Eukaryotic H/ACA box small nucleolar RNAs (snoRNAs) and their homologs in Archaea have been shown to guide pseudouridine modification in ribosomal RNAs and other essential noncoding RNAs. In this study, the authors use comparative genomics and deep RNA sequencing of hyperthermophilic Archaea to reveal a novel class of abbreviated H/ACA box small RNA that breaks previously believed invariant properties of this RNA family. The results shed new light on the minimal form of these guide RNAs and provide a comprehensive view of H/ACA-like small RNA from a crenarchaeal genus that is also noted for other atypical noncoding RNAs.
Keywords: H/ACA small RNA, Archaea, guide RNA, transcriptome sequencing, comparative genomics
Abstract
In the Eukarya and Archaea, small RNA-guided pseudouridine modification is believed to be an essential step in ribosomal RNA maturation. While readily modeled and identified by computational methods in eukaryotic species, these guide RNAs have not been found in most archaeal genomes. Using high-throughput transcriptome sequencing and comparative genomics, we have identified ten novel small RNA families that appear to function as H/ACA pseudouridylation guide sRNAs, yet surprisingly lack several expected canonical features. The new RNA genes are transcribed and highly conserved across at least six species in the archaeal hyperthermophilic genus Pyrobaculum. The sRNAs exhibit a single hairpin structure interrupted by a conserved kink-turn motif, yet only two of ten families contain the complete canonical structure found in all other H/ACA sRNAs. Half of the sRNAs lack the conserved 3′-terminal ACA sequence, and many contain only a single 3′ guide region rather than the canonical 5′ and 3′ bipartite guides. The predicted sRNA structures contain guide sequences that exhibit strong complementarity to ribosomal RNA or transfer RNA. Most of the predicted targets of pseudouridine modification are structurally equivalent to those known in other species. One sRNA appears capable of guiding pseudouridine modification at positions U54 and U55 in most or all Pyrobaculum tRNAs. We experimentally tested seven predicted pseudouridine modifications in ribosomal RNA, and all but one was confirmed. The structural insights provided by this new set of Pyrobaculum sRNAs will augment existing models and may facilitate the identification and characterization of new guide sRNAs in other archaeal species.
INTRODUCTION
Small RNA-guided modification of ribosomal RNA is an ancient, conserved process shared between two of the three domains of life, Archaea and Eukarya. First discovered and characterized in eukaryotic model organisms as C/D box small nucleolar RNAs (snoRNAs) (Tyc and Steitz 1989), the archaeal homologs, dubbed C/D box small RNAs (sRNAs), were later biochemically identified in both major branches of the Archaea: Pyrococcus species of Euryarchaeota (Gaspin et al. 2000), and Sulfolobus acidocaldarius of Crenarchaeota (Omer et al. 2000). In the latter study, sRNAs from two additional euryarchaeal species and a second crenarchaeon were computationally predicted and verified, indicating that RNA-guided modification is likely to be a general process common to all archaea. C/D box sRNAs function within ribonucleoprotein complexes (RNPs) that target 2′-O-methylation of specific positions within ribosomal RNA (rRNA) and, unique to the Archaea, transfer RNA (tRNA).
Soon after the discovery of C/D box snoRNAs, a second distinct class of guide RNAs in eukaryotes was identified and found to guide conversion of uridine to pseudouridine within rRNA (Balakin et al. 1996; Ganot et al. 1997; Ni et al. 1997). These guide RNAs, dubbed H/ACA snoRNAs, form distinct ribonucleoprotein complexes with four accessory proteins (Henras et al. 1998; Watkins et al. 1998). Homologs of those proteins were subsequently found among members of the Archaea (Watanabe and Gray 2000), suggesting that a homologous guide RNA mechanism also exists in Archaea to provide specificity to pseudouridine modification of rRNA.
A handful of the archaeal H/ACA-like guide sRNAs were later discovered by small RNA isolation and traditional Sanger sequencing of cDNAs: four in the euryarchaeon Archaeoglobus fulgidus (Tang et al. 2002) and just one in the crenarchaeon Sulfolobus solfataricus (Zago et al. 2005). More recently, computational methods have found other H/ACA sRNA genes predicted to guide rRNA modifications in Euryarchaea (Muller et al. 2007, 2008). Within the Crenarchaea, one H/ACA guide sRNA appears to direct pseudouridine formation in the anticodon of tRNATyr in Aeropyrum pernix and members of the Sulfolobales (Muller et al. 2009).
In the cases above, H/ACA sRNAs in archaea were identified based on models assuming feature similarity to their well-studied eukaryotic homologs. This is a reasonable assumption given that archaeal H/ACA sRNAs form ribonucleoprotein complexes with four proteins (L7Ae, Gar1, Nop10, and the pseudouridine synthase, Cbf5), each of which has a clear ortholog in eukaryotes. The canonical archaeal H/ACA sRNA (Rozhdestvensky et al. 2003; Baker et al. 2005; Gurha and Gupta 2008; Muller et al. 2008) is generally a 60–70 nt helical structure interrupted by two bulges (for example, see Fig. 1A). The upper bulge is part of a kink-turn (K-turn) motif that serves as the binding site for the L7Ae protein. Below the K-turn is an intermediate stem, followed by the second, larger internal bulge that forms the pseudouridylation pocket. The left and right arms of this bulge together create a bipartite guide sequence that is complementary to the target RNA sequence and positions the uridine to be modified at the base of the intermediate stem. A short lower stem closes the pocket, with short 5′ and 3′ extensions beyond the lower helix. A conserved ACA (or more generally, ANA) box sequence is located at the 3′ end, usually positioned 14 nt beyond the base of the intermediate stem (i.e., the pseudouridine pocket).
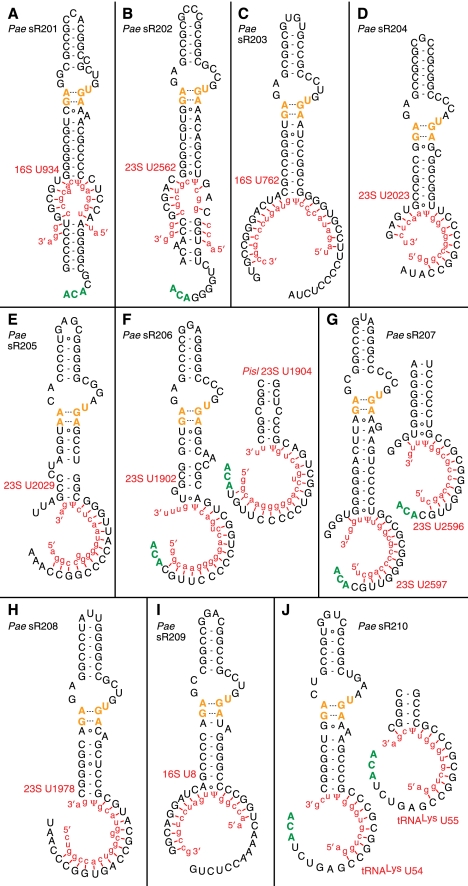
FIGURE 1.
Pseudouridylation guide sRNAs identified in Pyrobaculum. The predicted secondary structures of the Pyrobaculum pseudouridylation guide sRNAs and their predicted targets in ribosomal and transfer RNA (red sequences aligned to guides) are illustrated. (A,B) Canonical pseudouridylation guide sRNAs, containing an upper apical stem, internal kink-turn bulge (K-turn motif in orange), intermediate stem, pseudouridylation pocket, lower helix, and 3′ ACA sequences. The specific boundary between guides and the lower stem is uncertain in these two structures, as both target-pairing and stem-pairing interactions are plausible. Note that the uridine (U) modified in the target RNA is positioned at the base of the intermediate stem and that base-pairing between the guide and the target RNAs occurs both 5′ and 3′ to the site of modification. (C–J) Noncanonical pseudouridylation guides, all without a lower helix, some without the 5′ portion of the pseudouridylation pocket, and many without the terminal 3′ ACA sequence (green). The numbering for the predicted modified uridine, indicated at the base of each structure, uses the P. aerophilum rRNA numbering system.
It is assumed that H/ACA sRNA will be present in all archaeal species, although the paucity or complete lack of identified sRNA genes has been puzzling, particularly in the Crenarchaea. Biochemical evidence has demonstrated the presence of pseudouridine modifications in rRNA in several examined archaeal species (Ofengand and Bakin 1997; Massenet et al. 1999; Del Campo et al. 2005; Kirpekar et al. 2005). Furthermore, homologs of the four proteins known to associate with H/ACA sRNAs have been identified among all examined archaeal phylogenetic families (Finn et al. 2010). Taken together, we should expect to find H/ACA sRNAs among every archaeal clade.
Out of 119 currently available archaeal genomes, only seven have any published H/ACA sRNA predictions. Recent archaeal transcriptional studies (Jager et al. 2009; Wurtzel et al. 2010) have reported C/D box sRNAs (Wurtzel et al. 2010), but neither have reported H/ACA guide sRNAs. Computational search methods to find new H/ACA sRNA genes have had success in euryarchaeal species, where the identification depends on a canonical stem–loop, a bipartite bulge-guide structure, and the terminal ANA sequence found common to all known archaeal H/ACA sRNAs (Muller et al. 2007). Failure to identify archaeal H/ACA sRNAs in both genomic and transcriptional studies is likely due in part to insufficient automated annotation tools but may also be due to variation in the expected features of this sRNA class.
High-throughput transcriptome sequencing and comparative genomics, in combination, offer great promise to rapidly advance archaeal RNA biology. Our group has previously performed detailed genomic analyses of the crenarchaeon Pyrobaculum aerophilum, identifying more than 50 C/D box sRNAs (Fitz-Gibbon et al. 2002) using a strictly computational approach (Omer et al. 2000). However, attempts to identify H/ACA guide RNAs in P. aerophilum using similar methods have not been successful. Over the past several years, we have formed collaborations to sequence five additional Pyrobaculum genomes, as well as the small RNA transcriptomes of four of these organisms. Using these new resources, we provide compelling transcriptional and comparative evidence of ten highly conserved sRNAs that break the conventional view of H/ACA sRNAs in Archaea. Seven of these H/ACA-like sRNA transcripts have guide regions with strong complementarity to Pyrobaculum rRNA targets with equivalent pseudouridine modifications in other species. Significantly, another one of the sRNA appears to have the capacity to guide modification of a universally conserved site of pseudouridylation in every Pyrobaculum tRNA. This work significantly broadens our view of the minimal features likely necessary for RNA-guided pseudouridine modification in Archaea and provides ideal targets for future biochemical and structural studies.
RESULTS AND DISCUSSION
We have employed a high-throughput pyrosequencing strategy as a foundation to define and characterize the spectrum of small RNAs in the genus Pyrobaculum. Size-selected small RNA (16–70 nt) fractions were extracted from exponential and stationary phase cell cultures for Pyrobaculum aerophilum, P. arsenaticum, P. calidifontis, and P. islandicum, and cDNA libraries were prepared (Pak and Fire 2007). This method preserves RNA strand orientation, captures both the 5′ and 3′ ends of the small RNA, and is not biased by the 5′-phosphorylation state of the RNA. These eight barcoded cDNA libraries were concurrently sequenced and the data subsequently processed to yield a large collection of uniquely mapped sequence reads from their associated transcripts. We coupled this small RNA transcriptome data with multiple genome alignments constructed from the same four species, plus the genome sequences of Pyrobaculum oguniense and Thermoproteus neutrophilus (now considered a member of the Pyrobaculum genus).
Ten novel Pyrobaculum H/ACA-like sRNA are conserved and expressed
Motivated by the identification of genes for all four of the proteins involved in RNA-guided pseudouridylation within the Pyrobaculum genomes, computational searches were conducted for H/ACA-like small RNA genes using methods that have been successful in eukaryotic species (Schattner et al. 2004, 2006). These search attempts proved unsuccessful. In an unrelated computational search that screened only for conserved secondary structure, Weinberg and colleagues (Weinberg et al. 2010) identified two potential noncoding RNA elements in the Pyrobaculum genus, Pyrobac-HINT (a proposed cis regulatory element) and Pyrobac-1 (sRNA with no function suggested). We now recognize that these two structured RNA classes correspond to sR207 and sR202/sR203, identified independently and described in this work.
In contrast to previous computational screens, the analysis employed in this study focused strictly on genomic regions with conserved transcription as detected by deep sequencing. Among these transcribed regions, careful screening of transcripts for the typical sequence and structural features of H/ACA-like sRNAs revealed two strong candidates (sR201 and sR202) (Fig. 1A,B); these two sRNAs were found to be present in the transcript libraries from three or more Pyrobaculum species (Supplemental Table S1). Examination of the associated multiple genome alignments (Schneider et al. 2006) revealed syntenic conservation of these genes among all six of the Pyrobaculum genomes and allowed assignment into distinct sRNA families (Fig. 2A; Supplemental Fig. S1A).
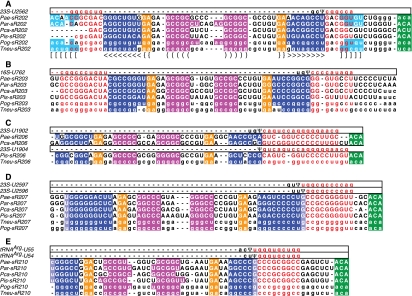
FIGURE 2.
Selected guide alignments from each of the sequenced Pyrobaculum. sR202 (panel A), sR203 (B), sR206 (C), sR207 (D), and sR210 (E) shown. Target RNA sequence shown in top sequence row of each panel. Predicted Watson-Crick and G-U base-pairing between target and sRNA guide regions shown (red). Stem-pairing of the lower stem (aqua, nested square brackets), intermediate stem (dark blue, nested arrows), and upper apical stem (purple, nested parentheses) are indicated. Lower stem/guide base-pairing is uncertain for sR202 (panel A) in region where target sequence (red) overlaps lower stem (aqua). Base-pairing of stems and kink-turn (orange) indicated with bracket notation (panel A, bottom row). Uncertain terminal base pair of intermediate stem (panels D,E) is indicated with lighter shading. RNA sequences are shown in uppercase for sequences recovered in RNA sequencing and in lowercase when sequence is inferred by conservation of aligned features in genome sequence.
Closer scrutiny of the sR201 and sR202 families revealed variability of features previously assumed to be essential. Surprisingly, the ACA sequence at the 3′ end of the sRNA was not conserved in the sR201 homolog from P. islandicum (Supplemental Fig. S1A), nor was it found in the sR202 homologs in P. calidifontis and T. neutrophilus (Fig. 2A). None of these sRNAs contained the anticipated apical loop sequence “GAG” found in several archaeal sRNAs (Baker et al. 2005) that is thought to interact with Cbf5, the pseudouridine synthetase in the complex. Furthermore, the boundary between the guide regions and the lower stem appeared unclear after mapping rRNA targets to these structures (Fig. 1A,B), suggesting that the lower stem may be either very short or nonexistent.
Because of the weak conservation of canonical features associated with sR201 and sR202 (Fig. 1A,B), our search was expanded to look for sRNAs exhibiting only a subset of the anticipated features: (1) a single helical structure interrupted by a K-turn; and (2) occurrence of conserved transcription in syntenic regions. Synteny between sRNA transcripts was inferred by linking their genomic positions to directly adjacent, predicted orthologous protein coding genes in three or more Pyrobaculum genomes. Most of the syntenic sRNA transcripts were found to encode candidate C/D box sRNAs. A small subset was identified using the UCSC Archaeal Genome Browser (Schneider et al. 2006), exhibiting two strongly conserved noncoding regions surrounding a short, weakly conserved segment corresponding to the apical loop. Further inspection of this candidate subset for the presence of a K-turn motif, a GC-rich stem above the K-turn, and an intermediate stem below the K-turn provided an enriched set of candidate H/ACA sRNAs. The putative guide regions (Fig. 2; Supplemental Fig. S1) of these novel sRNA candidates were then interrogated for complementarity to rRNA sequences with correctly positioned uridine targets.
Unexpected features of H/ACA-like sRNA in Pyrobaculum
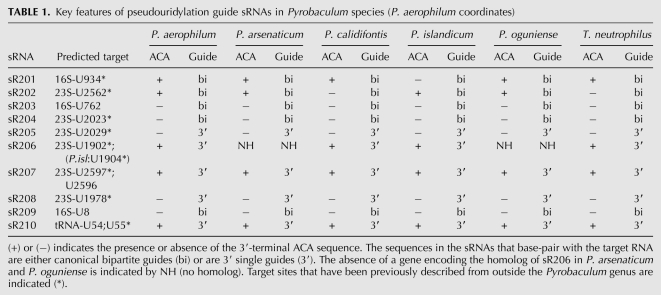
Our new search strategy yielded eight additional, somewhat atypical sRNA families for closer study. Based on their general similarity to sR201, sR202, and the H/ACA-like sRNAs found in other archaeal species, we hypothesized that they function as pseudouridylation guide sRNAs as well (sR203–sR210) (Fig. 1C–I). All eight of these sRNA families have GC-rich, fully paired stems of five to six base pairs above the K-turn and have an intermediate stem of seven to eight base pairs below the K-turn. Each candidate has a 10–17 nt guide complementarity to rRNA targets (allowing at most one bulged unpaired nucleotide). Notably, they all lack the short stem at the base of the helix that normally closes the pseudouridylation pocket (Table 1). In many instances, the sRNAs also lack the conserved ACA sequence at the 3′ end of the molecule (Table 1). Genes encoding homologs of sR201–sR205 and sR207–sR210 are present in all six of the Pyrobaculum genomes (Fig. 2; Supplemental Fig. S1), whereas the sR206 homolog is present in all but P. arsenaticum and P. oguniense, the two most closely related Pyrobaculum species with decoded genomes.
TABLE 1.
Key features of pseudouridylation guide sRNAs in Pyrobaculum species (P. aerophilum coordinates)
Surprisingly, the aligned sR205–sR208 and sR210 transcript families indicate that the 5′ end of these sRNAs is at or near the base of the intermediate stem (Fig. 1). In canonical H/ACA sRNAs, this region contains the 5′-portion of the guide. The remaining sRNAs (sR201–sR204, sR209) retain the expected 5′ portion of the guide. As would be expected, TATA-like promoter sequences (Palmer and Daniels 1995) are associated with both sets of sRNA genes, and are generally positioned 26 nt upstream of our observed start of sRNA transcription. Furthermore, there is no complementarity between the genomic region upstream of transcription-start in sR205–sR208 and sR210 (where the 5′ guide sequence would be expected in canonical sequences) and the extended sequence 3′ to the target pseudouridylation position, supporting the finding that a 5′ guide is not present in these sRNAs. It remains possible that some trans-encoded RNA might replace the “missing” 5′ guide in the mature RNP; however, such a 5′ splice is not found in any sequenced transcripts. Taken together, we conclude that sequencing reads reflect the native sRNA transcripts and that the “missing” 5′ guide portion and lower stem are not required for Pyrobaculum H/ACA-like pseudouridylation guide RNAs.
rRNA targets of H/ACA-like guides occur at conserved, structurally equivalent pseudouridine positions
In nine of the ten sRNAs (sR201–sR209), a guide region with extended complementarity to an rRNA position has been identified (Fig. 1; Table 1). Of these, six sRNAs (sR202, sR204, sR205, sR206, sR207, sR208) have at least one proposed modification target in the large subunit of ribosomal RNA (LSU) that is structurally equivalent to pseudouridine positions in one or more model species, including Escherichia coli, Bacillus subtilis, Arabidopsis thaliana, Saccharomyces cerevisiae, or human (Fig. 3; Ofengand and Bakin 1997; Piekna-Przybylska et al. 2008) Within small subunit (SSU) rRNA, we propose that sR201 guides modification of 16S-U934, the structural equivalent of the hypermodified 16S-966 (E. coli numbering), which is present in all three domains of life (Fig. 2; Ofengand and Bakin 1997; Kowalak et al. 2000; Piekna-Przybylska et al. 2008). Given that seven of nine sites predicted by Pyrobaculum H/ACA-like sRNAs occur at structurally conserved positions of pseudouridine modification in rRNA, we reasoned that these are highly likely to be functioning as H/ACA guide sRNA.
FIGURE 3.
Predicted pseudouridine modifications in ribosomal RNA based on complementary guide regions in Pyrobaculum H/ACA-like sRNAs (Pyrobaculum aerophilum domains shown). Structurally equivalent pseudouridine positions from model species shown (boxed) (Fournier website: people.biochem.umass.edu/fournierlab/3dmodmap/) (Muller et al. 2008). Position of complementary sRNA guide regions depicted by gray lines. Pseudouridine predictions supported only by conservation in other species shown as plain Ψ, and those supported by experimental evidence in Pyrobaclum as circled Ψ. The single predicted pseudouridine site with negative experimental data (16S-U762) depicted as circled U. Secondary structure map adapted from Thermoproteus tenax and Thermococcus celar, with permission (Gutell website: http://www.rna.icmb.utexas.edu/) (Cannone et al. 2002).
Experimental validation of pseudouridine positions in rRNA
To gain further support for the modification function of these novel sRNA families, we tested ribosomal RNA for the presence of pseudouridine residues at positions predicted by the sRNA guide sequences. We utilized a standard biochemical assay for this purpose; RNA treated with soluble carbodiimide (CMCT) preferentially forms bulky adducts at pseudouridine bases (Ofengand and Bakin 1997; Motorin et al. 2007) that are subsequently detected as strong stops in reverse-transcribed cDNA.
We found evidence of pseudouridine modifications at or adjacent to the predicted base in ribosomal RNA for all the sites we were able to assay. For sR201, sR202, sR204, sR205, and sR206, we found clear, unambiguous evidence for the predicted pseudouridine modifications at 16S-U934, 23S-U2562, 23S-U2023, 23S-U2029, and 23S-U1902, respectively (data not shown for 16S-U934 and 23S-U1902) (Fig. 4A,C).
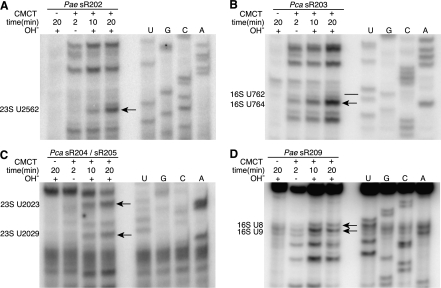
FIGURE 4.
Selected pseudouridine assays in Pyrobaculum aerophilum (Pae) and Pyrobaculum calidifontis (Pca) rRNA. In each experiment, four samples are treated with (+) or without (−) CMCT for either 2, 10, or 20 min followed with (+) or without (−) an alkaline (OH−) wash (lanes 1–4). RNA sequencing ladder is labeled in lanes U, G, C, A. The location of pseudouridine modification evidence is shown (black arrow), established by band absence in CMC− lane, faint banding at 2 min, and increasing band strength seen at 10- and 20-min intervals. Modification sites at 23S-U2582 (sR202), U2023 (sR204), and U2029 (sR205) show characteristic modification bands (panels A,C) located one position 3′ to the actual site of modification (lower in gel image) due to adduct formation. A potential site for sR203-guided modification (panel B) is U764 (black arrow), adjacent to the predicted U762 site that was negative for pseudouridine (black line). The modification site for sR209, 16S-U8 (panel D) is shown with the additional modification at U9.
For the target sites of sR207 and sR208, strong natural RT pauses impeded our reverse transcription assay; impediments of this type have been previously described (Motorin et al. 2007). In the cases of sR203 and sR209, experimental data altered or added to our computationally predicted targets. The predicted target of sR203 is 16S-U762, yet we find a dominant CMCT-induced stop supporting modification of 16S-U764 (Fig. 4B). This pattern is present in assays of rRNA from both P. aerophilum and P. calidifontis, yet we find no obvious alternate guide configuration that might explain the U764 modification. Potentially, an alternate fold of sR203 or some other mechanism may guide the U764 modification.
Our assay for the sR209-predicted pseudouridylation of 16S-U8 revealed strong reverse transcriptase stops associated with 16S-U9, U12, and U15 in both P. aerophilum and P. calidifontis rRNA, while evidence of the adjacent 16S-U8 induced stop was present but weaker (Fig. 4D). This result suggests a possible alternate target for sR209, and adducts at this position may be partially masking detection of the immediately upstream 16S-U8. We note numerous other examples of modification guide RNAs which were shown to direct modifications at bases directly adjacent to the predicted target by an unknown mechanism (U18, U24, snR13, snR52) (Kiss-Laszlo et al. 1996; Lowe and Eddy 1999). The apparent modifications at U12 and U15 do not appear consistent with an alternate configuration of sR209, though, as seen in studies of Pyrococcus abyssi (Pab35-1 targeting of 23S-U2672 [Muller et al. 2008]), surprising alternate targets may occur. When considering all available evidence, we consider 16S-U8 and U9 to be plausible targets of sR209.
Predicted variation in rRNA targets
The sR206 family members exhibit variable features in their guide-target properties. In two of the Pyrobaculum species, the predicted target for sR206 is 23S-U1902; the homologous sRNA in P. islandicum contains several nucleotide alterations in the intermediate stem such that the target site appears shifted to 23S-U1904 (Figs. 1F, 2C). For sR207 (Figs. 1G, 2D), the length of the intermediate stem is uncertain; if the stem were extended by an additional U:G base pair, the predicted site of modification would be shifted from 23S-U2597 to U2596. These adjacent uridines are located in a structurally conserved bulge in domain V (helix 89) of LSU rRNA (Fig. 3). In Bacillus subtilis (Ofengand and Bakin 1997), the structurally equivalent Bs-U2511 is modified, while in Pyrococcus abyssi (Muller et al. 2008), the adjacent Pab-U2585 is the modification target. In Pyrobaculum, it may be that either one or both members of the bulged uridine pair are targets for sR207.
Three H/ACA-like sRNA may have a role in 16S folding
In eukaryotes, the U3 snoRNA has been proposed to function as a chaperone in the endonucleolytic cleavage at the 5′ end of SSU rRNA, as well as the formation of a universally conserved pseudoknot structure (Hughes 1996; Gerczei et al. 2009). In Pyrobaculum, the sR209 guide may sequester the 5′ end of SSU rRNA when paired to its proposed target (16S-U8) (Fig. 3) in a manner analogous to that of U3 snoRNA and thus may play an important role in maturation and folding at the 5′ end of SSU RNA. In another instance, the guide region of sR203 exhibits an unusually long 17-nt complementarity to its 16S rRNA target. When paired to its target, the sRNA might be expected to suppress formation of the associated helix in SSU domain II (Fig. 3) through competitive base-pairing, potentially playing a role in rRNA folding and SSU maturation. An equally compelling case exists with sR201 and the associated 16S-U934 target, where the guide-target association overlaps a complete stem–loop (Fig. 3). All three of these examples appear to associate sRNAs with discrete rRNA secondary-structure elements that are invariant among studied Pyrobaculum species (Fig. 3) and may have a role in folding of those structure elements.
An H/ACA-like guide with a putative target in Pyrobaculum tRNA
One of the H/ACA-like sRNA families, sR210, may target all transfer RNAs within this genus (sR210) (Fig. 1J; Supplemental Fig. S2). In Pyrobaculum species, other archaea, and some higher eukaryotes, tRNAs contain adjacent pseudouridine modifications in both positions U54 and U55 instead of the ribothymidine and pseudouridine modifications that are expected at these positions for bacteria and most eukaryotes. Archaeal pseudouridine conversions at these sites are believed to be catalyzed by the proteins Cbf5 or Pus10 (Gurha and Gupta 2008) without a requisite guide RNA. The sR210 guide region exhibits extended complementarity to the sequence upstream of the Ψ-Ψ-C motif in all of the Pyrobaculum tRNAs; this suggests that the six Pyrobaculum species characterized here may utilize sR210 and the associated pseudouridylation guide machinery to modify tRNA at either positions U54 and U55 or both. The relative abundance of sR210 transcripts is among the highest for this class of Pyrobaculum sRNA (Supplemental Table S1) as might be necessary for a tRNA modification role. Once again, the absence or presence of the additional GC base pair at the base of the lower stem immediately above the pseudouridine pocket shifts the positioning of the target RNA such that either U54 or U55 could be accommodated at the active site (Fig. 1J). We view both of these as likely target sites.
The proposed guide region of the sR210-family exhibits near perfect conservation (Fig. 2E), and the extensive alignment (Supplemental Fig. S2) to tRNA provide compelling evidence of an association. The sR210 family guides align with all tRNAs in the region bounded by the variable loop and extending into the Ψ-Ψ-C motif, as illustrated in Supplemental Figure S2. With the abundance of noncanonical introns in the Pyrobaculum genus (Sugahara et al. 2008), it is remarkable that the tRNA region targeted by sR210 is free of introns in all species other than P. calidifontis, where 30% contain an intron in this region. Introns in the tRNA region used to position sRNA-guided modification would likely be under negative selection unless intron removal occurs before modification. The 2′-O methylation of C56 in P. aerophilum tRNA has been shown to be guided by the C/D sRNP machinery rather than an Ado-Met-dependent protein Trm56 family member, found in bacteria, S. cerevisiae, and other archaea (Renalier et al. 2005). This suggests that the entire tRNA Ψ-Ψ-Cm motif may be modified using sRNA-guided ribonucleoprotein complexes in all tRNA molecules in this genus.
CONCLUSIONS
The pseudouridine guide sRNAs that we have identified in Pyrobaculum differ in a number of important structural features when compared to those previously identified in other archaeal species. The lower helix closing the standard pseudouridylation pocket and the 3′-terminal ANA sequence at the end of the RNA do not appear to be required for Pyrobaculum RNA-guided pseudouridylation. We have examples of molecules with and without each of these features in all Pyrobaculum species examined. Moreover, they can make use of the classical bipartite guide structure as well as novel 3′ single guide forms that restrict base-pairing with the target to the region 5′ to the site of modification.
The single guide architecture would permit guide isomerization that positions either of two adjacent uridines into the active site of the RNP complex. This feature is seen with sR207, which is predicted to modify positions U2597 or U2596 in 23S rRNA, and with sR210, which is predicted to modify positions U54 or U55 in tRNA. In both of these instances, the gain or loss of a base pair from the guide target interaction is accompanied by a loss or gain of a base pair at the bottom of the helix above the pseudouridine pocket in the sRNA.
The novel features that we have identified in the H/ACA sRNAs of Pyrobaculum are likely to have a profound impact on the overall structure and mechanics of the RNP complex. In the crystal structure of a canonical H/ACA RNP from Pyrococcus furiosus, (Li and Ye 2006) the PAU domain (residues 1–38 and 239–333) of the Cbf5 protein makes specific contacts with the lower stem in the sRNA below the pseudouridylation pocket and the terminal ACA sequence. The interactions position the guide sequences so that they are primed to interact with the target to form a three-helix junction and position the uridine destined for modification near the active site of the Cbf5 protein. How this positioning and accompanying stabilization occur in the absence of the bipartite guide, the lower stem, or the terminal ACA sequence remains unclear.
To date, Pyrobaculum species are the only organisms observed to contain unusual pseudouridylation guide sRNAs that lack a well-formed pseudouridylation pocket and are predicted to use only a single 3′ guide sequence to target modification. If these unusual features are more widespread across the Archaea, it may account for the current dearth of recognized pseudouridylation guide sRNAs in these organisms and explain why the computational methods based on the conserved eukaryotic features have had limited success in their identification. Moreover, the overarching simplicity of the Pyrobaculum pseudouridylation guide sRNAs and their flexibility in target recognition may provide new insights into the function and evolutionary origins of this important class of guide RNPs.
MATERIALS AND METHODS
cDNA library preparation
Total RNA was extracted from exponential or stationary cultures; 100 μg of each preparation was loaded onto a 15% polyacrylamide gel, and size selected in the range 15–70 nt. The gel was post-stained with SYBR Gold, and the tRNA band was used as the upper exclusion point. The lower exclusion point was set at 75% of the region between xylene cyanol (XC) and bromophenol blue (BP) loading dye bands (Ambion protocol). Samples were eluted, EtOH-precipitated, and 3′ linker (5′-adenylated, 3′ ddC) was added as described by Lau (Lau et al. 2001) (IDTDNA, Linker1). A second gel purification was performed as above, excising the gel fragment above the XC dye band to remove excess 3′ linker. The recovered linked RNAs were reverse transcribed (RT) using Superscript III (Invitrogen) with a DNA primer complementary to Linker1. Following RT, Exonuclease I (EXO1, Thermo) was added to the RT reaction mixture and incubated for 30 min to remove excess primer. We utilized standard alkaline lysis treatment with NaOH-EDTA at 80°C for 15 min to remove any residual RNA as well as inactivate the reverse transcriptase and the EXO1 ssDNA nuclease. Neutralization and small fragment removal was performed with water-saturated G50 columns (Ambion NucAway). The recovered single strand cDNA was dried to near completion using a Servo SpeedVac, followed by a second 5′-adenylated linker addition (IDTDNA, Linker2) to the cDNA using T4 RNA ligase (Ambion).
A 2-μL volume of this reaction was amplified by PCR (20-μL reaction, 16 cycles). This was followed by a second amplification (20-μL reaction, 16 cycles) using 2 μL from the first amplification as template using Roche 454-specific hybrid adapters based on the method described by Hannon (Pak and Fire 2007). A four-base barcode was included in the 5′ hybrid primer. The final reaction was cleaned using the Zymo clean kit following the manufacturer's protocol.
Sequencing
Sequencing was performed using a Roche/454 GS FLX sequencer, and the GS emPCR Kit II (Roche). Sequencing reads in support of this work are included as supplemental files for each species. Barcodes and sequencing linkers have been removed, and identical sequences have been consolidated (indicated by counts in individual read headers -XX, where XX is the count).
Culture conditions
P. aerophilum cells were grown anaerobically in media containing 0.5 g/L yeast extract, 1× DSM390 salts, 10 g/L NaCl, 1× DSM 141 trace elements, 0.5 mg/L Fe(SO4)2(NH4)2, pH 6.5, with 10 mM NaNO3. P. islandicum and P. arsenaticum cells were grown anaerobically in media containing 10 g/L tryptone, 2 g/L yeast extract, 1× DSM390 salts, 1× DSM88 trace elements, and 20 mM Na2S2O3. P. calidifontis cells were grown aerobically in 1-L flasks using 500 ml of media containing 10 g/L tryptone, 2 g/L yeast extract, 1× DSM88 trace metals, 15 mM Na2S2O3, pH 6.8, loosely capped with moderate shaking at 125 rpm. Anaerobic cultures were grown in 2-L flasks with 1 L of media, prepared under nitrogen with resazurin as a redox indicator at 0.5 mg/L; 0.25 mM Na2S was added as a reductant. All cultures were grown at 95°C to late log or stationary phase, monitored at OD600.
The 10× DSM390 salts comprised (per L ddH2O) 1.3 g (NH4)2SO4, 2.8 g KH2PO4, 2.5 g MgSO4*7H2O. The 100× DSM88 trace metal solution comprised (per L 0.12N HCl), 0.9 mM MnCl2, 4.7 mM Na2B4O7, 76 μM ZnSO4, 25 μM CuCl2, 12.4 μM NaMoO4, 18 μM VOSO4, 6 μM CoSO4.
Computational prediction of orthologous gene clusters
Computational prediction of orthologous groups was established by computing Reciprocal Best BLASTP (RBB) (Altschul et al. 1990) protein coding gene-pairs among species. When at least three RBB pairs select the same interspecies gene cluster, this cluster is considered an orthologous gene cluster.
Pseudouridine mapping in Pyrobaculum rRNA
Pseudouridine residues were mapped using the CMCT method, initially described by Bakin and Ofengand (Bakin and Ofengand 1993). Total RNA (50 μg) from P. aerophilum and P. calidifontis was treated as described (Motorin et al. 2007), yielding a 30-μL preparation; 1 μL of this preparation was then used in 10-μL primer extension assays using Superscript III (Invitrogen) following the manufacturer's recommendations. Each of the four 10-μL RNA sequencing reactions (Superscript III) were conducted with 0.5 mM dNTP, supplemented with the appropriate 0.5 mm ddNTP. Primer sequences used in this study are included in Supplemental Material.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
ACKNOWLEDGMENTS
We thank members of the Joint Genome Institute for making 454 sequencing possible (P. Richardson and J. Bristow for providing resources, and E. Lindquist and N. Zvenigorodsky for sample preparation and analysis). This work was supported by National Science Foundation Grant EF-082277055 (T.L. and D.B.); the Graduate Research and Education in Adaptive Bio-Technology (GREAT) Training Program sponsored by the University of California Biotechnology Research and Education Program (D.B.); and by the National Science Foundation while P.P.D. was working at the Foundation. We are very appreciative of the thoughtful discussions and problem solving contributed by Aaron Cozen and Andrew Uzilov that were essential to this work and are especially grateful to Patricia Chan for assistance with figure preparation.
The opinions, findings, and conclusions expressed in this publication are ours and do not necessarily reflect the views of the National Science Foundation.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.031385.111.
REFERENCES
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ 1990. Basic local alignment search tool. J Mol Biol 215: 403–410 [DOI] [PubMed] [Google Scholar]
- Baker DL, Youssef OA, Chastkofsky MI, Dy DA, Terns RM, Terns MP 2005. RNA-guided RNA modification: Functional organization of the archaeal H/ACA RNP. Genes Dev 19: 1238–1248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakin A, Ofengand J 1993. Four newly located pseudouridylate residues in Escherichia coli 23S ribosomal RNA are all at the peptidyltransferase center: Analysis by the application of a new sequencing technique. Biochemistry 32: 9754–9762 [DOI] [PubMed] [Google Scholar]
- Balakin AG, Smith L, Fournier MJ 1996. The RNA world of the nucleolus: Two major families of small RNAs defined by different box elements with related functions. Cell 86: 823–834 [DOI] [PubMed] [Google Scholar]
- Cannone JJ, Subramanian S, Schnare MN, Collett JR, D'Souza LM, Du Y, Feng B, Lin N, Madabusi LV, Müller KM, et al. 2002. The comparative RNA web (CRW) site: an online database of comparative sequence and structure information for ribosomal, intron, and other RNAs. BMC Bioinformatics 3: 2 doi: 10.1186/1471-2105-3-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Campo M, Recinos C, Yanez G, Pomerantz SC, Guymon R, Crain PF, McCloskey JA, Ofengand J 2005. Number, position, and significance of the pseudouridines in the large subunit ribosomal RNA of Haloarcula marismortui and Deinococcus radiodurans. RNA 11: 210–219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn RD, Mistry J, Tate J, Coggill P, Heger A, Pollington JE, Gavin OL, Gunasekaran P, Ceric G, Forslund K, et al. 2010. The Pfam protein families database. Nucleic Acids Res 38: D211–D222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitz-Gibbon ST, Ladner H, Kim UJ, Stetter KO, Simon MI, Miller JH 2002. Genome sequence of the hyperthermophilic crenarchaeon Pyrobaculum aerophilum. Proc Natl Acad Sci 99: 984–989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganot P, Bortolin ML, Kiss T 1997. Site-specific pseudouridine formation in preribosomal RNA is guided by small nucleolar RNAs. Cell 89: 799–809 [DOI] [PubMed] [Google Scholar]
- Gaspin C, Cavaille J, Erauso G, Bachellerie JP 2000. Archaeal homologs of eukaryotic methylation guide small nucleolar RNAs: Lessons from the Pyrococcus genomes. J Mol Biol 297: 895–906 [DOI] [PubMed] [Google Scholar]
- Gerczei T, Shah BN, Manzo AJ, Walter NG, Correll CC 2009. RNA chaperones stimulate formation and yield of the U3 snoRNA-Pre-rRNA duplexes needed for eukaryotic ribosome biogenesis. J Mol Biol 390: 991–1006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurha P, Gupta R 2008. Archaeal Pus10 proteins can produce both pseudouridine 54 and 55 in tRNA. RNA 14: 2521–2527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henras A, Henry Y, Bousquet-Antonelli C, Noaillac-Depeyre J, Gelugne JP, Caizergues-Ferrer M 1998. Nhp2p and Nop10p are essential for the function of H/ACA snoRNPs. EMBO J 17: 7078–7090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes JM 1996. Functional base-pairing interaction between highly conserved elements of U3 small nucleolar RNA and the small ribosomal subunit RNA. J Mol Biol 259: 645–654 [DOI] [PubMed] [Google Scholar]
- Jager D, Sharma CM, Thomsen J, Ehlers C, Vogel J, Schmitz RA 2009. Deep sequencing analysis of the Methanosarcina mazei Go1 transcriptome in response to nitrogen availability. Proc Natl Acad Sci 106: 21878–21882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirpekar F, Hansen LH, Rasmussen A, Poehlsgaard J, Vester B 2005. The archaeon Haloarcula marismortui has few modifications in the central parts of its 23S ribosomal RNA. J Mol Biol 348: 563–573 [DOI] [PubMed] [Google Scholar]
- Kiss-Laszlo Z, Henry Y, Bachellerie JP, Caizergues-Ferrer M, Kiss T 1996. Site-specific ribose methylation of preribosomal RNA: A novel function for small nucleolar RNAs. Cell 85: 1077–1088 [DOI] [PubMed] [Google Scholar]
- Kowalak JA, Bruenger E, Crain PF, McCloskey JA 2000. Identities and phylogenetic comparisons of posttranscriptional modifications in 16 S ribosomal RNA from Haloferax volcanii. J Biol Chem 275: 24484–24489 [DOI] [PubMed] [Google Scholar]
- Lau NC, Lim LP, Weinstein EG, Bartel DP 2001. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science 294: 858–862 [DOI] [PubMed] [Google Scholar]
- Li L, Ye K 2006. Crystal structure of an H/ACA box ribonucleoprotein particle. Nature 443: 302–307 [DOI] [PubMed] [Google Scholar]
- Lowe TM, Eddy SR 1999. A computational screen for methylation guide snoRNAs in yeast. Science 283: 1168–1171 [DOI] [PubMed] [Google Scholar]
- Massenet S, Ansmant I, Motorin Y, Branlant C 1999. The first determination of pseudouridine residues in 23S ribosomal RNA from hyperthermophilic Archaea Sulfolobus acidocaldarius. FEBS Lett 462: 94–100 [DOI] [PubMed] [Google Scholar]
- Motorin Y, Muller S, Behm-Ansmant I, Branlant C 2007. Identification of modified residues in RNAs by reverse transcription-based methods. Methods Enzymol 425: 21–53 [DOI] [PubMed] [Google Scholar]
- Muller S, Charpentier B, Branlant C, Leclerc F 2007. A dedicated computational approach for the identification of archaeal H/ACA sRNAs. Methods Enzymol 425: 355–387 [DOI] [PubMed] [Google Scholar]
- Muller S, Leclerc F, Behm-Ansmant I, Fourmann JB, Charpentier B, Branlant C 2008. Combined in silico and experimental identification of the Pyrococcus abyssi H/ACA sRNAs and their target sites in ribosomal RNAs. Nucleic Acids Res 36: 2459–2475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller S, Urban A, Hecker A, Leclerc F, Branlant C, Motorin Y 2009. Deficiency of the tRNATyr:Ψ35-synthase aPus7 in Archaea of the Sulfolobales order might be rescued by the H/ACA sRNA-guided machinery. Nucleic Acids Res 37: 1308–1322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni J, Tien AL, Fournier MJ 1997. Small nucleolar RNAs direct site-specific synthesis of pseudouridine in ribosomal RNA. Cell 89: 565–573 [DOI] [PubMed] [Google Scholar]
- Ofengand J, Bakin A 1997. Mapping to nucleotide resolution of pseudouridine residues in large subunit ribosomal RNAs from representative eukaryotes, prokaryotes, archaebacteria, mitochondria and chloroplasts. J Mol Biol 266: 246–268 [DOI] [PubMed] [Google Scholar]
- Omer AD, Lowe TM, Russell AG, Ebhardt H, Eddy SR, Dennis PP 2000. Homologs of small nucleolar RNAs in Archaea. Science 288: 517–522 [DOI] [PubMed] [Google Scholar]
- Pak J, Fire A 2007. Distinct populations of primary and secondary effectors during RNAi in C. elegans. Science 315: 241–244 [DOI] [PubMed] [Google Scholar]
- Palmer JR, Daniels CJ 1995. In vivo definition of an archaeal promoter. J Bacteriol 177: 1844–1849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piekna-Przybylska D, Decatur WA, Fournier MJ 2008. The 3D rRNA modification maps database: With interactive tools for ribosome analysis. Nucleic Acids Res 36: D178–D183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renalier MH, Joseph N, Gaspin C, Thebault P, Mougin A 2005. The Cm56 tRNA modification in archaea is catalyzed either by a specific 2′-O-methylase, or a C/D sRNP. RNA 11: 1051–1063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozhdestvensky TS, Tang TH, Tchirkova IV, Brosius J, Bachellerie JP, Huttenhofer A 2003. Binding of L7Ae protein to the K-turn of archaeal snoRNAs: A shared RNA binding motif for C/D and H/ACA box snoRNAs in Archaea. Nucleic Acids Res 31: 869–877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schattner P, Decatur WA, Davis CA, Ares M Jr, Fournier MJ, Lowe TM 2004. Genome-wide searching for pseudouridylation guide snoRNAs: Analysis of the Saccharomyces cerevisiae genome. Nucleic Acids Res 32: 4281–4296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schattner P, Barberan-Soler S, Lowe TM 2006. A computational screen for mammalian pseudouridylation guide H/ACA RNAs. RNA 12: 15–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider KL, Pollard KS, Baertsch R, Pohl A, Lowe TM 2006. The UCSC Archaeal Genome Browser. Nucleic Acids Res 34: D407–D410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugahara J, Kikuta K, Fujishima K, Yachie N, Tomita M, Kanai A 2008. Comprehensive analysis of archaeal tRNA genes reveals rapid increase of tRNA introns in the order Thermoproteales. Mol Biol Evol 25: 2709–2716 [DOI] [PubMed] [Google Scholar]
- Tang TH, Bachellerie JP, Rozhdestvensky T, Bortolin ML, Huber H, Drungowski M, Elge T, Brosius J, Huttenhofer A 2002. Identification of 86 candidates for small non-messenger RNAs from the archaeon Archaeoglobus fulgidus. Proc Natl Acad Sci 99: 7536–7541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyc K, Steitz JA 1989. U3, U8 and U13 comprise a new class of mammalian snRNPs localized in the cell nucleolus. EMBO J 8: 3113–3119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe Y, Gray MW 2000. Evolutionary appearance of genes encoding proteins associated with box H/ACA snoRNAs: cbf5p in Euglena gracilis, an early diverging eukaryote, and candidate Gar1p and Nop10p homologs in archaebacteria. Nucleic Acids Res 28: 2342–2352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watkins NJ, Gottschalk A, Neubauer G, Kastner B, Fabrizio P, Mann M, Luhrmann R 1998. Cbf5p, a potential pseudouridine synthase, and Nhp2p, a putative RNA-binding protein, are present together with Gar1p in all H BOX/ACA-motif snoRNPs and constitute a common bipartite structure. RNA 4: 1549–1568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberg Z, Wang JX, Bogue J, Yang J, Corbino K, Moy RH, Breaker RR 2010. Comparative genomics reveals 104 candidate structured RNAs from bacteria, archaea, and their metagenomes. Genome Biol 11: R31 doi: 10.1186/gb-2010-11-3-r31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurtzel O, Sapra R, Chen F, Zhu Y, Simmons BA, Sorek R 2010. A single-base resolution map of an archaeal transcriptome. Genome Res 20: 133–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zago MA, Dennis PP, Omer AD 2005. The expanding world of small RNAs in the hyperthermophilic archaeon Sulfolobus solfataricus. Mol Microbiol 55: 1812–1828 [DOI] [PubMed] [Google Scholar]