Abstract
The p53 DNA damage response attenuated with age and we have evaluated downstream factors in the DNA damage response. In old animals p21 protein accumulates in the whole cell fraction but significantly declines in the nucleus, which may alter cell cycle and apoptotic programs in response to DNA damage. We evaluated the transcriptional response to DNA damage in young and old and find 2692 genes are differentially regulated in old compared to young in response to oxidative stress (p<0.005). As anticipated, the transcriptional profile of young mice is consistent with DNA damage induced cell cycle arrest while the profile of old mice is consistent with cell cycle progression in the presence of DNA damage, suggesting the potential for catastrophic accumulation of DNA damage at the replication fork. Unique sets of DNA repair genes are induced in response to damage in old and young, suggesting the types of damage accumulating differs between young and old. The DNA repair genes upregulated in old animals point to accumulation of replication-dependent DNA double strand breaks (DSB). Expression data is consistent with loss of apoptosis following DNA damage in old animals. These data suggest DNA damage responses differ greatly in young and old animals.
INTRODUCTION
A gradual and progressive decline in genomic integrity is observed with age (Warner and Price, Bohr and Anson, Walter et al., Vijg et al.). We and others have reported that DNA repair capacity, specifically base excision repair (BER) capacity is reduced with age (Cabelof et al. 2002a, Intano et al.), and have suggested a causal role for this reduced repair capacity in the age-dependent decrease in genomic stability. BER resolves the spontaneous DNA damage that accumulates with age, but also resolves lesions induced by agents that produce small, non-helix distorting DNA damage. In addition to loss of baseline repair capacity with age, we and others have shown that the DNA damage response is altered with age, evidenced by loss of inducibility of a rate-determining protein in BER, DNA polymerase β (β-pol) (Cabelof et al., 2006b; Kaneko et al.). As such, aging reduces both endogenous BER and the ability to respond to exogenous stressors by the BER pathway. We have suggested that loss of the BER response to damage with age results from a reduced p53 response with age. In support, loss of the p53 response with has been reported in aged human dermal fibroblasts exposed to UV (Goukassian et al.), and in old mouse liver exposed to oxidative stress (Cabelof et al., 2002b). Specifically, we showed that in response to oxidative stress induced by 2Nitropropane (2NP), young animals exhibited a robust induction in p53 protein in liver, while old animals completely lost this response and actually slightly reversed it (Cabelof et al., 2002b). In a more comprehensive evaluation of the p53 response, Feng et al. investigated the impact of aging on the p53 response in several inbred strains using a variety of stressors and also found a loss of the p53 response with age in a variety of tissues, including spleen, kidney and brain.
In this study, we are interested in evaluating the role that altered p53 functionality may play in determining the phenotypic responses to DNA damage in young and old animals. A potential loss of p53 functionality with age is interesting, as various p53 mouse models suggest a direct role for this gene product in the aging process (Campisi). As a key player in the regulation of DNA damage responses, the loss of a p53 response in aged animals would lead us to anticipate broad effects of age on DNA damage signaling and processing. Because of the pleitropic nature of p53, loss of functionality with age should induce significant differential expression of p53 target genes in old animals compared to young animals in response to DNA damage. We are interested in DNA repair, cell cycle regulation and apoptosis as these are all p53-dependent pathways known to be altered with aging. We have utilized transcriptional profiling as a nonbiased approach to elucidate the impact of aging on the DNA damage response and on p53 functionality. In addition, we have developed an approach by which to compare our transcript data against databases of p53-target genes (Riley et al.) and p53-responsive genes (Kho et al.). Both the transcriptional signatures and the database comparisons point strongly to loss of p53 as an important driving factor in the altered DNA damage responses with age. Accordingly, these data point to loss of p53 functionality as an important source of genomic instability with age. Future studies should be aimed at identification of the upstream source(s) of p53 dysfunction, as those gene products may prove to be important molecular targets in the preservation of genomic integrity over time.
METHODS
Animals
Experiments were performed in young (4–6 months) and old (24–28 months) C57BL/6 male specific pathogen-free mice in accordance with the NIH guidelines for the use and care of laboratory animals. Old mice were aged at the University of Texas Health Science Center, San Antonio and shipped to Wayne State University for treatment exposure and termination. Animals were acclimatized over two weeks prior to initiation of exposure studies. The animal protocols were approved by the Wayne State University and the University of Texas Health Science Center Animal Investigation Committees. Mice were maintained on a 12-hour light/dark cycle and were fed a standard mouse lab chow and water ad libitum. Animals were subjected to one i.p. injection of 100 mg/kg body weight 2NP (Sigma-Aldrich, CAS registry number 79-46-9, St. Louis, MO), or olive oil vehicle. Mice were sacrificed 24 hours post exposure by CO2 asphyxiation and cervical dislocation. Organs were flash frozen in liquid nitrogen and stored at −70°C.
RNA Isolation and Quality Assessment
RNA was isolated from livers of untreated and 2NP treated young and old mice using the VersaGene RNA Isolation Kit (Manufacturer). Electorpherograms were run for all samples to assess RNA quality and RNA integrity (RIN) values were determined using the Agilent Bioanalyzer. RNAs with RIN values between 8.2 and 9.1 were used for the array hybridizations.
Gene Expression Profiling
Gene expression profiles comparing the response to carcinogen in young mice to the response to carcinogen in old mice was determined using the Agilent Whole Mouse Genome oligonucleotide micorarray containing probes for over 41,000 well characterized genes and transcripts. Sample labeling was done using the Agilent Low Input Fluorescent Linear Amplification Kit. For each microarray, 500 ng of total RNA for each sample was used in the labeling reaction which uses a single amplification step. In the initial cDNA synthesis from total RNA, a primer containing poly dT and a T7 polymerase promoter was used to anneal to the poly A RNA. The first and second strand of cDNA was synthesized using MMLV Reverse transcriptase. The labeled cRNA is purified using Qiagen RNeasy (Qiagen, Valencia, CA). Concentrations of cRNA and dye labeling efficiency were determined using the Nanodrop spectrophotometer. In the hybridization reaction, equal quantities of the Alexa 647 and Alexa 555 labeled samples were mixed together and allowed to co-hybridize on the array for 17 hours at 60° C.
A balanced block experimental design was used: each microarray included a labeled sample from a 2NP treated animal co-hybridized with a labeled sample from an untreated animal. Four microarrays were completed using liver RNA from the young group, and four microarrays were completed using liver RNA from the old group. Samples on a given array were oppositely labeled with Alexa 647 and Alexa 555 dyes. The four microarrays for a given group (i.e., young or old) represent samples from eight separate mice, providing consideration of biological variation. In total, 8 arrays representing 16 mice (8 young, 8 old) were completed. Dye swaps were used to account for dye bias effects such that of the four arrays in a given phenotype group, two had 2NP treated samples labeled with Alexa 647 co-hybridized with control samples labeled with Alexa 555 while the other two arrays within the same phenotype group had opposite dye orientations.
Microarrays were scanned using the Agilent dual laser DNA micorarray scanner, model G2565AA, with 10 micron resolution. Microarray image analysis was performed with Agilent Feature Extraction software, version A.5.1.1. Outlier features (probes) having aberrant image characteristics were flagged and excluded from subsequent analysis, and the percent of “good” features was determined. Fluorescent intensity values were adjusted using local background subtraction. Lowess intensity-dependent normalization was used to correct for systematic dye bias. For each probe on the array a log ratio was calculated, representing the relative abundance of transcript in the treated sample to untreated. Changes in gene expression are presented as fold change values, calculated as follows: where T is the expression value for a 2NP treated sample and C is the expression value for the respective control.
The comparison between ΔO and ΔY is also presented as a fold change, calculated as:
Identification of biological processes and pathways differentially affected by 2NP treatment in old compared to young was performed using GenMapp, KEGG and DAVID.
Hierarchical clustering was performed using GeneSpring version 7.3. Samples (microarrays) representing biological replicates were clustered with the Euclidean distance metric. Cluster analysis was performed independently using the data for genes belonging to each of three gene ontologies: apoptosis, cell cycle arrest, and cell cycle progression. Red heatmap coloring indicates increased gene expression in response to 2NP treatment, relative to control, while blue represents a decrease in expression.
Real-time RT-PCR validation of array
Total RNAs were isolated from the livers of untreated and 2NP treated young and aged mice using VersaGene as described above. Validation was completed with RNA from mice used in the array hybridizations as well as with RNA from mice that were not used in the array hybridizations. cDNAs were synthesized from 2µg RNA using random hexamer primers as described previously (Cabelof et al.. 2006a) and purified with the QIAquick PCR Purification Kit (Qiagen, Valencia, CA). Transcripts were quantitated with a LightCycler real time PCR machine (Roche). PCR reactions contained 2µl purified cDNA, 4mM MgCl2, 0.5 µM each of sense and antisense primers, and 2µl FastStart DNA Master SYBR Green I enzyme-SYBR reaction mix (Roche). For all amplifications, PCR conditions consisted of an initial denaturing step of 99°C for 10 min, followed by 35–55 cycles of 96°C for 10s, optimal annealing temperatures based on specific primer pairs for 10s and 72°C for 5s, with a melting curve analysis from 40°C to 99°C to confirm specificity. External standards were prepared by amplification of cDNAs for all genes validating the array. The amplicons were cloned into pGEM-T Easy vector, the vectors were linearized and used to prepare external standard curves. Transcripts were normalized to β-actin. Results are expressed as mean values obtained from 10 animals per experimental group.
Western analysis
Protein was obtained using the CelLytic NuclearExtraction kit (Sigma, St. Louis, MO) per manufacturer protocol. Whole cell extracts and crude nuclear extracts from liver tissues of young and old, control and treated animals were subjected to SDS-PAGE and transferred to nitrocellulose using a BioRad semi-dry transfer apparatus according to the manufacturer's protocol. Equal loading of proteins was determined by gel staining (GelCode, Pierce, Rockford, IL) and equal transfer was determined by reversible staining of the membrane with MemCode (Pierce, Rockford, IL). Western analysis was accomplished using affinity purified monoclonal antisera developed against mouse p53 (pAb240 GenWay Biotech, SanDiego, CA) and mouse p21 (7960 Abcam, Cambridge, MA). p21 bands did not migrate according to molecular weight, so a positive control was run on every gel to ensure identification of the accurate band (SAPK/JNK Control cell extracts, #9253 Cell Signaling Technology, Danvers, MA). The positive control cells were 293 cells exposed to UV and harvested 30 minutes after exposure. In some instances, the positive control signal was significantly lighter than the experimental samples and required additional exposure. In this eventuality two exposures were obtained and the images were spliced side by side (indicated by break in the gel image). B-tubulin (6046 Abcam, Cambridge, MA) and TATA binding protein (818 Abcam, Cambridge, MA) were used as loading controls. Bands were detected by ChemiImager after incubation in SuperSignal Chemiluminescent Substrate luminol/enhancer and SuperSignal chemiluminescent Substrate stable peroxide solution (Pierce, Rockford, IL). Intensity of the bands was quantified by ChemiImager, and the data are expressed as the integrated density of the band per µg protein loaded.
Identification of p53 target genes and p53-responsive genes
In order to evaluate the role that p53 dysregulation may play in the response of old animals to oxidative stress we evaluated the expression of p53-target genes and p53-regulated genes in response to 2NP in young and old animals. Our objective was to complete an unbiased screen of the array data against genes known to be either direct targets of p53 or indirectly regulated by p53 in response to carcinogen exposure. We generated two lists of p53-related genes from two publications in which these genes were identified. The first list names genes that have been experimentally demonstrated to be direct p53-target genes by at least 3 of the following 4 criteria: presence of a p53 response element close to or in the gene; up- or downregulation of the mRNA and protein by activated wildtype p53 but not by mutant p53; p53 activation of a cloned promoter for the gene; and/or chromatin immunoprecipitation using p53 antibody showing presence of p53 protein on the response element site of the gene (Riley et al.). Genes identified in the Riley et al. study are human genes, so mouse homologues were identified using NCBI Homologene. Briefly, homo sapiens accession numbers and gene names were cross-referenced and used to identify the homologous mus musculus accession number. When accounting for human genes without identifiable mouse homologues, this generated 115 mouse genes to be analyzed. The array data was then screened for each mouse accession number as follows: all statistically significant expression changes in young mice exposed to 2NP were screened against each p53 target gene; any gene identified as being differentially expressed in response to 2NP was noted as up- or down-regulated; these genes were then evaluated in old mice exposed to 2NP; impact of treatment on expression was noted as up- or downregulated or unchanged. The second list names genes that have been shown by genetic means to be responsive to carcinogen exposure in a p53-dependent manner (Kho et al.). As described above, mouse homologues were identified, generating a list of 34 genes upregulated in a p53-dependent manner, and 149 genes downregulated in a p53-dependent manner. As above, these genes were screened against the array data.
Statistical analysis
Microarray data analysis was done using Rosetta Resolver (Rosetta Biosoftware) and GeneSpring (Silicon Genetics). Analysis of variance (ANOVA) was used to identify genes that have a statistically significant difference when comparing the log ratios of the young and old groups. The false discovery rate (FDR) was controlled by using the Benjamini & Hochberg multiple test correction in the analysis. The FDR was also controlled by setting p<0.001 when determining differences in log ratios in response to 2NP (FDR 2.4%), and p<0.005 when determining ΔO:ΔY (FDR 7.7%). Data analysis for the real-time RT-PCR validation and western analyses was accomplished by ANOVA, with significance set at a p-value of <0.05.
RESULTS
Western analyses
In this study the DNA damage response in old animals is evaluated following exposure to a well-characterized DNA oxidizing agent, 2-nitropropane (2NP). We have previously demonstrated that exposure to a 100mg/kg dose of 2NP followed by sacrifice at 24 hours induces a 5-fold increase in levels of 8-hydroxydeoxyguanosine (Cabelof et al., 2002b), and equivalent stress responses in both young and old animals (Cabelof et al., 2006b). To evaluate whether approximately equivalent damage was induced in both young and old animals, we evaluated expression of GRO (growth-related oncogene), a gene strongly induced by exposure to diesel exhaust (which includes 2NP) (Salvi et al., 2000). Importantly, GRO is induced in a dose-dependent manner in response to DNA damage (Hamadeh et al., 2002), and we see equivalent induction of GRO in both young and old mice exposed to 2NP (Cabelof et al., 2006b), suggesting similar DNA damage loads in both young and old. This is important, because 2NP requires activation by an aryl sulfotransferase (Leakey et al.). However activity of this aryl sulfotransferase has been shown to be unaffected by aging (Guo et al.,Tarloff et al., Runge-Morris) such that the initial DNA damage load should be similar between young and old mice.
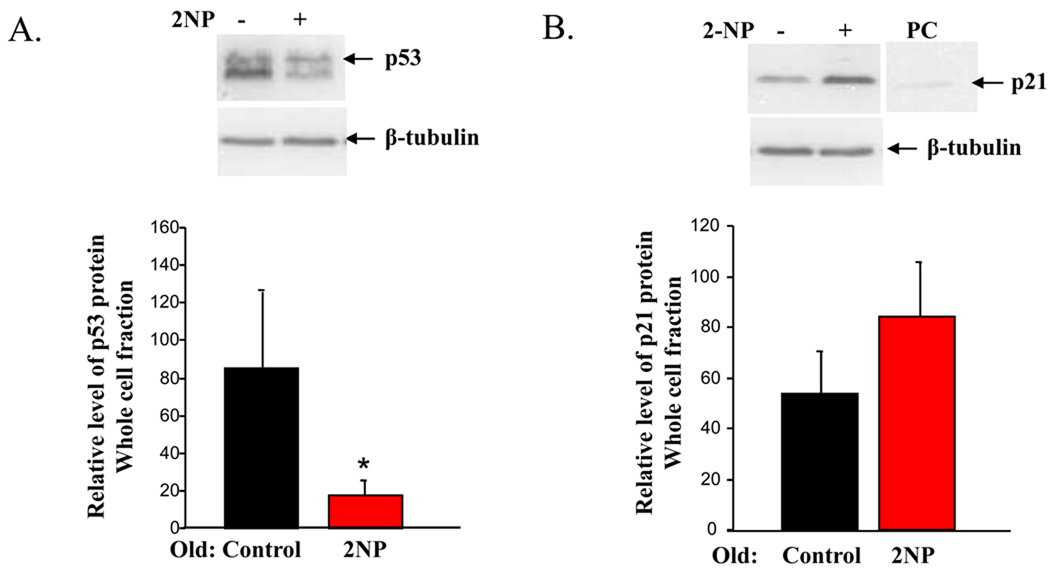
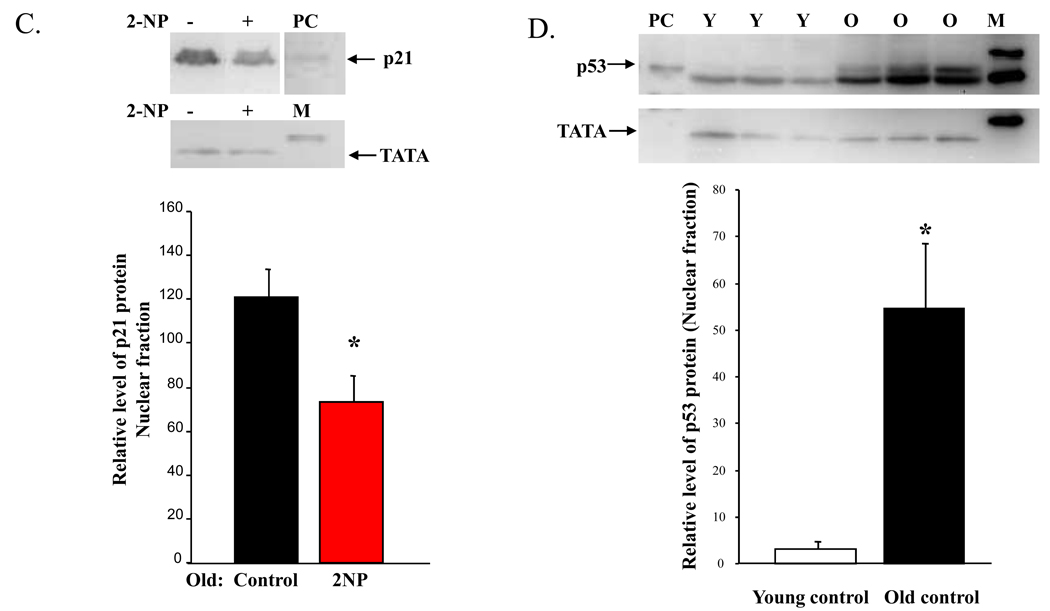
We previously demonstrated that in the nuclear fraction, p53 protein was reduced in old mice exposed to 2NP (Cabelof et al., 2006b). In this paper, we were interested in determining whether we would see a similar effect of aging throughout the cell. We looked at p53 protein accumulation in the whole cell fraction and found that p53 protein levels are significantly reduced in old, treated animals (Figure 1A, 79% reduction, p<0.05), validating the decline in p53 previously observed in the nuclear fraction. This reduction in p53 protein in response to DNA damage is in sharp contrast to the induction of p53 seen in young animals treated with DNA damaging agents, including 2NP. Whether this loss of p53 would impact downstream target genes was evaluated through investigation of p21 protein levels. We anticipated a p53-dependent loss of p21 induction in response to DNA damage in old animals, but did not see this in the whole cell fraction. Surprisingly we found that in the whole cell fraction p21 levels were unchanged, and trended higher in old, 2NP treated animals (Figure 1B), suggesting either that p53 transcriptional activation of p21 is not altered with age, or that p21 transcriptional activation was p53-independent. Interestingly, we did observe a significant decline in p21 protein in the nuclear fraction from old, treated animals (Figure 1C, 40% reduction, p<0.02). As only nuclear p21 can inhibit cell cycle progression (Rodríguez-Vilarrupla et al.), these data suggest a p53-independent induction of transcript and altered post-translational regulation of p21 with age.
Figure 1. Differential induction and cellular localization of p53 and p21 protein in response to 2NP in young and old animals.
Whole cell and crude nuclear extracts were isolated from liver tissue of young (4–6 months) and old (24–28 months) mice injected with 2NP and their non-injected counterparts as described in Materials and Methods. Protein levels were normalized to µg protein loaded on gel and verified by gelCode and memCode staining. Representative images are shown. A. Loss of p53 response in old animals exposed to 2NP. Levels of p53 protein were determined in whole cell extracts from old animals untreated and treated with 2NP. β-tubulin serves as loading control. Values represent an average (±SEM) for p53 data obtained from five animals in each group. −, no 2NP; +, 2NP injected; (*) value significantly different from control at p<0.05. B. Induction of p21 protein in old animals exposed to 2NP. Levels of p21 protein were determined in whole cell extracts from old animals untreated and treated with 2NP. β-tubulin serves as loading control. Values represent an average (±SEM) for p21 data obtained from five animals in each group. −, no 2NP; +, 2NP injected. C. Loss of nuclear p21 protein in old animals exposed to 2NP. Levels of p21 protein were determined in crude nuclear extracts from old animals untreated and treated with 2NP. TATA1 binding protein serves as loading control and marker of subcellular fractionation. Values represent an average (±SEM) for p21 data obtained from five animals in each group. −, no 2NP; +, 2NP injected. Positive control: (*) value significantly different from control at p<0.05. D. Aging results in accumulation of p53 in the nucleus. Levels of p53 protein were determined in crude nuclear extracts from young and old control animals (untreated). TATA1 binding protein serves as loading control and marker of subcellular fractionation. Values represent an average (±SEM) for p53 data obtained from three animals in each group. PC= positive control; M=marker; Y=young; O=old.
While we continue to hypothesize that loss of the p53 DNA damage response with age is mechanistically important in the aberrant DNA damage response that occurs with age, clearly not all p53-target genes are negatively impacted by age. Based on our data and corroborated by data from Feng et al. and Goukassian et al, it is, nonetheless apparent that in response to a variety of stressors and in a variety of tissues and strains, the p53 DNA damage response is attenuated, or even lost, with age. We suggest that loss of the p53 DNA damage response has an important and broad impact on downstream DNA damage responses. One unaddressed question is the impact of aging on p53 protein accumulation in the absence of DNA damaging agents in the target organ of these studies, liver. To address this, we have measured p53 protein levels in livers of control (untreated) young and old mice. We find a very large increase in p53 accumulation in old liver (Figure 1D, 17-fold increase, p<0.01). This difference is maintained, but attenuated to an 8-fold increase with age if we normalize the p53 to TATA (IDV p53/IDV TATA). Additionally, the positive control points to the top band as the p53 band, which is what we have quantified. However, it is possible that both bands represent p53, as all the p53 protein may have been phosphorylated in the UV-treated positive control cells. In either event, it is clear from the image that both bands are significantly increased with age. This impact of aging might provide an explanation for the inability of old animals to induce the critical, acute p53 response when exposed to DNA damaging agents. Our aim in this work is to investigate which genes are impacted by aging in response to DNA damage.
Gene expression profiling and real time RT-PCR validation
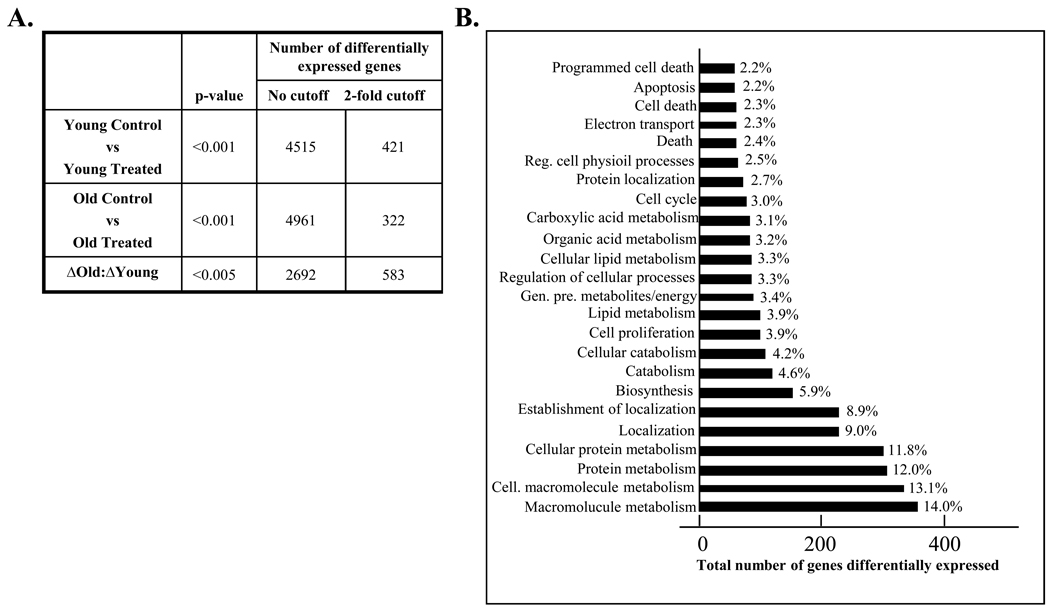
To address this question, we have used a transcriptional profiling approach, comparing the DNA damage response in young to the DNA damage response in old animals. To achieve the greatest statistical power from our study design, we have directly evaluated the DNA damage response, as described in methodology, and find that the gene expression profile in response to treatment is very different in old animals as compared to young. The number of genes differentially expressed between experimental groups is presented in Figure 2A. We have presented the data with information on statistically significant, differentially expressed genes irrespective of fold change, and the number of genes differentially expressed by 2-fold or more. We find that in aging studies small, persistent changes can induce significant phenotypes, and thus we resist use of an arbitrary cutoff for gene selection. However, the importance of very small fold changes is arguably minimal and we avoid further interpretation of genes with very small fold changes. In comparing the response in young to the response in old, we find significant differential responses in 454 (2-fold) genes, or 2692 genes (no fold restriction), demonstrating a differential response to carcinogen exposure in old compared to young animals. The microarray platform provided excellent feature morphology, low background signal, and a high signal to noise ratio. Over 99.5% of all array features passed quality filtering, and less than 0.5% were flagged as outliers. In an initial assessment of the differentially expresses genes, we used DAVID to evaluate the distribution of genes across biological processes. This allows us to determine which biological pathways have an enrichment of differentially expressed genes in the array, as shown in Figure 2B. As anticipated, we see an overrepresentation of genes from categories involved in cell cycle and apoptosis/cell death.
Figure 2. Gene expression profiling of DNA damage response in young and old animals.
RNA was isolated from liver tissue of young (4–6 months) and old (24–28 months) animals injected with 2NP and their non-injected counterparts and gene expression profiles were determined using the Agilent Whole Mouse Genome oligonucleotide microarray as described in Materials and Methods. A. Enumeration of differentially expressed genes. Determination of differentially expressed genes was accomplished using Rosetta Resolver (Rosetta Biosoftware) and GeneSpring (Silicon Genetics). Analysis of variance (ANOVA) was used to identify genes that have a statistically significant difference when comparing the log ratios of the young and old groups (change in old relative to change in young, ΔOld:ΔYoung). For each probe on the array a log ratio was calculated, representing the relative abundance of transcript in the treated sample to untreated. The false discovery rate was controlled by using the Benjamini & Hochberg multiple test correction in the analysis and a p-value cutoff of <0.005. B. Identification of biological processes impacted by 2NP treatment in old relative to young. Genes differentially expressed in response to treatment in old relative to young were organized by biological pathway using DAVID. The total number of genes differentially expressed (ΔOld:ΔYoung) is organized by biological process. The percent annotation indicates the percent of all genes within each pathway that are differentially expressed between treatment groups, demonstrating overrepresentation of these pathways in the response to treatment in old animals. Non-informative pathways were omitted from presentation.
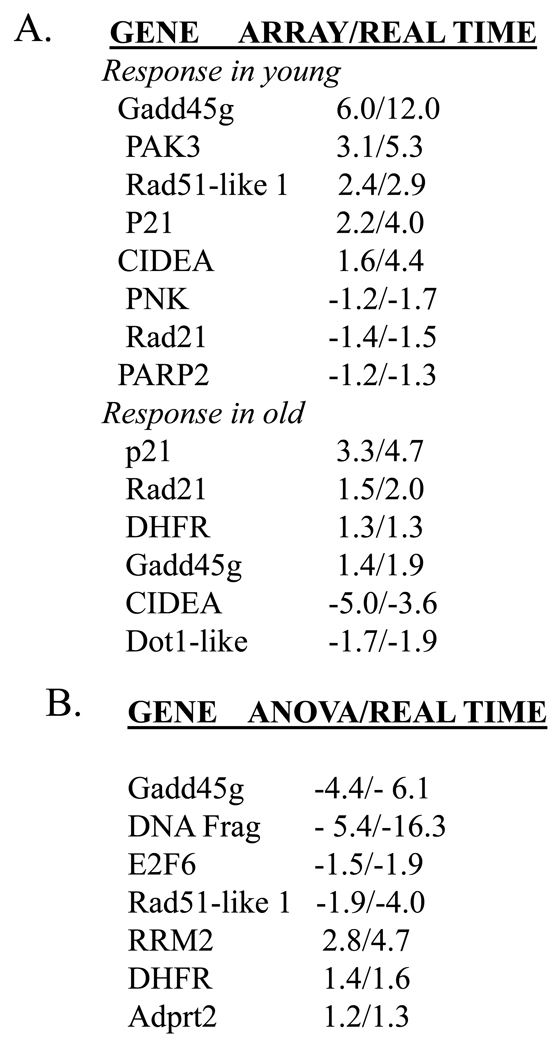
In order to validate the microarray data, we have conducted real-time RT-PCR on a number of genes differentially expressed (as determined in the microarray). We have validated both the samples used in the array hybridizations (4 young control/4 young treated/4 old control/4 old treated), as well as additional samples not used in array hybridizations (external validation). We find, in every case, that real-time RT-PCR data validates the microarray data (Figure 3A). Further, we experimentally validated the statistically derived differences comparing the response in young to the response in old, and again all array data was validated (Figure 3B).
Figure 3. Validation of array data by real-time quantitative RT-PCR.
cDNAs were prepared from RNA isolated from liver tissue of young (4–6 months) and old (24–28 months) mice injected with 2NP and their non-injected counterparts as described in Materials and Methods. A. Validation of genes differentially expressed in response to 2NP in young and in response to 2NP in old animals. Differential expression as determined by array represents average fold change from 4 arrays for each group (p<0.005). Real-time RT-PCR data represents the average fold change for 10 animals from each group (±SEM, p<0.05). Data is presented as fold change array/fold change real-time. B. Experimental validation of statistically derived differences comparing response in old to response in young (ΔOld:ΔYoung). Differential expression in response to treatment in old relative to young (ΔOld:ΔYoung) was determined by ANOVA (p<0.005). Real-time RT-PCR data represents the average fold change for 10 animals from each group (±SEM, p<0.05). Data is presented as fold change ANOVA/fold change real-time.
Genes/pathways differentially expressed in response to treatment in old and young animals
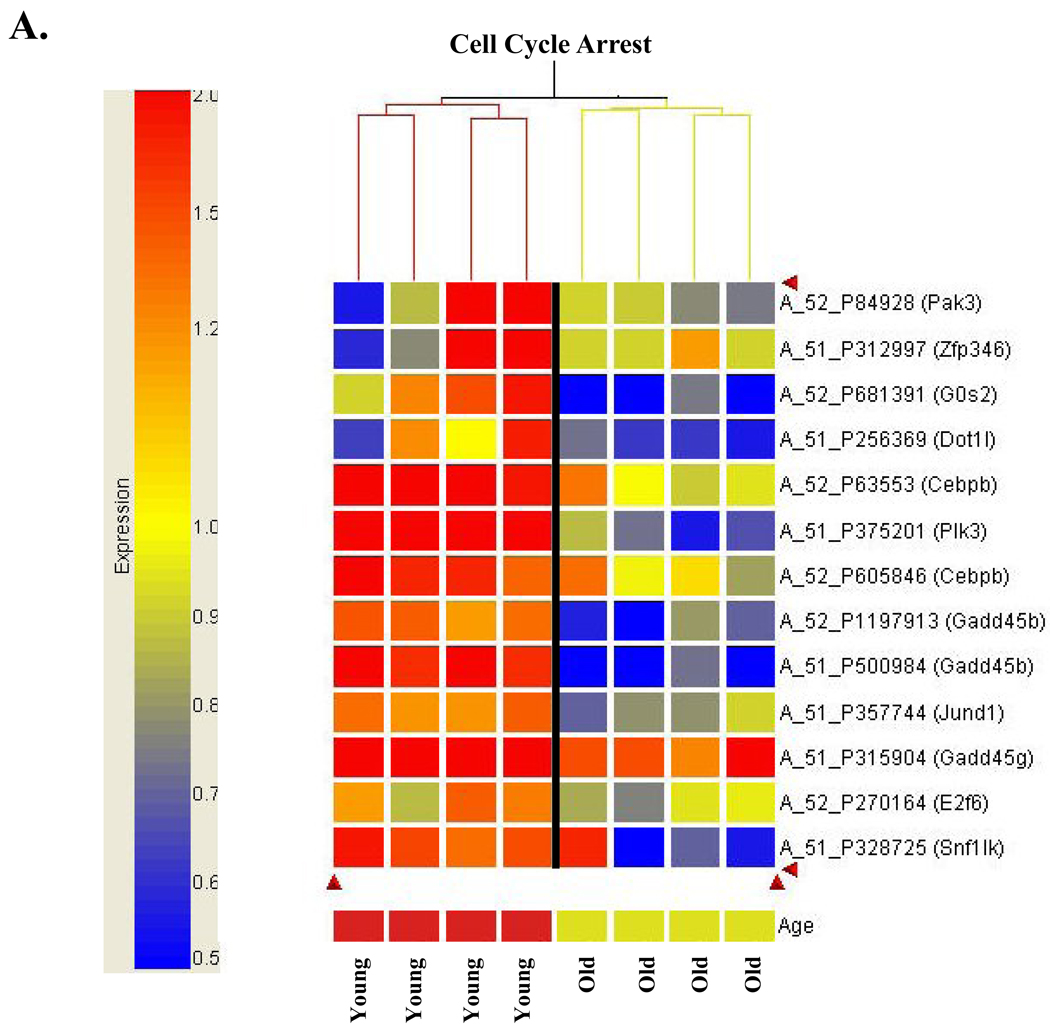
Based on reports that the p53 response to DNA damage is attenuated/absent in old animals, we looked to genes involved in three major pathways known to be regulated by p53: cell cycle regulation, apoptosis and DNA repair. In Table 1A we present a list of genes involved in cell cycle arrest that are differentially expressed in old compared to young animals exposed to DNA damage. The data is presented as the change in old relative to the change in young (ΔO:ΔY) alongside the change in young alone (ΔY) and the change in old alone (ΔO). This allows us to evaluate how the overall differences in response (ΔO:ΔY) arise. Figure 4A presents these data in heat map format (with an expression scale), and it is immediately clear that young animals have increased expression of genes involved in cell cycle arrest, while old animals do not. The heat map data are presented so that the variability in response is apparent. Also, this presentation allows determination of how the ΔO:ΔY values (presented in Table 1A) arise. For example, we can see that the substantial reduction in PLK3 expression (↓4.9 fold, ΔO:ΔY, Table 1A) results from loss of the induction seen in young animals. This gene is of particular interest because of its potential role in senescence. Unlike PLK1, which functions in mitosis, PLK3 is an immediate early gene functioning at G1 to S-phase to arrest cell cycle progression (Zimmerman and Erikson). A few additional genes are downregulated in ΔO:ΔY as a result solely of loss of induction seen in young (Gadd45g, Jaz, G0/G1 switch 2 and Cebpβ), while several result both from loss of induction in young and repression in old (Gadd45b, PAK3, SNF1-like kinase, and Jund1). One gene of interest that is not induced in young, but is substantially repressed in old (↓2.4 fold, ΔO:ΔY, Table 1A) is Dot1-like. The S. cerevisiae homologue, Dot1, is a histone H3 methyltransferase known to play a critical role in G1 and S-phase checkpoints in yeast (Wysocki et al.). In yeast, Dot1 is required for methylation of approximately 90% of histone H3 on lysine 79 (van Leeuwen et al.), and loss of H3-Lys(79) methylation prevents activation of the DNA damage response.
Table 1.
Aging alters expression of cell cycle-related genes in response to oxidative stress
| A. Cell Cycle Arrest | ||||
|---|---|---|---|---|
| Gene name | Assession # | ΔO: ΔY | ΔY | ΔO |
| PLK3 | NM_013807 | −4.9 | 3.7 | |
| Gadd45g | NM_011817 | −4.4 | 6.0 | 1.4 |
| Gadd45b | NM_008655 | −4.3 | 2.0 | −2.1 |
| PAK3 | NM_008778 | −3.7 | 3.1 | −2.1 |
| Zpf346/Jaz | NM_012017 | −3.3 | 3.0 | |
| G0/G1 switch 2 | NM_008059 | −2.9 | 1.5 | |
| SNF1-like kinase | NM_010831 | −2.8 | 1.5 | −1.8 |
| Dot1-like | NM_199322 | −2.4 | −1.7 | |
| Cebpβ | NM_009883 | −2.2 | 2.0 | |
| Jund1 | NM_010592 | −1.7 | 1.3 | −1.3 |
| E2F6 | NM_033270 | −1.5 | ||
| B. Cell Cycle Progression | ||||
|---|---|---|---|---|
| Gene name | Assession # | ΔO: ΔY | ΔY | ΔO |
| RRM2 | NM_009104 | 2.8 | −2.5 | |
| Thymidine kinase | NM_009387 | 2.0 | −1.4 | 1.4 |
| RRM1 | NM_009103 | 1.9 | −1.3 | 1.4 |
| Kras2 | NM_021284 | 1.7 | −1.4 | 1.2 |
| BRCA2 | NM_009765 | 1.7 | 1.6 | |
| Ask/BBF4 | NM_013726 | 1.7 | ||
| Thioredoxin | NM_021284 | 1.6 | −1.2 | 1.2 |
| Cyclin H | NM_023243 | 1.5 | −1.4 | |
| DHFR | NM_010049 | 1.5 | 1.3 | |
| PCNA | NM_011045 | 1.4 | −1.3 | |
| Poldelta 2 | NM_008894 | 1.4 | 1.6 | |
The differential response to 2NP in old relative to young is presented as ΔO:ΔY. Additionally, response to 2NP in young (ΔY) and old (ΔO) are presented. All differences are significant at p<0.005.
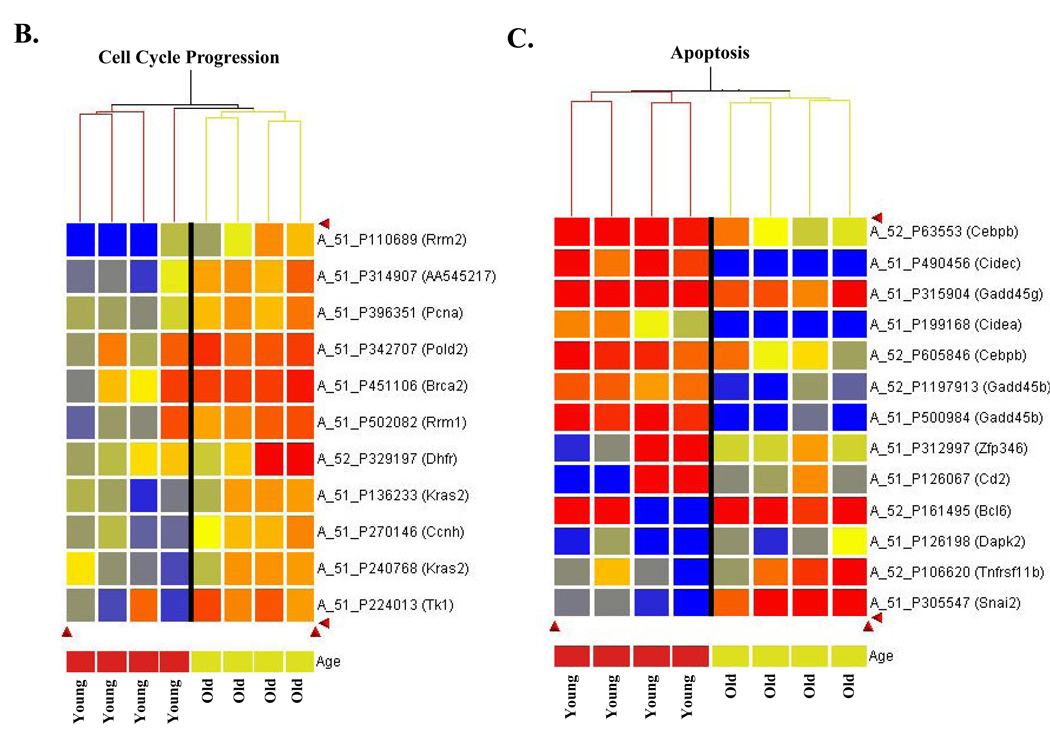
Figure 4. Cluster analysis of differentially expressed genes demonstrates differential response to oxidative stress in old compared to young animals.
Hierarchical clustering was performed as described in Materials and Methods. A. Aging alters expression of cell cycle arrest genes in response to oxidative stress. Agilent probe number and gene name are indicated. Expression scale is presented to left of cluster analysis to indicate magnitude of expression differences. B. Aging alters expression of cell cycle progression genes in response to oxidative stress. Agilent probe number and gene name are indicated. C. Aging alters expression of genes involved in apoptosis following oxidative stress. Agilent probe number and gene name are indicated. Expression scale: Red heatmap coloring indicates increased gene expression in response to 2NP treatment, relative to control, while blue represents a decrease in expression. Presentation of a gene more than once indicates that distinct probes for the same gene showed the dame
In addition to genes involved in cell cycle arrest, we also find differential expression in those involved in cell cycle progression (Table 1B). As above, the data are presented both in tabular format and in heat map format (Figure 4B). Here is it clear that these genes involved in cell growth are either unchanged or downregulated in young animals exposed to DNA damage, but are upregulated in the old animals. For example, in response to treatment, cyclin H expression is upregulated in old relative to young (↑1.5 fold, Table 1B), but is downregulated (↓1.4 fold) in young, showing that the ΔO:ΔY is due to loss of repression seen in young animals. In another example BRCA2 expression is upregulated in response to treatment in old relative to young (↑1.7 fold), this time not resulting from loss of the response seen in young, but rather as a result of direct upregulation in old animals (↑1.6 fold). Many of the S-phase progression genes upregulated in ΔO:ΔY that are involved in DNA synthesis (thioredoxin, RRM1, RRM2, thymidine kinase, PCNA) are differentially expressed in old relative to young as a result of loss of repression seen in young, treated animals, and in several cases in conjunction with induced expression in old, treated animals. That is, in young animals a protective response to carcinogen exposure may be to reduce the availability of DNA synthesis precursors and accessory factors, a response that may be lost in old, treated animals. The data in Figures 4A and 4B point to checkpoint escape in the presence of DNA damage, and suggest loss of the DNA damage response in old animals.
In addition to induction of cell cycle arrest, stabilization of p53 by DNA damage also induces apoptosis. Further, apoptosis increases with age in the liver (Kujoth et al.), consistent with our finding that p53 protein accumulates in old liver. As such we were interested in determining whether loss of the p53 response with age would translate to loss of apoptosis in response to damage with age. We find that there is a significant downregulation in expression of proapoptotic genes in old animals in response to DNA damage (Table 2). The data is presented as the change in old relative to the change in young (ΔO:ΔY) alongside the change in young alone (ΔY) and the change in old alone (ΔO). We have also included a brief description of the role each gene plays in apoptosis, since presentation of up- or downregulation alone isn’t necessarily informative, as genes within this pathway are often negative regulators. The heat map for apoptotic genes (Figure 4C) clearly shows that the proapoptotic genes are strongly upregulated in young animals exposed to DNA damage, and strongly downregulated or unchanged in the old animlas. For the antiapoptotic genes, the expression pattern is reversed such that young animals show strong downregulation while the old show no change or upregulation. Thus, the expression profile is consistent with loss of apoptosis in response to DNA damage in old animals. In light of reports that cytoplasmic localization of p21 inhibits apoptosis (Besson et al), the apparent loss of apoptotic response as determined by expression profiling is consistent with cytoplasmic p21 accumulation.
Table 2.
Aging alters expression of genes involved in apoptosis following oxidative stress
| Accession# | Gene name | ΔO:ΔY | ΔY | ΔO | Role in Apoptosis |
|---|---|---|---|---|---|
| NM_007702 | Cidea | −5.4 | −5.1 | Activates apoptosis | |
| NM_178373 | Cidec | −4.5 | 1.6 | −2.8 | Promotes apoptosis |
| NM_011817 | Gadd45g | −4.4 | 6.0 | 1.4 | Strongly potentiates apoptosis; also cdc2/cyclinB1 kinase inhibitor |
| NM_008655 | Gadd45b | −4.3 | 2.0 | −2.1 | Effector of TGFbeta-induced apoptosis |
| NM_012017 | Zfp346 | −3.3 | 3.0 | Mediates apoptosis (and cell cycle arrest) as positive regulator of p53; alias, JAZ | |
| NM_013486 | Cd2 | −2.6 | Induces caspase-independent apoptosis | ||
| NM_009883 | Cebpb | −2.2 | 2.0 | Shown both to promote and inhibit apoptosis | |
| NM_009744 | Bcl6 | 2.1 | Anti-apoptotic | ||
| NM_010019 | Dapk2 | 2.2 | −2.5 | Pro-apoptotic | |
| NM_008764 | Tnfrsf11b | 2.2 | 1.5 | Anti- apoptotic | |
| NM_011415 | Slugh | 3.1 | −1.3 | 2.3 | Anti-apop totic |
The differential response to 2NP in old relative to young is presented as ΔO:ΔY. Additionally, response to 2NP in young (ΔY) and old (ΔO) are presented. All differences are significant at p<0.005.
Another p53-dependent response to DNA damage is induction of DNA repair pathways. The DNA repair data is arranged differently than that that for cell cycle and apoptosis, as here we are primarily interested in seeing what genes/pathways are induced in response to damage, and whether this differs from young to old. So in Figure 3A we present DNA repair genes that are upregulated by oxidative stress and in Figure 3B, DNA repair genes that are downregulated by oxidative stress. What is immediately apparent is that, with the exception of Ung, there is no overlap between genes that are induced or repressed in young and those that are induced or repressed in old in response to 2NP. This suggests that the types of DNA damage that accumulate in response to 2NP are different between young and old. As above, the data is presented as the change in old relative to the change in young (ΔO:ΔY) alongside the change in young alone (ΔY) and the change in old alone (ΔO). Here it is informative to evaluate genes specifically upregulated in old (ΔO; Figure 5A), as many genes exhibit an active upregulation in expression. On the presumption that genes are induced in response to specific stimuli, we looked to the functions of genes actively upregulated in old animals following induction of DNA damage. Timeless (↑1.7 fold) functions to prevent replication fork collapse in the presence of DNA damage (Chou and Elledge) and protects against the formation of DNA double strand breaks (DSB) (Gotter et al.). XRCC5 (↑1.4 fold; aka Ku80; aka Ku86) is also a DSB repair gene. In particular, XRCC5 plays a role in microhomology-mediated end-joining (Katsura et al.). Rad52 (↑1.3 fold) mediates faithful resolution of DSB, but in cooperation with Rad51 (which is significantly downregulated with age (Rad51-like 1, ↓1.9 fold ΔO:ΔY; Rad51-like3, ↓1.3 fold, ΔO; Figure 5B). Further, loss of Rad51 results in G2/M accumulation and increased chromosome breakage (Sonoda et al.). MLH1 (↑1.4 fold), is a MMR gene that also plays a role in mitotic recombination (Wang et al). Rad1 (↑1.3 fold), part of the 9-1-1 checkpoint response, also plays a role in DSB repair and G2 checkpoint control (Udell et al.). Further, Rad1 may play a role in translesion synthesis (TLS), and Pol eta (↑1.4 fold) is a TLS polymerase. An increased requirement for TLS points to the possibility that increased DNA damage exists at the replication fork in old animals. The expression profile of old, exposed animals points to DSB and the replication fork as a source of the DNA damage response in old animals. In order to present this scenario more clearly, we have generated a list of genes that function in replication fork stability and DSB processing. Further, we looked to genes important at the G2 checkpoint on the assumption that damage at the replication fork might induce a G2 arrest. This list (Table 4) includes a brief description of the genes’ functions, including several genes also presented in other categories, and builds a story that the DNA repair/genome maintenance needs differ greatly between young and old animals.
Table 4.
Aging alters expression of genes involved in G2 arrest folowing oxidative stress and induces genes involved in double strand break repair at the replication fork
| Gene | Fold change | Function |
|---|---|---|
| STK2 | ↓5.6 | Required for progression through G2. Loss of STK2 prevents progression through the G2 block to mitosis. |
| MCM8 | ↑ 3.5 | Prevents accumulation of replication intermediates; localizes to replication forks |
| GEMENIN | ↑ 2.1 | Prevents re-replication in cells arrested in G2 |
| MAD2 | ↑ 1.9 | Inhibits mitosis in presence of misaligned chromosomes. |
| BRCA2 | ↑ 1.7 | Stabilizes blocked replication forks. |
| TIMELESS | ↑ 1.7 * | Involved at replication fork: coordinates replication with genotoxic stress |
| PRIMASE1 | ↑ 1.5 * | Stabilizes lagging strand synthesis; important in replication fork stability and DSB repair. |
| BLM | ↑ 1.5 | RecQ helicase: recombination helicase. Repairs damaged replication forks; required for fork restart. |
| HAT1 | ↑1.5* | Acetyltransferase involved in DSB repair, both NHEJ and HR. |
| RAD21 | ↑ 1.5 | Involved in sister chromatid alignment; DSB repair; component of cohesin |
| MSH3 | ↑ 1.5 | Involved in MMR; also involved in crossover recombination |
| BRCA1 | ↑ 1.4 * | Involved in DSB repair; localizes to replication forks in response to replication stress. |
| MLH1 | ↑ 1.4* | Involved in MMR; also involved in crossover recombination |
| MSH6 | ↑ 1.4 | Involved in MMR; also involved in crossover recombination |
| MSH2 | ↑ 1.4 | Involved in MMR; also involved in crossover recombination |
| RPA | ↑1.4 | Single strand binding protein; critical in DSB repair. Prevents chromosome breaks and aneuploidy. |
| RAD52 | ↑ 1.3 * | Critical initiator of DSB repair; recruits Rad51 onto RPA-bound single strand DNA. |
Fold change represents the difference in response to oxidative stress between old and young animals (ΔOld:ΔYoung) unless annotated by (*) which indicates a statistically significant change seen in old, treated animals only (ΔOld). All differences are significant at p<0.005.
Artemis (PSO2 homologue) deserves special attention by virtue both of its functions and the magnitude of its change in response to DNA damage with age. We observe a 5.4-fold decrease in Artemis expression in old animals (Figure 5B; ΔO:ΔY), primarily resulting from loss of induction seen in young animals exposed to 2NP. Artemis is a component of the NHEJ machinery and has a defined role in DSB repair and genome maintenance as evidenced by the impact of its loss of chromosomal instability (Rooney et al.). Importantly, mice deficient in both Artemis and p53 develop tumors and DNA amplifications through DSB intermediates (Rooney et al.). Our data point to the possibility that subtle changes in Artemis and p53 functionality could promote genomic instability with age. These data (cell cycle, apoptosis, DNA repair) all point to alteration in the DNA damage response with age. Further, they all support the hypothesis that the p53 response is lost with age.
Identification of p53 target genes
As such we aimed to objectively identify genes responding to p53, either directly or in p53-dependent manner. As described in methodology, we compared our array data against known p53 target genes. In Table 5A we present the genes shown by Riley et al. to be direct p53 target genes that were impacted by 2NP treatment in young animals. The original list from Riley et al. included 129 genes, 14 of which had no known mouse homologue, generating a list of 115 p53 target genes from which to make our analysis. Of these 115 genes, we find that 30 genes (26%) were impacted by 2NP treatment, demonstrating that 2NP treatment effectively induced a p53 response in our animals. Of the p53 target genes that are impacted by 2NP treatment in young animals, 19/30 (63%) are differentially expressed in old animals (Figure 5A), signifying a substantial loss of the p53 response to DNA damage with age. To further investigate this dysregulation, we sorted the data according to whether the genes were upregulated or repressed by 2NP. Here we find that 14/19 genes demonstrated loss of induction in old animals, while 5/19 demonstrated loss of repression.
Table 5.
Aging results in loss of the p53 DNA damage response
| A. Dysregulation of direct p53-target genes in response to 2NP in old animals | |||
|---|---|---|---|
| Gene name | Assession # | ΔYoung | ΔOld |
| AIFM2, AMID | NM_153779 | up | nc |
| BAX | NM_007527 | up | up |
| BTG2, TIS21 | NM_007570 | up | nc |
| CCNG1 | NM_009831 | up | up |
| CDKN1A, p21 | NM_007669 | up | up |
| COL18A1 | NM_009929 | down | nc |
| DDIT4, REDD1 | NM_029083 | up | up |
| DUSP1, MKP1 | NM_013642 | up | nc |
| EGFR | NM_007912 | up | up |
| FAS, CD95 | NM_007987 | up | nc |
| GDF15, MIC-1 | NM_011819 | up | nc |
| GPX1 | NM_008160 | up | nc |
| KRT8, CK8 | NM_031170 | up | up |
| LGALS3, galectin-3 | NM_010705 | down | nc |
| mdm2 | NM_010786 | up | up |
| ODC1 | NM_013614 | up | nc |
| PCNA | NM_011045 | down | nc |
| PLK2, SNK | NM_152804 | up | nc |
| PLK3 | NM_013807 | up | nc |
| PML | NM_008884 | up | up |
| PPM1J, MGC19531 | NM_027982 | up | nc |
| RABGGTA | NM_019519 | up | up |
| RB1 | NM_009029 | down | nc |
| RFWD2 | NM_011931 | down | nc |
| RPS27L | NM_026467 | up | up |
| SERPINE1 | NM_008871 | up | nc |
| SLC38A2 | NM_175121 | up | nc |
| STEAP3, TSAP6 | NM_133186 | up | nc |
| TP53INP1 | NM_021897 | up | nc |
| UBD, FAT10 | NM_023137 | up | up |
| B. Dysregulation of p53-responsive genes in response to 2NP in old animals | |||
|---|---|---|---|
| Gene | Accession# | ΔYoung | ΔOld |
| Cdkn1a | NM_007669 | up | up |
| Btg2 | NM_007570 | up | nc |
| Tnsrsf6 | NM_172571 | up | nc |
| Epha2 | NM_010139 | up | nc |
| Ier5 | NM_010500 | up | nc |
| Tob1 | NM_009427 | up | nc |
| Ninj1 | NM_013610 | up | nc |
| Ses2 | NM_144907 | up | nc |
| Gdf15 | NM_011819 | up | nc |
| Plk3 | NM_013807 | up | nc |
| Ddit4 | NM_030143 | up | up |
| Tp53inp1 | NM_021897 | up | nc |
| Rps27l | NM_026467 | up | up |
| Plk2 | NM_152804 | up | nc |
| fas/apo-1 | NM_007987 | up | nc |
| Nek2 | NM_010892 | down | nc |
| Gamt | NM_010255 | down | up |
| Lrba | NM_030695 | down | nc |
| Mcm3 | NM_008563 | down | nc |
| Sms | NM_009214 | down | nc |
| Dck | NM_007832 | down | nc |
| Sp3 | NM_001018042 | down | nc |
| Chrna4 | NM_015730 | down | nc |
| Ap3s1 | NM_009681 | down | down |
| Cbs | NM_144855 | down | nc |
| Ets1 | NM_011808 | down | nc |
| Birc3 | NM_007464 | down | up |
| Col18a1 | NM_009929 | down | nc |
| Ifi16 | NM_008329 | down | nc |
| Ca14 | NM_011797 | down | nc |
| Siglec9 | NM_031181 | down | nc |
| Cyc1 | NM_025567 | down | nc |
| Pmf1 | NM_025928 | down | nc |
| Nucks | NM_175294 | down | nc |
| Sfrs6 | NM_026499 | down | down |
| Trap1 | NM_026508 | down | nc |
| Mcm7 | NM_008568 | down | down |
| Ppa2 | NM_146141 | down | nc |
A. p53 target genes were identified by Riley et al (REF). Accession numbers identify the mouse genes. B. p53-regulated genes were identified by Kho et al. (REF). Accession numbers identify mouse genes. up=upregulated in response to 2NP, p<0.005; down=downregulated in response to 2NP, p<0.005; nc=no change in expression in response to 2NP.
We carried out an identical analysis with another gene set. Here genes were identified as p53 responsive if induction/repression in response to treatment required the presence of p53 (Kho et al.). Of the 38 genes shown to be upregulated, 34 had mouse homologues. In young animals, 2NP upregulated 15/34 genes (44%), while only 3 genes (8%) were upregulated in old animals (Table 5B). These data confirm that we have induced a significant p53 response in young animals, and as above, signifies a substantial loss of p53-dependent gene upregulation with age. With respect to p53-repressed genes, of the 171 genes shown to be downregulated in a p53-dependent manner, 22 genes had no identifiable mouse homologue, generating a list of 149 p53-dependent repressible genes. Twenty-three of these 149 genes (15%) were downregulated in our young animals, while only 3 (2%) were downregulated in old animals (Table 5B). As above, these data demonstrate both loss of p53 induction and loss of p53 repression in old animals exposed to a DNA damaging agent. The impact of this magnitude of loss of the p53 response seems a likely mechanism by which to explain the differential gene expression between young and old in response to a DNA damaging agent.
DISCUSSION
It is clear that the p53 DNA damage response is absent or attenuated with age. We previously showed that exposure to 2NP resulted in similar stress responses in young and old animals, but that nuclear accumulation of p53 was reduced with age (Cabelof et al., 2006 b). Here we show that this loss of p53 is not specific only to the nucleus. We anticipated loss of p53 would result in p21 downregulation, and were surprised to see similar upregulation of p21 transcript in young and old treated animals. As such, we looked to see whether p21 protein localization might impact functionality, and found an age-dependent loss of nuclear p21 in response to oxidative stress. Only nuclear p21 protein can lead to cell cycle arrest (Rodríguez-Vilarrupla et al.), pointing to a potential upstream explanation for the apparent loss of the G1 and/or S-phase checkpoints in this study. Additionally, accumulation of p21 in the cytoplasm may be oncogenic through inhibition of apoptosis (Besson et al.). Thus, sequestration of p21 outside the nucleus is consistent with both loss of G1 arrest and anti-apoptosis that are suggested by the transcript data.
The transcriptional profile of old mice is consistent with cell cycle progression in the presence of DNA damage, akin to radio-resistant DNA synthesis, suggesting the potential for catastrophic accumulation of DNA damage at the replication fork. The switch in relative DNA repair mechanisms in old animals likewise suggests this, and could point to generation of replication-dependent DNA double strand breaks (DSB). Accordingly, the unique sets of DNA repair genes induced in young compared to old in response to the same DNA damage exposure suggests that old animals are more prone to DSB formation, which has been previously demonstrated. Cells from old mice contain more DNA DSB (Singh and Martin). An accumulation of gammaH2AX foci, a marker for DSB, is seen both in senescent cells (Bakkenist et al., d'Adda di Fagagna et al., Sedelnikova et al) and in aging mice (Sedelnikova et al.). It is now understood DSB can induce senescence, a process that requires the presence of functional p53 (Kim et al.). Our finding that p53 accumulates in liver with age in the absence of exogenous stimuli suggests the possibility that a percentage of cells in liver of old mice may be undergoing p53-dependent senescence. An important question becomes whether senescence, if in fact happening, prevents the ability to respond to exogenous stressors.
Double strand breaks, potent inducers of senescence, are processed by nonhomologous end-joining (NHEJ) and homologous recombination (HR). In drosophila HR is the least frequent mechanism for DSB repair in young (13.6%), but the most frequent mechanism for DSB repair in old (60.9%) (Preston et al.). This is a dramatic switch in DSB repair mechanisms with age that is likewise reflected in the transcript data presented here. The dramatic loss of Artemis (↓5.4 fold) in response to oxidative stress in old mice points to reduced NHEJ with age, and the increase we observe in HR-related genes suggest that the findings in drosophila might also apply to a mammalian system. We also find evidence that DSB resolution becomes more error prone with age, as shown by Selunaov et al. Rad52 mediates faithful resolution of DSB, but does so in cooperation with Rad51 (Wu et al.). What we see at the transcript level in old animals is a significant upregulation in Rad52 alongside a significant downregulation in the Rad51 genes (Rad51-like 1 and Rad51-like 3). Another gene product that might promote error-prone DSB repair is XRCC5 (↑1.4 fold) which plays a role in microhomology-mediated end-joining (Katsura et al.). Repair of DSB in the absence of adequate NHEJ results in imprecise DNA repair that is frequently generated by microhomology-mediated joining. These primarily subtle changes in gene expression suggest a shift in the predominant types of DNA damage that accumulate with age, and point to an attempt by old animals to preserve genomic integrity and/or cell viability through upregulation of genes involved in DSB repair, albeit perhaps error-prone repair.
We hypothesize that DSB occur with age as a result of accumulation of DNA single strand breaks (SSB). It has been elegantly established that SSB are the predominant source of replication stress and collapsed replication forks in vivo, and that accumulation of SSB drives recombination (Saleh-Gohari et al.). It is also established that SSB accumulate with age (Ivancsits et al), in response to 2NP (Cabelof et al., 2002b), and in response to BER deficiency (Saleh-Gohari et al.), which occurs with age (Cabelof et al., 2002a). Thus we have a model system uniquely predisposed to accumulation of SSB. What is surprising is the possibility presented by this data that these cells are escaping initial checkpoints and proceeding through DNA synthesis in the presence of accumulating DNA damage. The upregulation of genes with functions at the replication fork and in G2 arrest (Figure 6) suggests that old animals may respond to accumulation of DNA damage at the replication fork through an adaptive response that forces mitotic arrest. Evidence that damage is accumulating at the replication fork comes in several forms. Increased expression of translesion polymerases, of genes involved in recombination, and of genes involved in replication fork stability and restart are all consistent with more damage at the replication fork in old relative to young animals. In response to this type of catastrophic damage in the cell, a genome-stabilizing response would be prevention of mitosis. Evidence that old animals may induce a mitotic block is seen in expression of STK2 (STE20-like kinase), which is significantly and substantially reduced (5.6-fold) in old animals in response to oxidative stress. Loss of this gene product prevents progression through the G2 block to mitosis (O’Reilly et al.,), suggesting that its downregulation in old may signify G2 block. In support, Gemenin, which prevents rereplication in cells arrested in G2 (Zhu et al.), and MAD2 which inhibits mitosis in the presence of misaligned chromosomes (Walters et al.) are significantly upregulated in old animals in response to oxidative stress. Further, increased MAD2 results in mitotic arrest (Fang et al.), once more pointing to accumulation of old cells in G2. These data raise many important questions about how the DNA damage response proceeds in old animals. Whether early checkpoint escape occurs, whether damage is accumulating at the replication fork, and whether old animals are able to induce mitotic arrest cannot be answered here. (Although there is evidence that multinucleated/polyploid cells accumulate in old liver, reviewed in Medvedev).
In this work we have identified an important effect of aging on the p53 response using a novel approach. Using two independent databases of experimentally determined human p53-target genes, we have developed a database of mouse p53-target genes against which we can complete a nonbiased screen of our differentially expressed genes. Using this approach we were able to clearly demonstrate a significant induction of the p53 response by 2NP in young animals, and a significant attenuation of this response with age. Coppe et al. have found that loss of p53 increases the risk that cells exposed to genotoxic stress will escape senescence, and have demonstrated that these cells express a highly amplified secretory phenotype, which can increase cancer risk through a cell-nonautonomous mechanism. What is uncertain in this study is whether loss of the p53 response, in the face of endogenous accumulation of p53 with age, would induce a similar phenotype. Based on the cell cycle-related transcript profile of the old animals exposed to damage, it seems the liver cells are not senescing in response to DNA damage, and thereby not providing a barrier to tumorigenesis. It is difficult to use aging as a model to prove or disprove data generated through genetic studies, or in senescent cells, because the changes occurring with age are typically subtle and occur over the life of the animal. Further, these changes occur within the context of many age-dependent factors for which we cannot control. But we can look to data from these “all-or-none” systems to evaluate the relevance these factors might play within the context of a biologically relevant system, like aging. In so doing, we propose that our data is consistent loss of the p53 response to DNA damage with age that may be responsible for the differential cell cycle, apoptotic and DNA repair programs we observe with age.
Supplementary Material
Table 3.
Impact of oxidative damage on DNA repair response in young and old mice
| A. Upregulated DNA Repair Genes | ||||
|---|---|---|---|---|
| Gene name | Assession # | ΔO: ΔY | ΔY | ΔO |
| BRCA2 | NM_009765 | 1.7 | 1.6 | |
| Rad21 | NM_009009 | 1.5 | ||
| Blm | NM_007550 | 1.5 | ||
| Msh3 | BC040784 | 1.5 | ||
| Msh6 | NM_010830 | 1.4 | ||
| Msh2 | NM_008628 | 1.4 | ||
| Rpa1 | NM_026653 | 1.4 | ||
| PSO2 homologue/artemis |
NM_146114 | 3.3 | ||
| Rad51-like 1 | NM_009014 | 2.4 | ||
| Apex2 | NM_029943 | 1.9 | ||
| Ung | NM_011677 | 1.4 | 1.7 | |
| Rad51-like 3 | NM_011235 | 1.3 | ||
| Mpg | NM_010822 | 1.3 | ||
| Ercc3 | NM_133658 | 1.3 | ||
| Timeless | NM_011589 | 1.7 | ||
| Ercc3 | NM_133658 | 1.5 | ||
| Xrcc5 | NM_009533 | 1.4 | ||
| Pol eta | BC034004 | 1.4 | ||
| Mlh1 | NM_026810 | 1.4 | ||
| Rad52 | NM_011236 | 1.3 | ||
| Rad1 | NM_011232 | 1.3 | ||
| B. Downregulated DNA Repair Genes | ||||
|---|---|---|---|---|
| Gene name | Assession # | ΔO: ΔY | ΔY | ΔO |
| PSO2 homologue/artemis |
NM_146114 | −5.4 | ||
| Rad51-like 1 | NM_009014 | −1.9 | ||
| Wrnip | NM_030215 | −1.4 | −1.7 | |
| Tdg | NM_172552 | −1.3 | ||
| Xrcc1 | NM_009532 | −1.5 | ||
| PMS1 | NM_153556 | −1.5 | ||
| Rad21 | NM_009009 | −1.4 | ||
| Rev3-like | NM_011264 | −1.4 | ||
| Msh3 | BC040784 | −1.4 | ||
| Primase 2 | NM_008922 | −1.4 | ||
| Blm | NM_007550 | −1.3 | ||
| Rad23b | AK084917 | −1.5 | ||
| Rad51-like 3 | NM_011235 | −1.3 | ||
| Stk2 | NM_009289 | −1.6 | ||
| Xpa | NM_011728 | −1.3 | ||
The differential response to 2NP in old relative to young is presented as ΔO:ΔY. Additionally, response to 2NP in young (ΔY) and old (ΔO) are presented. All differences are significant at p<0.005.
Acknowledgements
This work was supported by NIH 1F32-ES013643 (DCC) and American Federation for Aging Research (DCC). Microarray and bioinformatics work was facilitated by the Microarray and Bioinformatics Facility Core of the Environmental Health Sciences Center at Wayne State University (NIEHS Center Grant P30 ES06639). We would like to thank Dr. Thierry Soussi for his input on p53 target genes. We would also like to thank Anna Unnikrishnan, Sridevi Salagrama and Daniela Cukovic for their technical assistance.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Bakkenist CJ, Drissi R, Wu J, Kastan MB, Dome JS. Disappearance of the telomere dysfunction-induced stress response in fully senescent cells. Cancer Res. 2004;64:3748–3752. doi: 10.1158/0008-5472.CAN-04-0453. [DOI] [PubMed] [Google Scholar]
- Besson A, Assoian RK, Roberts JM. Regulation of the cytoskeleton: an oncogenic function for CDK inhibitors? Nat Rev Cancer. 2004;4:948–955. doi: 10.1038/nrc1501. [DOI] [PubMed] [Google Scholar]
- Bohr VA, Anson RM. DNA damage, mutation and fine structure DNA repair in aging. Mutation Res. 1995;338:25–34. doi: 10.1016/0921-8734(95)00008-t. [DOI] [PubMed] [Google Scholar]
- Cabelof DC, Ikeno Y, Nyska A, Busuttil RA, Anyangwe N, Vijg J, Matherly LH, Tucker JD, Wilson SH, Richardson A, Heydari AR. Haploinsufficiency in DNA polymerase beta increases cancer risk with age and alters mortality rate. Cancer Res. 2006a;66:7460–7465. doi: 10.1158/0008-5472.CAN-06-1177. [DOI] [PubMed] [Google Scholar]
- Cabelof DC, Raffoul JJ, Ge Y, van Remmen H, Matherly LH, Heydari AR. Age-related loss of the DNA repair response following exposure to oxidative stress. J Gerontol A Biol Sci Med Sci. 2006b;61:427–434. doi: 10.1093/gerona/61.5.427. [DOI] [PubMed] [Google Scholar]
- Cabelof DC, Raffoul JJ, Yanamadala S, Ganir C, Guo Z, Heydari AR. Attenuation in DNA polymerase β-dependent base excision repair pathway and increased DMS induced mutagenicity in aged mice. Mutation Research. 2002a;500:135–145. doi: 10.1016/s0027-5107(02)00003-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabelof DC, Raffoul JJ, Yanamadala S, Guo Z, Heydari AR. Induction of DNA polymerase beta-dependent base excision repair in response to oxidative stress in vivo. Carcinogenesis. 2002b;23:1419–1425. doi: 10.1093/carcin/23.9.1419. [DOI] [PubMed] [Google Scholar]
- Campisi J. Senescent cells, tumor suppression, and organismal aging: good citizens, bad neighbors. Cell. 2005;12:513–522. doi: 10.1016/j.cell.2005.02.003. [DOI] [PubMed] [Google Scholar]
- Chou DM, Elledge SJ. Tipin and Timeless form a mutually protective complex required for genotoxic stress resistance and checkpoint function. Proc Natl Acad Sci USA. 2006;103:18143–18147. doi: 10.1073/pnas.0609251103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coppe JP, Patil CK, Rodier F, Sun Y, Munoz DP, Goldstein J, Nelson PS, Desprez PY, Campisi J. Senescence-associated secretory phenotypes reveal cell-nonautonomous functions of oncogenic RAS and the p53 tumor suppressor. PLoS Biol. 2008;6:e301. doi: 10.1371/journal.pbio.0060301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- d’Adda di Fagagna F, Reaper PM, Clay-Farrace L, Fiegler H, Carr P, Von Zglinicki T, Saretzki G, Carter NP, Jackson SP. A DNA damage checkpoint response in telomere-initiated senescence. Nature. 2003;426:194–198. doi: 10.1038/nature02118. EMBO J 20, 914-923. [DOI] [PubMed] [Google Scholar]
- Fang G, Yu H, Kirschner MW. The checkpoint protein MAD2 and the mitotic regulator CDC20 form a ternary complex with the anaphase-promoting complex to control anaphase initiation. Genes Dev. 1998;12:1871–1883. doi: 10.1101/gad.12.12.1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Z, Hu W, Teresky AK, Hernando E, Cordon-Cardo C, Levine AJ. Declining p53 function in the aging process: a possible mechanism for the increased tumor incidence in older populations. Proc Natl Acad Sci USA. 2007;104:16633–16638. doi: 10.1073/pnas.0708043104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotter AL, Suppa C, Emanuel BS. Mammalian TIMELESS and Tipin are evolutionarily conserved replication fork-associated factors. J Mol Biol. 2007;366:36–52. doi: 10.1016/j.jmb.2006.10.097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goukassian D, Gad F, Yaar M, Eller MS, Nehal US, Gilchrest BA. Mechanisms and implications of the age-associated decrease in DNA repair capacity. FASEB J. 2000;14:1325–1334. doi: 10.1096/fj.14.10.1325. [DOI] [PubMed] [Google Scholar]
- Guo Z, Wang M, Tian G, Burger J, Gochfeld M, Yang CS. Age- and gender-related variations in the activities of drug-metabolizing and antioxidant enzymes in the white-footed mouse (Peromyscus leucopus) Growth Dev Aging. 1993;57:85–100. [PubMed] [Google Scholar]
- Hamadeh HK, Trouba KJ, Amin RP, Afshari CA, Germolec D. Coordination of altered DNA repair and damage pathways in arsenite-exposed keratinocytes. Toxicol Sci. 2002;69:306–316. doi: 10.1093/toxsci/69.2.306. [DOI] [PubMed] [Google Scholar]
- Herbig U, Sdeivy JM. Regulation of growth arrest in senescence: telomere damage is not the end of the story. Mech Ageing Dev. 2006;127:16–24. doi: 10.1016/j.mad.2005.09.002. [DOI] [PubMed] [Google Scholar]
- Intano GW, Cho EJ, McMahan CA, Walter CA. Age-related base excision repair activity in mouse brain and liver nuclear extracts. J Gerontol A Biol Sci Med Sci. 2003;58:205–211. doi: 10.1093/gerona/58.3.b205. [DOI] [PubMed] [Google Scholar]
- Ivancsits S, Diem E, Jahn O, Rudiger HW. Age-related effects on induction of DNA strand breaks by intermittent exposure to electromagnetic fields. Mech Ageing Dev. 2003;124:847–850. doi: 10.1016/s0047-6374(03)00125-8. [DOI] [PubMed] [Google Scholar]
- Kaneko T, Tahara S, Tanno M, Taguchi T. Age-related changes in the induction of DNA polymerases in rat liver by gamma-ray. Mech Ageing Dev. 2002;123:1521–1528. doi: 10.1016/s0047-6374(02)00119-7. [DOI] [PubMed] [Google Scholar]
- Katsure Y, Sasaki S, Sato M, Yamaoka K, Suzukawa K, Nagasawa T, Yokota J, Kohno T. Involvement of Ku80 in microhomology-mediated end joining for DNA double-strand breaks in vivo. DNA Repair. 2007;6:639–648. doi: 10.1016/j.dnarep.2006.12.002. [DOI] [PubMed] [Google Scholar]
- Kho PS, Wang Z, Zhuang L, Li Y, Chew JL, Ng HH, Liu ET, Yu Q. p53-regulated transcriptional program associated with genotoxic stress-induced apoptosis. J Biol Chem. 2004;279:21183–21192. doi: 10.1074/jbc.M311912200. [DOI] [PubMed] [Google Scholar]
- Kim KS, Kang KW, Seu YB, Baek SH, Kim JR. Interferon-gamma induces cellular senescence through p53-dependent DNA damage signaling in human endothelial cells. Mech Ageing Dev. 2008 doi: 10.1016/j.mad.2008.11.004. in press. [DOI] [PubMed] [Google Scholar]
- Kujoth GC, Hiona A, Pugh TD, Someya S, Panzer K, Wohlgemuth SE, Hofer T, Seo AY, Sullivan R, Jobling WA, Morrow JD, Van Remmen H, Sedivy JM, Yamasoba T, Tanokura M, Weindruch R, Leeuwenburgh C, Prolla TA. Mitochondrial DNA mutations, oxidative stress, and apoptosis in mammalian aging. Science. 2005;309:481–484. doi: 10.1126/science.1112125. [DOI] [PubMed] [Google Scholar]
- Leakey JA, Cunny J, Bazasre J, Webb PJ, Feuers RJ, Duffy PH, Hart RW. Effects of aging and caloric restriction on hepatic drug metabolizing enzymes in the fisher 344 rat: I. the cytochrome P-450 dependent monooxygenase system. Mech.Ageing Dev. 1989;48:145–155. doi: 10.1016/0047-6374(89)90046-8. [DOI] [PubMed] [Google Scholar]
- Medvedev ZA. Age-related polyploidization of hepatocytes: the cause and possible role. A mini-review. Exp Gerontol. 1986;21:277–282. doi: 10.1016/0531-5565(86)90037-9. [DOI] [PubMed] [Google Scholar]
- O’Reilly PG, Wagner S, Franks DJ, Cailliau K, Browaeys E, Dissous C, Sabourin LA. The Ste20-like kinase SLK is required for cell cycle progression through G2. J Biol Chem. 2005;280:42383–42390. doi: 10.1074/jbc.M510763200. [DOI] [PubMed] [Google Scholar]
- Preston CR, Flores C, Engels WR. Age-dependent usage of double-strand-break repair pathways. Curr Biol. 2006;16:2009–2015. doi: 10.1016/j.cub.2006.08.058. [DOI] [PubMed] [Google Scholar]
- Riley T, Sontag E, Chen P, Levine A. Transcriptional control of human p53-regulated genes. Nat Rev Mol Cell Biol. 2008;9:401–412. doi: 10.1038/nrm2395. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Vilarrupla A, Diaz C, Canela N, Rahn HP, Bachs O, Agell N. Identification of the nuclear localization signal of p21(cip1) and consequences of its mutation on cell proliferation. FEBS Lett. 2002;531:319–323. doi: 10.1016/s0014-5793(02)03549-4. [DOI] [PubMed] [Google Scholar]
- Rooney S, Sekiguchi J, Whitlow S, Eckersdorff M, Manis JP, Lee C, Ferguson DO, Alt FW. Artemis and p53 cooperate to suppress oncogenic N-myc amplification in progenitor B cells. Proc Natl Acad Sci USA. 2004;101:2410–2415. doi: 10.1073/pnas.0308757101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rooney S, Sekiguchi J, Zhu C, Cheng HL, Manis J, Whitlow S, DeVido J, Foy D, Chaudhuri J, Lombard D, Alt FW. Leaky Scid phenotype associated with defective V(D)J coding end processing in artemis-deficient mice. Mol Cell. 2002;10:1379–1390. doi: 10.1016/s1097-2765(02)00755-4. [DOI] [PubMed] [Google Scholar]
- Runge-Morris MA. Sulfotransferase gene expression in rat hepatic and extrahepatic tissues. Chem Biol Interact. 1994;92:67–76. doi: 10.1016/0009-2797(94)90054-x. [DOI] [PubMed] [Google Scholar]
- Saleh-Gohari N, Bryant HE, Schultz N, Parker KM, Cassel TN, Helleday T. Spontaneous homologous recombination is induced by collapsed replication forks that are caused by endogenous DNA single-strand breaks. Mol Cell Biol. 2005;25:7158–7169. doi: 10.1128/MCB.25.16.7158-7169.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salvi SS, Nordenhall C, Blomberg A, Rudell B, Pourazar J, Kelly FJ, Wilson S, Sandstrom T, Holgate ST, Frew AJ. Acute exposure to diesel exhaust increases IL-8 and GRO-alpha production in healthy human airways. Am J Respir Crit Care Med. 2000;161:550–557. doi: 10.1164/ajrccm.161.2.9905052. [DOI] [PubMed] [Google Scholar]
- Sedelnikova OA, Horikawa I, Zimonjic DB, Popescu NC, Bonner WM, Barrett JC. Senescing human cells and ageing mice accumulate DNA lesions with unrepairable double-strand breaks. Nat Cell Biol. 2004;6:168–170. doi: 10.1038/ncb1095. [DOI] [PubMed] [Google Scholar]
- Seluanov A, Mittelman D, Pereira-Smith OM, Wilson JH, Gorbunova V. DNA end joining becomes less efficient and more error-prone during cellular senescence. Proc Natl Acad Sci USAs. 2004;101:7624–7629. doi: 10.1073/pnas.0400726101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo YR, Fishel ML, Amundson S, Kelley MR, Smith ML. Implication of p53 in base excision DNA repair: in vivo evidence. Oncogene. 2002;21:731–737. doi: 10.1038/sj.onc.1205129. [DOI] [PubMed] [Google Scholar]
- Singh NP, Ogburn CE, Wolf NS, van Belle G, Martin GM. DNA double-strand breaks in mouse kidney cells with age. Biogerontology. 2001;2:261–270. doi: 10.1023/a:1013262327193. [DOI] [PubMed] [Google Scholar]
- Sonoda E, Sasaki MS, Buerstedde JM, Bezzubova O, Shinohara A, Ogawa H, Takata M, Yamaguchi-Iwai Y, Tekeda S. Rad51-deficient vertebrate cells accumulate chromosomal breaks prior to cell death. EMBO J. 1998;17:598–608. doi: 10.1093/emboj/17.2.598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarloff JB, Goldstein RS, Sozio RS, Hook JB. Hepatic and renal conjugation (Phase II) enzyme activities in young adult, middle-aged, and senescent male Sprague-Dawley rats. Proc Soc Exp Biol Med. 1991;197:297–303. doi: 10.3181/00379727-197-43259. [DOI] [PubMed] [Google Scholar]
- Udell CM, Lee SK, Davey S. HRAD1 and MRAD1 encode mammalian homologues of the fission yeast rad1(+) cell cycle checkpoint control gene. Nucleic Acids Res. 1998;26:3971–3976. doi: 10.1093/nar/26.17.3971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Leeuwen F, Gafken PR, Gottschling DE. Dot1p modulates silencing in yeast by methylation of the nucleosome core. Cell. 2002;109:745–756. doi: 10.1016/s0092-8674(02)00759-6. [DOI] [PubMed] [Google Scholar]
- Vijg J, Dole ME, Martus HJ, Boerrigter ME. Transgenic mouse models for studying mutations in vivo: applications in aging research. Mech Ageing Dev. 1997;98:189–202. doi: 10.1016/s0047-6374(97)00107-3. [DOI] [PubMed] [Google Scholar]
- Walter CA, Grabowski DT, Street KA, Conrad CC, Richardson A. Analysis and modulation of DNA repair in aging. Mech Ageing Dev. 1997;98:203–222. doi: 10.1016/s0047-6374(97)00108-5. [DOI] [PubMed] [Google Scholar]
- Wang Q, Ponomareva ON, Lasarev M, Turker MS. High frequency induction of mitotic recombination by ionizing radiation in Mlh1 null mouse cells. Mutat Res. 2006;594:189–198. doi: 10.1016/j.mrfmmm.2005.09.005. [DOI] [PubMed] [Google Scholar]
- Warner HR, Price AR. Involvement of DNA repair in cancer and aging. J Gerontol. 1989;44:45–54. doi: 10.1093/geronj/44.6.45. [DOI] [PubMed] [Google Scholar]
- Waters JC, Chen RH, Murray AW, Gorbsky GJ, Salmon EE, Niklas RB. Mad2 binding by phosphorylated kinetochores links error detection and checkpoint action in mitosis. Curr Biol. 1999;9:649–652. doi: 10.1016/s0960-9822(99)80287-5. [DOI] [PubMed] [Google Scholar]
- Wu Y, Kantake N, Sugiyama T, Kowalczykowski SC. Rad51 protein controls Rad52-mediated DNA annealing. J Biol Chem. 2008;283:14883–14892. doi: 10.1074/jbc.M801097200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wysocki R, Javaheri A, Allard S, Sha F, Cote J, Kron SJ. Role of Dot1-dependent histone H3 methylation in G1 and S phase DNA damage checkpoint functions of Rad9. Mol Cell Biol. 2005;25:8430–8443. doi: 10.1128/MCB.25.19.8430-8443.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Ahn J, Wilson SH, Prives C. A role for p53 in base excision repair. 2001 doi: 10.1093/emboj/20.4.914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu W, Chen Y, Dutta A. Rereplication by depletion of gemenin is seen regardless of p53 status and activates a G2/M checkpoint. Mol Cell Biol. 2004;24:7140–7150. doi: 10.1128/MCB.24.16.7140-7150.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmerman WC, Erikson RL. Polo-like kinase 3 is required for entry into S phase. Proc Natl Acad Sci USA. 2007;104:1847–1852. doi: 10.1073/pnas.0610856104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.