEN method22 or a modified protocol that uses antibiotic selection.24 The OPEN selections we performed required the use of several additional OPEN pools for new 3 bp subsites (J.K. Joung et al., manuscript in preparation). Candidates from OPEN selections were assayed for DNA-binding activity in the bacterial-two-hybrid system as previously described.23 Selected zinc fingers were cloned on an XbaI-BamHI fragment into a ZFN expression vector that expresses heterodimeric ZFNs (either EL/KK31 or ELD/KKR32 fused by a T2A self-cleaving peptide. The CMV promoter driving ZFN expression in the resulting plasmids was then replaced with an EF1α promoter. This ZFN expression plasmid also contains a PGK promoter-GFP reporter gene cassette that can be used to assess transfection efficiency.
Transient transfection for Gene Targeting and Loop-out in iPS cells
For gene targeting, 1×106 iPSCs were transfected with 2 μg of ZFN-encoding plasmid and 10 μg of donor template plasmid by Amaxa nuclefection. After four days, cells were selected in media containing puromycin (1 μg/ml) for 2 weeks. To excise selection and/or reprogramming cassettes, cells were transfected with an EF1a-Cre-ires-puro plasmid (a gift kindly provided by Gustavo Mostoslavsky). Alternatively infection with adenovirus expressing a Cre-recombinase-GFP fusion (Vector Biolabs) was used. FACS was used to isolate GFP-positive cells that were seeded as single cells on Matrigel in mTeSR1 medium containing 2 μM Thiazovivin. Individual colonies were manually picked and screened by PCR.
Southern Blotting
Genomic DNA was separated on a 0.8% agarose gel following an overnight restriction digest with indicated restriction enzymes, transferred to a nylon membrane (Amersham), and hybridized with 32P-labeled probes as indicated made by random priming following the manufacturer’s instructions (Agilent Prime-It II kit).
Teratoma Formation and Analysis
Cells from a confluent 10 cm plate were harvested by Accutase digestion, resuspended in phosphate buffered saline, and injected subcutaneously into immuno-compromised NOG-SCID mice (obtained from the Jackson Laboratories). Four to eight weeks after injection, teratomas were dissected, fixed in 4% PFA, and processed for H&E.
Expression analysis
Immunofluorescence and RT-PCR analysis was performed following standard protocols as described in the Supplementary Materials. Please see Supplementary Methods also for a list of primer sequences and antibodies used in this study.
Karyotyping
Metaphase spreads were prepared after following treatment with 10 ug/mL of Colcemid (Invitrogen) and processed for SKY-FISH hybridization and image analysis according to the manufacturer’s instructions (Applied Spectral Imaging).
Results
Derivation and characterization of iPS cells from two sickle cell anemia patients
Many different non-integrating methods have been developed for the safe generation of human iPS cells.5, 33–37 While these methods have great potential, we employed an integrating lentivirus that contains a polycistronic cassette encoding the four required reprogramming factors Oct4, Sox2, Klf4, and c-myc that is flanked by loxP sites.30, 38 This approach enables high reprogramming efficiencies together with the generation of transgene-free iPS cells following Cre recombinase-mediated excision of the reprogramming cassettes.
Primary fibroblast cultures were established using skin punch biopsies from two patients diagnosed with sickle cell anemia (hereafter Patient 1 and 2). DNA sequencing revealed that Patient 1 has a sickle cell E6V mutation in one beta-globin allele and a splice donor site mutation at the end of exon 1 in the other allele while Patient 2 is homozygous for the E6V mutation (data not shown). Fibroblasts from each patient were infected with a polycistronic reprogramming factor virus and seeded onto mitomycin C-treated feeder cells (Materials and Methods). Multiple colonies appeared after 4 to 6 weeks and these were picked and further expanded in separate dishes. Four lines (hereafter I and II for the clones derived from Patient 1 and III and IV for the clones derived from Patient 2) were selected for continued expansion. After several passages on feeders, these clones could also be successfully propagated under feeder-free conditions (mTeSR1 medium on matrigel-coated plates). All four clones expressed multiple pluripotency markers at the mRNA and protein level, showed telomerase activity similar to control human H9 embryonic stem cells, possessed a normal karyotype, and could be differentiated in vitro into derivatives of all three germ layers (Figure 1A–D and Table S1). Clones I, II and III were injected under the skin of immuno-compromised mice and formed tumors containing various tissue types representing cells from all three germ layers (Figure 1E, Figure S1 and Table S1). Thus, surprisingly, the lentiviral transgenes appeared to be silenced allowing the cells to differentiate. Southern blot analysis revealed the presence of three proviral integrations in clone I, two in clone II, one in clone III and one in clone IV (Figure 1F and Table S1).
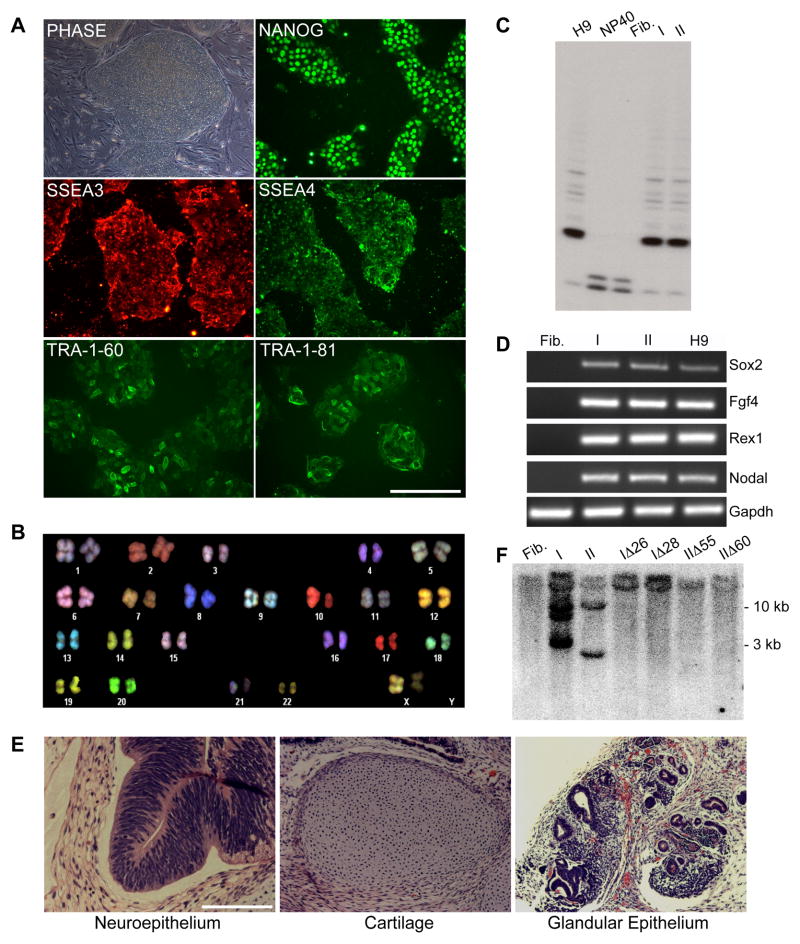
Figure 1. Characterization of sickle cell iPS clones I and II.
A. Phase contrast image of iPS clone I grown on feeder cells and immunofluorescence staining images of feeder-free cultures.
B. SKY-FISH analysis of a representative normal diploid metaphase spread of iPS clone I.
C. Telomerase activity assay: Fibroblasts show no telomerase activity as in the negative control (NP40); conversely iPS clones I and II show a pattern similar to the one observed in H9 ES cells (positive control).
D. RT-PCR analysis of pluripotency-associated genes in patient parental fibroblasts, iPS clones I and II, and H9 ES cells.
E. H&E staining of teratoma derived from iPS clone I showing tissues representative of all three germ layers.
F. Southern Blot analysis using a c-myc probe showing the number of STEMCCA cassette integrations in parental iPS clones I and II and four derivative clones transfected with Cre recombinase. Clones I and II show 3 and 2 integrations, respectively, while clones transfected with Cre recombinase (IΔ26, IΔ28, IΔ55, and IΔ60) show no integrations, similar to what is observed in uninfected fibroblasts.
Abbreviations: Fib.: Fibroblasts from sickle cell anemia patient 1; I: iPS clone I; II: iPS clone II; NP40: negative control for telomerase activity.
We next verified that the iPS cell lines we generated were stable in the absence of the reprogramming factors. To remove the proviral sequences, clones I and II were transfected with a plasmid encoding Cre recombinase and the puromycin-resistance gene and then selected in the presence of puromycin for 48 hours. Resulting colonies were picked, expanded and their genomic DNA was isolated. A PCR assay detecting specific sequences within the viral vector was performed to screen for potential clones in which all of the reprogramming factors had been removed (data not shown). Two subclones each for clones I and II that were negative for reprogramming factor sequences by PCR were analyzed by Southern blotting to verify loss of the integrated vector sequences (clones IΔ26 IΔ28, IIΔ55, IIΔ60) (Figure 1F). All four clones retained a fully undifferentiated state as judged by the expression of a battery of pluripotency markers and a normal karyotype (Figure S1 and Table S2).
Engineering of OPEN ZFNs targeted to the human beta-globin gene
Using our web-based Zinc Finger Targeter (ZiFiT) program,39 we identified two target sites occurring near the sickle cell anemia mutation in our patient-derived iPS cell clones for which ZFNs could be potentially engineered using the publicly available Oligomerized Pool ENgineering (OPEN) protocol previously described by our group (Figure 2A).23 These two sites fall within 25 bps of the sickle mutation (Figure 2B): one is present ~24 bps downstream (ZFN target site 1) and the other is directly at the site of the mutation (ZFN target site 2). OPEN selections for ZFN target sites 1 and 2 were performed and successfully yielded multiple different zinc finger arrays for all four half-sites. Quantitative testing in a bacterial two-hybrid (B2H) reporter assay revealed that the majority of these arrays activated transcription by threefold or more (data not shown), a threshold we have previously shown is a strong predictor for efficient function as ZFNs in human cells.11, 22 Based on these results, we chose four and two ZFN pairs for target sites 1 and 2, respectively, to carry forward for additional testing. The sequences of the recognition helices and the activities of these zinc finger arrays in the B2H reporter system are shown in Figure 2C.
Figure 2. Engineering and characterization of ZFNs.
A. Schematic overview of Oligomerized Pool Engineering (OPEN) strategy for constructing zinc finger arrays. Individual zinc fingers are depicted as spheres and DNA target sites as rectangles.
B. Schematic of the human β-globin locus showing the position of the two ZFN target sites in relation to the sickle cell anemia mutation (asterisk) and to the insertion site of the selectable marker (arrow). Exon represented by thick line, introns by thin lines. Not drawn to scale.
C. Recognition helix sequences and bacterial two-hybrid (B2H) fold-activation values of selected zinc finger proteins engineered by OPEN. Amino acid sequence of the recognition helices are shown below their respective three base pair target DNA sites. B2H fold-activation values are shown as blue bars on the right.
D. Cleavage efficiency of ZFN pairs as measured by induction of mutations caused by non-homologous end-joining (NHEJ) repair in HEK293 cells (for site 1) or primary human fibroblasts (for site 2). Wild-type sequence is shown with zinc finger binding sites highlighted in yellow. Mutant sequences are shown below with deletions indicated by dashes and insertions shown in red, lower-case letters. The frequency with which each sequence was detected is indicated to the right.
To test the cleavage activities of our OPEN-selected zinc finger arrays as ZFNs in human cells, we assessed their capabilities to induce site-specific insertion/deletion mutations (indels) via non-homologous end joining. Previous work has shown that, in the absence of a donor template, active ZFNs will induce indels with high efficiency at their site of cleavage in somatic human cells.40 For ZFN target site 1, two of the four pairs tested showed activity in human HEK293 cells, with one pair (hereafter designated ZFN Pair A) exhibiting a substantially higher mutation frequency (12.9%) than the others (Figure 2D). For ZFN target site 2, both pairs (hereafter ZFN Pairs B and C) tested yielded roughly equivalent mutation frequencies in primary human fibroblasts derived from Patient 1 (Figure 2D). Based on these experiments, we carried forward ZFN pairs A, B, and C for gene correction experiments in our sickle cell anemia iPS cells.
Construction of a “donor template” required for ZFN-mediated correction of the sickle cell anemia mutation
Our ZFN-based strategy to correct the sickle cell anemia mutation also requires a homologous “donor template” for introducing the desired correction. We therefore constructed a donor template that harbors ~1.6 kb of beta-globin gene sequence centered approximately on the location of the sickle cell anemia E6V mutation but which codes for the wild-type E6 codon (Figure 3A). In addition, we introduced two translationally silent mutations into the donor construct to reduce binding and cleavage of the donor by ZFN Pair A. Based on previous experience,9, 11 we presumed that ZFN-induced gene targeting in iPS cells would require drug-selection to identify desired recombinants. Therefore, we modified the donor template by inserting a puromycin or neomycin-resistance gene cassette into the intron located downstream of the E6V mutation. The drug-resistance cassette was flanked by loxP sites to enable its subsequent excision using Cre recombinase. In this configuration, the 5′ end of the puromycin or neomycin-resistance gene insertion is positioned ~58 or ~82 bps downstream of the cleavage sites for ZFN Pairs A or B/C, respectively (Figures 2B and 3A).
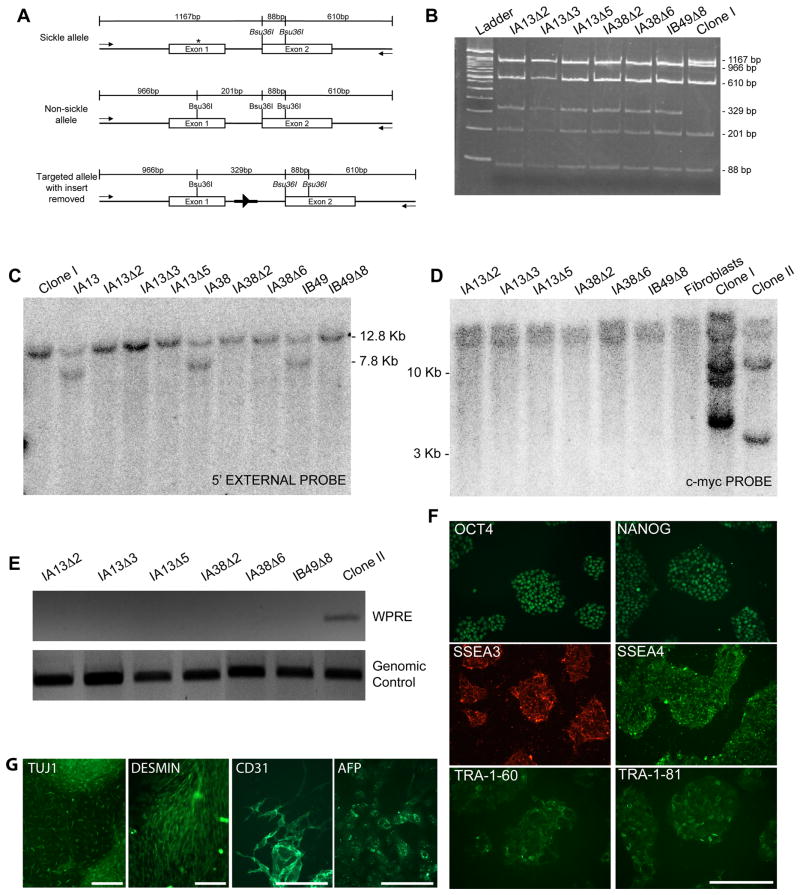
Figure 3. Gene targeting of the endogenous human beta-globin locus.
A. Schematic overview of gene targeting strategy for the human beta-globin locus. The sickle cell mutation is labeled by an asterisk and ZFN target sites are indicated by arrows. The desired recombination event inserts a PGK promoter-puromycin-resistance cassette or PGK promoter-neomycin-resistance cassette flanked by loxP sites (black triangles) into the intron between exons 1 and 2. Southern blot probes are indicated by grey bars and PCR primers are indicated by arrows. Not drawn to scale.
B. Representative PCR analysis of puromycin-resistant clones. PCR of gene targeted clones gives a band of 1.4 kb when amplified with 3′ primers and 1.6 kb with 5′ primers. Non-targeted clones fail to give PCR product.
C. Southern blot using a 5′ external probe on genomic DNA digested with PvuII. A beta-globin allele that has not undergone gene targeting gives a 12.8kb band while a targeted allele gives a 7.8kb band due to the presence of a PvuII site in the puromycin-resistance gene. Note that after expansion in culture, four clones previously revealed to be mixed populations by sequencing no longer possess the gene targeted allele.
D. Sequence of gene targeted allele detected by PCR assay. The allele that has undergone homologous recombination with the donor template is wild-type at the E6 codon that is mutated in sickle cell disease, contains translationally silent mutations incorporated into ZFN target site 1, and has incorporated the puromycin or neomycin-resistance cassette.
E. Sequence of the non-gene targeted allele. The untargeted allele in clones IA12, IA13, IA38, IB20, IB48, IB49, IB50, IC54 and IIBI is the allele that does not harbor the sickle cell mutation, indicating that the sickle cell allele underwent gene targeting. In clone IA5, the untargeted allele is the sickle cell allele. It was not possible to determine which allele underwent gene targeting in clones IA54, IB54, IC43 or IC52 presumably because these were mixed populations of both targeted and untargeted clones.
F. Table showing the number of puromycin or neomycin-resistant colonies picked for each experiment and the percentage of these that were determined to be correctly targeted by the PCR screening assay. Asterisk indicates that ZFNs were expressed as fusions to the heterodimeric ELD/KKR FokI nuclease domain as opposed to the heterodimeric EL/KK FokI nuclease domain (no asterisk).
Efficient correction of the sickle cell anemia mutation in human iPS cells using ZFN-mediated gene targeting
To perform gene correction in our sickle cell iPS cells, we transfected plasmids expressing each of the ZFN pairs together with the donor template into clone I (Materials and Methods). We performed two to four independent experiments for each of the three ZFN Pairs (A, B, and C) and obtained puromycin-resistant colonies for all transfections (Figure 3F). We manually picked and expanded drug-resistant clones, isolated genomic DNA, and performed PCR reactions to screen for the presence of the puromycin-resistance gene in the endogenous beta-globin gene using independent sets of primers expected to amplify the 5′ and the 3′ junctions in a correctly targeted beta-globin locus (Figure 3B). We achieved targeting efficiencies as high as 37.9% (mean of 9.8 %) as judged by this PCR screening assay (Figure 3B and 3F). DNA sequencing of 13 PCR-positive lines revealed the presence of a successfully targeted allele in all of these cells (Figure 3D). The presence of a polymorphism on the non-sickle allele allowed us to identify which of the two alleles had undergone targeting in each clone: seven of the 13 candidates (IA5, IA12, IA13, IA38, IB20, IB49 and IC54) were clonal populations with all but one (IA5) having undergone gene targeting of the sickle cell allele while six of the 13 candidates were mixed populations containing both targeted and non-targeted cells (IA54, IB48, IB50, IB54, IC43 and IC52). Following further expansion in culture, these six mixed populations resolved into clonal lines: two consisting of cells successfully targeted in the sickle E6V allele (IB48 and IB50) as judged by DNA sequencing (Figure 3D and 3E) and four consisting of non-targeted cells (IA54, IB54, IC43 and IC52) as determined by PCR analysis (Table S3). Importantly, it is worth noting that DNA sequence analysis showed that for all of the corrected clonal lines the non-targeted allele did not show evidence of ZFN-mediated indels introduced by non-homologous end joining (Figure 3E).
To provide further evidence of successful homologous recombination at the beta-globin locus, we also performed Southern blots on seven correctly targeted clones (IA13, IA38, IB20, IB48, IB49, IB50, IC54) using both 5′ external and 3′ external probes. These experiments confirmed stable presence of the puromycin-resistance insert in the beta-globin locus (Figure 3C and data not shown). In addition, Southern blots performed with these same probes on three of the four clones that had become negative for the gene targeting event as judged by PCR confirmed that these cells (IA54, IB54, and IC52) no longer contain a targeted clone following their expansion in culture (Figure 3C). Finally, Southern blots performed using a 5′ homology arm (internal) probe confirmed the absence of random insertions of the donor template in all seven correctly targeted clones (Figure S2B).
We next sought to demonstrate whether ZFN-induced gene correction could be performed in additional iPS cell lines. We therefore isolated puromycin or neomycin-resistant colonies from targeting experiments with iPS cell lines II and IV and identified successful gene correction in both lines, as judged by PCR, Southern blot and DNA sequencing (clone IIB1 in Figure 3B–F; clones IVA3 and IVC1 in Figure 3F and Figure S2A). DNA sequencing of the non-targeted allele in all three of these clones also revealed the absence of ZFN-induced indel mutations (Figure 3E and data not shown). Taken together, our results with iPS cell clones I, II and IV demonstrate that all three ZFN Pairs A, B, and C can efficiently induce stable gene correction of the sickle cell mutation in human iPS cells without inducing additional mutagenesis of the other beta-globin allele in the same cell.
Evaluation of ZFN off-target effects at other closely related globin genes
An important question to address for eventual therapeutic application of ZFNs is the range of unwanted “off-target” mutagenic effects caused by these reagents 16. For treatment of sickle cell disease, it is particularly important to ensure that the highly related paralogous genes encoding gamma-globin and delta-globin are not inadvertently mutagenized by the ZFNs targeted to beta-globin. The single delta-globin and two gamma-globin genes each possess DNA sequences that are very similar to the target sites for ZFN pairs A, B, and C (Figure S3A). DNA sequencing revealed that none of these sites were altered in any of the nine corrected iPS cell clones derived from the three different ZFN pairs in iPS lines I and II (Figure S3A). Additionally, we computationally identified 12 sites in human genome sequence that differed from the ZFN target sites by only a single base pair change (Supplementary Methods). We reasoned that these 3 sites for ZFN A and 9 sites for ZFNs B and C would be the most likely off-target sites for ZFN-induced mutations. Sequencing of these off-target sites in three targeted clones (IA13, IA38 and IB49) showed that none of them harbored ZFN-induced mutations (Figure S3B). These results show that our ZFNs did not affect very closely related off-target sites in both paralogous genes and the most closely related potential off-target sites in the genome during the process of correction, demonstrating the high degree of specificity these nucleases possess for their intended target sites.
Generation of transgene-free, gene-corrected iPS cell lines
We next sought to simultaneously excise both the floxed puromycin-resistance gene incorporated into the beta-globin intron and the randomly integrated floxed reprogramming factors by transiently expressing Cre recombinase in our corrected iPS cells. Three independent corrected lines derived from Clone I (IA13, IA38, and IB49) were infected with an adenovirus expressing a Cre recombinase-GFP fusion protein. Single cells with high level GFP expression were isolated by FACS 48 hours post-infection and plated. After two to three weeks, single colonies were picked and clonally expanded. PCR and restriction digest with the enzyme Bsu36I, which cleaves the wild-type but not sickle cell sequence, as well as DNA sequencing, confirmed excision of the puromycin-resistance cassette and the presence of the expected residual 34 bp loxP site “scar” with several associated restriction site sequences in the intron of the corrected beta-globin gene for six clones (IA13Δ2, IA13Δ3, IA13Δ5, IA38Δ2, IA38Δ6, and IB49Δ8) derived from the three corrected lines (Figures 4A, 4B and data not shown). Southern blotting verified excision of the puromycin-resistance cassette from these six clones (Figure 4C). PCR and Southern blot also demonstrated removal of the reprogramming factor cassette for these six clones (Figures 4D, 4E and data not shown). These successfully corrected, transgene-free iPS cell lines remained pluripotent as judged by expression of multiple pluripotency markers and in vitro differentiation into derivatives of the three germ layers (Figure 4F, 4G and Figure S4). Finally, SKY-FISH analysis showed that all six correctly targeted clones were karyotypically normal (Table S2). Thus, no translocations were observed despite transient ZFN and Cre recombinase expression in our cell lines harboring multiple loxP sequences.
Figure 4. Characterization of gene targeted iPS cell clones.
A. Schematic illustrating location of primers and restriction sites in the beta-globin locus used to identify gene-targeted clones that have undergone removal of the puromycin-resistance cassette. PCR primers external to donor template homology arms are depicted by arrows. The restriction enzyme Bsu36I will cleave sequence with a wild-type E6 codon but not sequence containing the E6V sickle cell mutation (asterisk). Residual “scar” sequence containing a single loxP site (black triangle) and flanking restriction sites (bold dark line) are left in the intron sequence. Expected sizes of bands are labeled. Not drawn to scale.
B. PCR and restriction digest analysis for removal of puromycin-resistance insert. PCR products were digested with Bsu36I and run on a 5% polyacrylamide gel to yield fragments of expected sizes as depicted in A.
C. Southern blot analysis to confirm removal of the puromycin-resistance insert. Blot was performed using an external probe that hybridizes upstream of the 5′ homology arm on genomic DNA digested with PvuII (as show in Figure 3A).
D. Southern blot analysis to confirm removal of reprogramming factors from gene targeted clones. Blot was performed using a probe that hybridizes to the c-myc gene on genomic DNA digested with BamHI. Genomic DNA from the sickle cell anemia patient primary fibroblasts was used as a control to define the endogenous c-myc allele. Note that the last three lanes are the same as the first three lanes in Figure 1F.
E. PCR screening assay for the WPRE sequence to verify successful excision of the STEMCCA reprogramming cassette
F. Immunofluorescence images of feeder-free cultures (clone IA13Δ2) stained for pluripotency-associated transcription factors and surface markers.
G. Immunofluorescence images of clone IA13Δ2 differentiated in vitro.
Discussion
In this study, we used publicly available engineered ZFN technology to effect facile, efficient, and stable correction of the E6V mutation in the beta-globin gene in three independent iPS cell lines derived from two patients with sickle cell anemia. Our data show that ZFN technology allows efficient gene targeting of disease-causing mutations with no evidence for off-target cleavage in closely related DNA sequences. Our results demonstrate that reprogramming and ZFN-induced gene targeting can be used in combination to create corrected patient-derived iPS cells and provide an important initial proof-of-principle that such an approach might be used in the future to develop therapies for monogenic diseases.
In contrast to previous studies including those with human iPS cells,9, 11, 29, 41 we introduced transgenic sequences up to 82 base pairs away from the ZFN cut site – a prerequisite for correcting disease-causing mutations. Indeed, previous work in mouse ES cells has demonstrated that the efficiency of alterations introduced by homologous recombination decreases substantially as distance from the nuclease-induced DSB increases.42, 43 Our studies illustrate that several configurations of the ZFN-induced DSB, the genetic correction event, and the drug-resistance cassette insertion point can be successfully used when performing ZFN-enhanced HR in human iPS cells. In one case, the ZFN-induced DSB (for ZFN target site 2) directly overlapped the mutation to be corrected with the drug-resistance cassette insertion positioned 82 bps downstream. This demonstrates that ZFNs can mediate cassette insertion even when it occurs nearly 100 bps away from the DSB. In a second case, the ZFN-induced DSB was positioned between the correction event and insertion point -- 24 bps downstream of the correction and 58 bps upstream of the insertion. Success in this latter configuration shows that two events, a correction and an insertion, can occur efficiently even when they are positioned some distance away from each other and on opposite sides of the DSB. These results allow us to draw the important conclusion that the gene correction event and the drug-resistance cassette insertion point do not need to be positioned precisely at the location of the ZFN-induced DSB.
A particularly important area for future investigation will be to define the full extent of genetic alterations introduced into iPS cells during generation and repair. Karyotype analyses showed that the reprogramming, ZFN-induced gene correction and Cre recombinase excision processes we performed did not introduce any gross chromosomal translocations or alterations into the patient-derived iPS cells. However, we know that one or more loxP “scar” sequences were left behind at undefined locations in the genome following excision of reprogramming factors by Cre recombinase. Furthermore, following correction of the sickle cell mutation, we also recognize that one residual loxP scar remains in the first intron of the beta-globin gene. Presumably, additional somatic mutations were either present in fibroblasts of the biopsied skin or introduced during the reprogramming process as suggested by recent studies examining genome-wide coding sequences and copy number variations 44, 45. Finally, although it remains theoretically possible that additional “off-target” mutations may have been introduced by the ZFNs, the absence of ZFN-induced indel mutations at the uncorrected beta-globin allele (with a perfect ZFN target site) as well as at highly similar sites in the paralogous delta- and gamma-globin genes and at closely matched, computationally identified potential off-target sites in any of our gene-corrected iPS clones strongly suggests that off-target effects are likely to be minimal.
Previous studies have used other methods such as adeno-associated virus (AAV) vector-based,46, 47 bacterial artificial chromosome-based,13, 48 and helper-dependent adenovirus-based14 donor templates to enhance gene-targeting efficiencies in human iPS cells. Although these technologies, like ZFNs, improve the targeting efficiency, the absolute efficiencies of successful recombination events are still sufficiently low enough to require the use of drug selection. In addition, the construction of the donor templates for these methods is more complicated than those required for ZFN-induced HR. However, we note that any of these three methods could also, in principle, be used together with ZFNs to effect correction in iPS cells. It will be of interest to determine if combined approaches could lead to further enhancements in the efficiencies of gene correction in human iPS cells. We also note that a recent report demonstrated the successful use of ZFNs together with donors lacking drug-selection cassettes (both plasmids and single-stranded oligonucleotides) to effect efficient gene correction or gene editing in human pluripotent stem cells.49 However, the efficiencies of gene alteration in these experiments were notably lower than those performed using donors harboring a drug-selection cassette.
Our efficient and reproducible correction of the sickle cell mutation in human iPS cells provides an important proof-of-principle and defines a framework for the use of engineered ZFN technology to correct a disease-causing mutation by HR. This result advances the field one important step closer to clinical realization of combined gene and cell therapy based on human iPS cells especially if such methods are used in combination with integration-free methods (e.g. mRNA- and or protein-based approaches) for the derivation of iPSCs.35, 37
Supplementary Material
Acknowledgments
We thank the patients who volunteered to donate a skin biopsy for this project, Rob Cho for consenting the patients and collecting the biopsies, Pei Wang for help with the Adeno-Cre infections and FACS sorting, Stacey Thibodeau-Beganny for help with plasmid construction, and Gerlinde Wernig for providing critical technical advice. This work was supported by National Institutes of Health R01 GM069906 (J.K.J.) and Pioneer Award DP1 OD006862 (J.K.J.); by National Science Foundation Graduate Research Fellowships (M.L.M. & C.L.R.); by Start-up funds of the Institute for Stem Cell Biology and Regenerative Medicine, Stanford University School of Medicine (M.W.); by the Donald E. and Delia B. Baxter Foundation (M.W.); and by the CIRM Training Grant for postdoctoral fellows (V.S.).
Footnotes
Author Contributions: Vittorio Sebastiano: Collection and/or assembly of data, data analysis and interpretation, manuscript writing, final approval of manuscript
Morgan L. Maeder: Collection and/or assembly of data, data analysis and interpretation, manuscript writing, final approval of manuscript
James F. Angstam: Collection and/or assembly of data, final approval of manuscript
Bahareh Haddad: Collection and/or assembly of data, final approval of manuscript
Cyd Khayter: Collection and/or assembly of data, final approval of manuscript
Dana T. Yeo: Collection and/or assembly of data, data analysis and interpretation, final approval of manuscript
Mathew J. Goodwin: Collection and/or assembly of data, final approval of manuscript
John S. Hawkins: Collection and/or assembly of data, final approval of manuscript
Cheire L. Ramirez: Collection and/or assembly of data, final approval of manuscript
Luis F. Z. Batista: Collection and/or assembly of data, final approval of manuscript
Steven E. Artandi: Financial support, data analysis and interpretation, final approval of manuscript
Marius Wernig: Conception and design, financial support, data analysis and interpretation, manuscript writing, final approval of manuscript
J. Keith Joung: Conception and design, financial support, data analysis and interpretation, manuscript writing, final approval of manuscript
References
- 1.Yamanaka S. Strategies and new developments in the generation of patient-specific pluripotent stem cells. Cell Stem Cell. 2007;1:39–49. doi: 10.1016/j.stem.2007.05.012. [DOI] [PubMed] [Google Scholar]
- 2.Saha K, Jaenisch R. Technical challenges in using human induced pluripotent stem cells to model disease. Cell Stem Cell. 2009;5:584–595. doi: 10.1016/j.stem.2009.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lowry WE, Richter L, Yachechko R, et al. Generation of human induced pluripotent stem cells from dermal fibroblasts. Proc Natl Acad Sci U S A. 2008;105:2883–2888. doi: 10.1073/pnas.0711983105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Park IH, Zhao R, West JA, et al. Reprogramming of human somatic cells to pluripotency with defined factors. Nature. 2008;451:141–146. doi: 10.1038/nature06534. [DOI] [PubMed] [Google Scholar]
- 5.Takahashi K, Tanabe K, Ohnuki M, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 6.Yu J, Vodyanik MA, Smuga-Otto K, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- 7.Hanna J, Wernig M, Markoulaki S, et al. Treatment of sickle cell anemia mouse model with iPS cells generated from autologous skin. Science. 2007;318:1920–1923. doi: 10.1126/science.1152092. [DOI] [PubMed] [Google Scholar]
- 8.Hockemeyer D, Jaenisch R. Gene Targeting in Human Pluripotent Cells. Cold Spring Harb Symp Quant Biol. 2011 doi: 10.1101/sqb.2010.75.021. [DOI] [PubMed] [Google Scholar]
- 9.Hockemeyer D, Soldner F, Beard C, et al. Efficient targeting of expressed and silent genes in human ESCs and iPSCs using zinc-finger nucleases. Nat Biotechnol. 2009;27:851–857. doi: 10.1038/nbt.1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Irion S, Luche H, Gadue P, et al. Identification and targeting of the ROSA26 locus in human embryonic stem cells. Nat Biotechnol. 2007;25:1477–1482. doi: 10.1038/nbt1362. [DOI] [PubMed] [Google Scholar]
- 11.Zou J, Maeder ML, Mali P, et al. Gene targeting of a disease-related gene in human induced pluripotent stem and embryonic stem cells. Cell Stem Cell. 2009;5:97–110. doi: 10.1016/j.stem.2009.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zwaka TP, Thomson JA. Homologous recombination in human embryonic stem cells. Nat Biotechnol. 2003;21:319–321. doi: 10.1038/nbt788. [DOI] [PubMed] [Google Scholar]
- 13.Howden SE, Gore A, Li Z, et al. Genetic correction and analysis of induced pluripotent stem cells from a patient with gyrate atrophy. Proc Natl Acad Sci U S A. 2011 doi: 10.1073/pnas.1103388108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu GH, Suzuki K, Qu J, et al. Targeted Gene Correction of Laminopathy-Associated LMNA Mutations in Patient-Specific iPSCs. Cell Stem Cell. 2011 doi: 10.1016/j.stem.2011.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Urnov FD, Rebar EJ, Holmes MC, et al. Genome editing with engineered zinc finger nucleases. Nat Rev Genet. 2010;11:636–646. doi: 10.1038/nrg2842. [DOI] [PubMed] [Google Scholar]
- 16.Handel EM, Cathomen T. Zinc-finger nuclease based genome surgery: it’s all about specificity. Curr Gene Ther. 2011;11:28–37. doi: 10.2174/156652311794520120. [DOI] [PubMed] [Google Scholar]
- 17.Carroll D. Zinc-finger nucleases: a panoramic view. Curr Gene Ther. 2011;11:2–10. doi: 10.2174/156652311794520076. [DOI] [PubMed] [Google Scholar]
- 18.Kim YG, Cha J, Chandrasegaran S. Hybrid restriction enzymes: zinc finger fusions to Fok I cleavage domain. Proc Natl Acad Sci U S A. 1996;93:1156–1160. doi: 10.1073/pnas.93.3.1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bae KH, Kwon YD, Shin HC, et al. Human zinc fingers as building blocks in the construction of artificial transcription factors. Nat Biotechnol. 2003;21:275–280. doi: 10.1038/nbt796. [DOI] [PubMed] [Google Scholar]
- 20.Mandell JG, Barbas CF., 3rd Zinc Finger Tools: custom DNA-binding domains for transcription factors and nucleases. Nucleic Acids Res. 2006;34:W516–523. doi: 10.1093/nar/gkl209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Carroll D, Morton JJ, Beumer KJ, et al. Design, construction and in vitro testing of zinc finger nucleases. Nat Protoc. 2006;1:1329–1341. doi: 10.1038/nprot.2006.231. [DOI] [PubMed] [Google Scholar]
- 22.Maeder ML, Thibodeau-Beganny S, Osiak A, et al. Rapid “open-source” engineering of customized zinc-finger nucleases for highly efficient gene modification. Mol Cell. 2008;31:294–301. doi: 10.1016/j.molcel.2008.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maeder ML, Thibodeau-Beganny S, Sander JD, et al. Oligomerized pool engineering (OPEN): an ‘open-source’ protocol for making customized zinc-finger arrays. Nat Protoc. 2009;4:1471–1501. doi: 10.1038/nprot.2009.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sander JD, Dahlborg EJ, Goodwin MJ, et al. Selection-free zinc-finger-nuclease engineering by context-dependent assembly (CoDA) Nat Methods. 2011;8:67–69. doi: 10.1038/nmeth.1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Doyon Y, McCammon JM, Miller JC, et al. Heritable targeted gene disruption in zebrafish using designed zinc-finger nucleases. Nat Biotechnol. 2008;26:702–708. doi: 10.1038/nbt1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang F, Maeder ML, Unger-Wallace E, et al. High frequency targeted mutagenesis in Arabidopsis thaliana using zinc finger nucleases. Proc Natl Acad Sci U S A. 2010;107:12028–12033. doi: 10.1073/pnas.0914991107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Townsend JA, Wright DA, Winfrey RJ, et al. High-frequency modification of plant genes using engineered zinc-finger nucleases. Nature. 2009;459:442–445. doi: 10.1038/nature07845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Foley JE, Yeh JR, Maeder ML, et al. Rapid mutation of endogenous zebrafish genes using zinc finger nucleases made by Oligomerized Pool ENgineering (OPEN) PLoS ONE. 2009;4:e4348. doi: 10.1371/journal.pone.0004348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zou J, Sweeney CL, Chou BK, et al. Oxidase deficient neutrophils from X-linked chronic granulomatous disease iPS cells: functional correction by zinc finger nuclease mediated safe harbor targeting. Blood. 2011 doi: 10.1182/blood-2010-12-328161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sommer CA, Stadtfeld M, Murphy GJ, et al. Induced pluripotent stem cell generation using a single lentiviral stem cell cassette. Stem Cells. 2009;27:543–549. doi: 10.1634/stemcells.2008-1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Miller JC, Holmes MC, Wang J, et al. An improved zinc-finger nuclease architecture for highly specific genome editing. Nat Biotechnol. 2007;25:778–785. doi: 10.1038/nbt1319. [DOI] [PubMed] [Google Scholar]
- 32.Doyon Y, Vo TD, Mendel MC, et al. Enhancing zinc-finger-nuclease activity with improved obligate heterodimeric architectures. Nat Methods. 2011;8:74–79. doi: 10.1038/nmeth.1539. [DOI] [PubMed] [Google Scholar]
- 33.Okita K, Nakagawa M, Hyenjong H, et al. Generation of mouse induced pluripotent stem cells without viral vectors. Science. 2008;322:949–953. doi: 10.1126/science.1164270. [DOI] [PubMed] [Google Scholar]
- 34.Yu J, Hu K, Smuga-Otto K, et al. Human induced pluripotent stem cells free of vector and transgene sequences. Science. 2009;324:797–801. doi: 10.1126/science.1172482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim D, Kim CH, Moon JI, et al. Generation of human induced pluripotent stem cells by direct delivery of reprogramming proteins. Cell Stem Cell. 2009;4:472–476. doi: 10.1016/j.stem.2009.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fusaki N, Ban H, Nishiyama A, et al. Efficient induction of transgene-free human pluripotent stem cells using a vector based on Sendai virus, an RNA virus that does not integrate into the host genome. Proc Jpn Acad Ser B Phys Biol Sci. 2009;85:348–362. doi: 10.2183/pjab.85.348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Warren L, Manos PD, Ahfeldt T, et al. Highly efficient reprogramming to pluripotency and directed differentiation of human cells with synthetic modified mRNA. Cell Stem Cell. 2010;7:618–630. doi: 10.1016/j.stem.2010.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Somers A, Jean JC, Sommer CA, et al. Generation of transgene-free lung disease-specific human induced pluripotent stem cells using a single excisable lentiviral stem cell cassette. Stem Cells. 2010;28:1728–1740. doi: 10.1002/stem.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sander JD, Maeder ML, Reyon D, et al. ZiFiT (Zinc Finger Targeter): an updated zinc finger engineering tool. Nucleic Acids Res. 2010;38:W462–468. doi: 10.1093/nar/gkq319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Santiago Y, Chan E, Liu PQ, et al. Targeted gene knockout in mammalian cells by using engineered zinc-finger nucleases. Proc Natl Acad Sci U S A. 2008;105:5809–5814. doi: 10.1073/pnas.0800940105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.DeKelver RC, Choi VM, Moehle EA, et al. Functional genomics, proteomics, and regulatory DNA analysis in isogenic settings using zinc finger nuclease-driven transgenesis into a safe harbor locus in the human genome. Genome Res. 2010;20:1133–1142. doi: 10.1101/gr.106773.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Donoho G, Jasin M, Berg P. Analysis of gene targeting and intrachromosomal homologous recombination stimulated by genomic double-strand breaks in mouse embryonic stem cells. Mol Cell Biol. 1998;18:4070–4078. doi: 10.1128/mcb.18.7.4070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stark JM, Pierce AJ, Oh J, et al. Genetic steps of mammalian homologous repair with distinct mutagenic consequences. Mol Cell Biol. 2004;24:9305–9316. doi: 10.1128/MCB.24.21.9305-9316.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hussein SM, Batada NN, Vuoristo S, et al. Copy number variation and selection during reprogramming to pluripotency. Nature. 2011;471:58–62. doi: 10.1038/nature09871. [DOI] [PubMed] [Google Scholar]
- 45.Gore A, Li Z, Fung HL, et al. Somatic coding mutations in human induced pluripotent stem cells. Nature. 2011;471:63–67. doi: 10.1038/nature09805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mitsui K, Suzuki K, Aizawa E, et al. Gene targeting in human pluripotent stem cells with adeno-associated virus vectors. Biochem Biophys Res Commun. 2009;388:711–717. doi: 10.1016/j.bbrc.2009.08.075. [DOI] [PubMed] [Google Scholar]
- 47.Khan IF, Hirata RK, Wang PR, et al. Engineering of human pluripotent stem cells by AAV-mediated gene targeting. Mol Ther. 2010;18:1192–1199. doi: 10.1038/mt.2010.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Song H, Chung SK, Xu Y. Modeling disease in human ESCs using an efficient BAC-based homologous recombination system. Cell Stem Cell. 2010;6:80–89. doi: 10.1016/j.stem.2009.11.016. [DOI] [PubMed] [Google Scholar]
- 49.Soldner F, Laganiere J, Cheng AW, et al. Generation of isogenic pluripotent stem cells differing exclusively at two early onset Parkinson point mutations. Cell. 2011;146:318–331. doi: 10.1016/j.cell.2011.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.