Deep-sequencing of endothelial cells exposed to hypoxia reveals the complexity of known and novel microRNAs. MicroRNAs are regulators of gene expression with an impact on many different cellular processes. In this article, the authors use deep-sequencing techniques to profile microRNAs and miRNA variants expressed by endothelial cells exposed to hypoxia. The authors present a novel customized bioinformatic analysis pipeline for miRNA annotation. Results presented here revealed the high degree of variability of the miRNAs expressed in endothelial cells, giving an insight into the role of miRNAs in vascular physiopathology.
Keywords: microRNA, isomiR, deep-sequencing, hypoxia, endothelium
Abstract
In order to understand the role of microRNAs (miRNAs) in vascular physiopathology, we took advantage of deep-sequencing techniques to accurately and comprehensively profile the entire miRNA population expressed by endothelial cells exposed to hypoxia. SOLiD sequencing of small RNAs derived from human umbilical vein endothelial cells (HUVECs) exposed to 1% O2 or normoxia for 24 h yielded more than 22 million reads per library. A customized bioinformatic pipeline identified more than 400 annotated microRNA/microRNA* species with a broad abundance range: miR-21 and miR-126 totaled almost 40% of all miRNAs. A complex repertoire of isomiRs was found, displaying also 5′ variations, potentially affecting target recognition. High-stringency bioinformatic analysis identified microRNA candidates, whose predicted pre-miRNAs folded into a stable hairpin. Validation of a subset by qPCR identified 18 high-confidence novel miRNAs as detectable in independent HUVEC cultures and associated to the RISC complex. The expression of two novel miRNAs was significantly down-modulated by hypoxia, while miR-210 was significantly induced. Gene ontology analysis of their predicted targets revealed a significant association to hypoxia-inducible factor signaling, cardiovascular diseases, and cancer. Overexpression of the novel miRNAs in hypoxic endothelial cells affected cell growth and confirmed the biological relevance of their down-modulation. In conclusion, deep-sequencing accurately profiled known, variant, and novel microRNAs expressed by endothelial cells in normoxia and hypoxia.
INTRODUCTION
MicroRNAs (miRNAs) are endogenous, short, 21- to 23-nucleotide (nt), nonprotein-coding RNAs that act as negative regulators of gene expression, modulating the translational efficiency and/or the stability of target messenger RNAs (mRNAs). These RNA molecules are transcribed as a long primary transcript, termed pri-miRNA, sequentially processed by Drosha and Dicer ribonucleases, first to a precursor-miRNA with a characteristic hairpin structure and then to the mature miRNA species. Then, the single-stranded, mature miRNA is loaded together with Argonaute (Ago) proteins into the RNA-induced silencing complex (RISC), a ribonucleoprotein complex mediating post-transcriptional gene silencing. Sequence-specific interaction with the functional miRNA leads to destabilization or translational inhibition of the target mRNAs by the Ago protein (Winter et al. 2009). For several pre-miRNAs, both hairpin arms are processed and give rise to a functional miRNA. In this case, mature miRNAs are designated with the suffix “-3p” or “-5p,” depending on the originating hairpin arm. Alternatively, if either one is expressed at much lower levels, it is designated as miRNA* (star form) (Winter et al. 2009; Krol et al. 2010).
MiRNAs have essential functions in physiological processes, including cellular differentiation, development, and apoptosis, but are also involved in pathological processes, like cancer (Garzon et al. 2009; McCormick et al. 2010) or cardiovascular diseases (Urbich et al. 2008; Dorn 2010; Fasanaro et al. 2010).
Hypoxia describes the insufficient oxygen supply to tissues and occurs during several physiopathological circumstances such as rapid tissue growth, acute and chronic ischemia, organ and tumor development, and high altitude (Semenza 2010b). Decreased oxygen tension triggers a distinct program of responses aimed at relieving tissue hypoxia and removing irreversibly damaged cells. These responses include endothelial cell proliferation, migration, and angiogenesis and also growth arrest and apoptotic cell death (Semenza 2010b). Several reports have demonstrated that specific miRNAs are involved in the hypoxic response (Ivan et al. 2008). In particular, we and others found that the expression of miR-210 is induced in hypoxic endothelial cells (Fasanaro et al. 2008; Chan et al. 2009), stimulating their vascular endothelial growth factor (VEGF)–driven migration and tubulogenesis (Fasanaro et al. 2008).
In order to understand how miRNAs regulate gene expression in endothelial cells exposed to low-oxygen tension, it is essential to accurately profile the entire miRNA population. Currently, there are three widely used platforms for miRNA profiling: quantitative polymerase chain reaction (qPCR), microarray hybridization (Shendure 2008), and deep-sequencing (also named next-generation sequencing or massively parallel sequencing). While qPCR and microarrays rely on the availability of miRNA sequences in public and proprietary databases for designing probes or primers (Griffiths-Jones et al. 2008), deep-sequencing offers the possibility to comprehensively assess miRNA expression in a timely and cost-effective fashion. Along with unprecedented scale and linear range, the sequencing approach does not have thermodynamic biases usually associated with hybridization-based qPCR and microarray platforms (Shendure 2008). Moreover, deep-sequencing allows discovery of previously uncharacterized miRNA species. These include not only new miRNAs that eluded previous cloning but also a panoply of miRNA variants, also known as isomiRs (Landgraf et al. 2007; Morin et al. 2008; Fernandez-Valverde et al. 2010).
In this study the power of deep-sequencing technology was adopted to determine the complete repertoire of miRNAs expressed by human endothelial cells and to identify their modulations induced by low-oxygen tension.
RESULTS
Small RNA library generation, sequencing, and primary analysis
Human umbilical vein endothelial cells (HUVECs) were cultured either in normoxic conditions or at 1% oxygen for 24 h. Then, small RNA libraries were prepared and deep-sequenced by using a SOLiD3 Plus Sequencer platform. As customary for these kinds of studies, two independent populations were analyzed, each both in normoxia and in hypoxia. The corresponding four cDNA libraries yielded a total of 114 million sequenced reads, and 78% (89 million, an average of 22.2 million per library) of these reads passed the quality control filter (quality value [QV] ≥ 9). Reads mapping across genomic sequence by >25 nt were removed owing to the low probability of being miRNA derived. Then, all remaining reads were trimmed to 25 nt, the maximum expected length for a miRNA (Winter et al. 2009; Krol et al. 2010), and used for mapping against various coding and noncoding transcript and genomic data sets.
Identification and quantification of annotated miRNAs
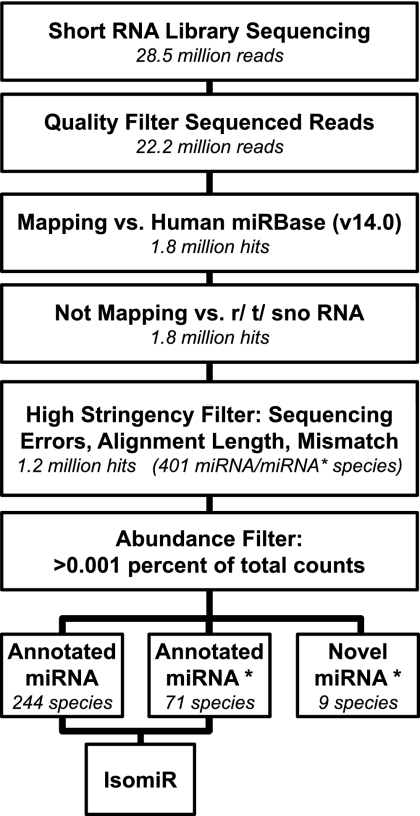
The customized analysis pipeline represented in Figure 1 was used to quantify previously known (annotated) miRNAs. The reads corresponding to annotated miRNAs were identified by selecting all the reads mapping against the human sequences contained in miRBase v14.0, excluding any read mapping also against a collection of other small RNAs databases (transfer RNAs [tRNAs], small nucleolar RNAs [snoRNAs], ribosomal RNAs [rRNAs]). Then, further selection criteria based on alignment length, absence of mismatches in the seed region, and absence of sequencing errors were applied: An average of 1.2 million reads per library were identified as annotated miRNAs, representing 305 mature and 96 miRNA* species. As expected, a very significant correlation between the results obtained analyzing the two normoxic and the two hypoxic libraries was observed (Supplemental Fig. S1). The counts per miRNA were then normalized with respect to the total library size, and all species with percentage counts above 0.001% in both normoxic libraries (244 miRNAs and 71 miRNA*s) were investigated further (see Supplemental Table S1). Large differences in expression levels were observed: 32% of all identified miRNA sequences belonged to miR-21, while 7% belonged to miR-126 (Fig. 2A,B; Supplemental Table S1).
FIGURE 1.
Analysis pipeline of annotated miRNAs. Flowchart describing data analysis pipeline designed for the identification of mature and star sequences of annotated miRNAs, their isoforms, and novel miRNA star sequences. The number of remaining reads/hits/miRNAs after each filtering step, averaged across all four libraries, is indicated in italics. The high-stringency filter of alignment (step 5) consisted in minimum alignment length of 17 nt, with a maximum of one mismatch and three sequencing errors. Reads with sequencing errors in the first 15 nt or mismatches in the seed region were excluded.
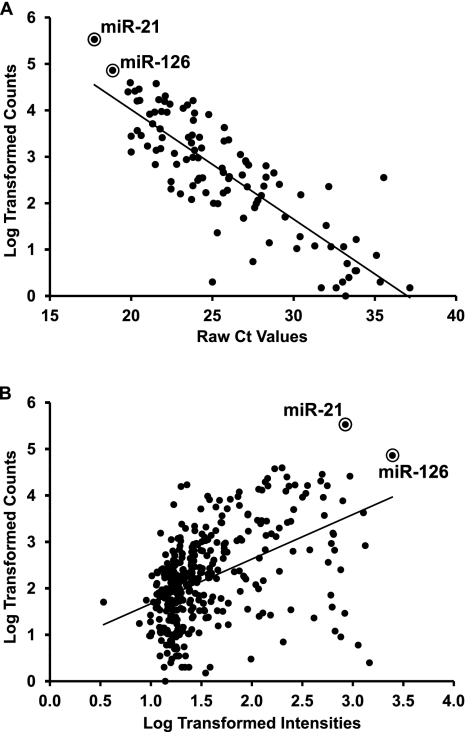
FIGURE 2.
Similar miRNA expression profiles of normoxic HUVECs obtained with independent techniques. (A) Average Ct of 114 miRNA species detected by qPCR (n = 4) were compared with the read counts, expressed in a Log10 scale, obtained by deep-sequencing (Pearson's r = −0.86, P < 0.0001). The circles highlight miR-21 and miR-126, the two most abundant miRNAs among all identified species in HUVECs. (B) Average Log10-transformed Hy3 intensities of 269 miRNA probes of miRCURY LNAmicroRNA Arrays version 11.0, Exiqon (n = 7), were compared to the Log10-transformed read counts obtained by deep-sequencing (Spearman r = 0.64, P < 0.0001). The circles highlight miR-21 and miR-126.
Then, deep-sequencing data were compared to the HUVEC expression profiles we obtained by qPCR (Fasanaro et al. 2008) and microarray (C Voellenkle, J van Rooij, and F Martelli, unpubl.) in independent experimental settings. A highly significant correlation was observed between deep-sequencing and qPCR data (see Fig. 2A). A lower, albeit still highly significant, level of correlation was observed between the deep-sequencing and microarray data. Intriguingly, highly abundant miR-21 and miR-126 were associated with fluorescence levels lower than expected, possibly due to a lower dynamic range of microarray analysis compared with deep-sequencing (see Fig. 2B).
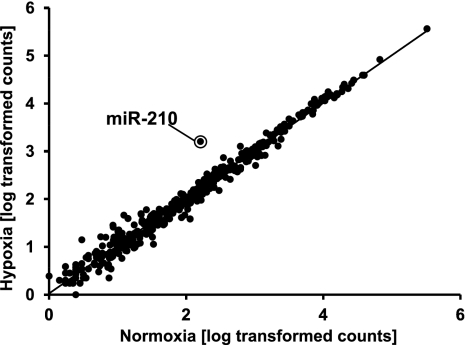
Finally, normoxic and hypoxic HUVEC samples were compared (Fig. 3). As expected, the majority of the miRNA species displayed no significant differences (Audic and Claverie test) (Audic and Claverie 1997). However, in accordance with previous observations (Fasanaro et al. 2008, 2009; Chan et al. 2009), miR-210 was significantly induced more than eightfold, further validating our deep-sequencing analysis.
FIGURE 3.
Expression levels of annotated miRNAs in HUVECs exposed to hypoxic and normoxic conditions. Average Log10-transformed read counts obtained by deep-sequencing of normoxic versus hypoxic HUVECs. The circle highlights hypoxia-induced miR-210.
Alternative duplex strand selection of annotated miRNA/miRNA* pairs
During typical miRNA biogenesis, one strand of the ∼22-nt RNA duplex is preferentially selected for entry into a silencing complex, whereas the other strand of the hairpin, known as the passenger strand or miRNA* strand, is generally degraded (Winter et al. 2009; Krol et al. 2010). However, several studies demonstrate that this preference in hairpin strand selection can vary in a tissue-specific manner (Ro et al. 2007; Chiang et al. 2010). Thus, the ratios between expression levels of annotated mature miRNAs and their miRNA* siblings were determined. Indeed, for three different miRNA species, a clear bias toward the miRNA* sequence was observed, showing more than five times higher expression compared with the relevant mature sequence (Supplemental Fig. S2). These observations suggest that in endothelial cells, there may be a preferred selection of the passenger strand for the miRNA/miRNA* pairs of miR-33a, miR-214, and miR-616.
Identification of novel miRNA* species
Increasing evidence indicates that miRNAs* have a widespread regulatory activity in vertebrates (Mah et al. 2010). To identify novel miRNA* species, we investigated all miRNA stem–loops that harbored reads from both arms but had only one arm already annotated in miRBase. Reads mapping to an unannotated arm were accepted as novel miRNA* species only if their counts reached the abundance of 0.001%, showed >50% complementarity to the annotated mature species, and showed an overhang of no more than 3 nt on either the 3′ or 5′ arm, compared with the annotated mature sequence. The application of these stringent selection criteria resulted in the discovery of nine novel miRNA* species, none of which was annotated in miRBase version 18.0. It is noteworthy that with the exception of novel miR-216* (/miR-216-3p), these sequences were also found by others and are displayed in the deep-sequencing data view of miRBase (Supplemental Fig. S3).
Mature miRNA variants
Mature sequence variants (isomiRs) (Landgraf et al. 2007) were analyzed, and reads aligning to annotated miRNA species displaying an occurrence of editing events, single nucleotide polymorphisms (SNPs), and variations in length were further investigated. In total, 3153 different isomiRs with abundance of at least 0.001%, representing 230 different miRNA and miRNA* species, were identified. Roughly 45% of the reads aligned precisely to the annotated mature sequences. The characteristics of the identified isomiRs are summarized in Supplemental Table S2. We found that the most common isomiR species were represented by sequence length variations, at either the 5′ (Supplemental Fig. S4A) or 3′ end (Supplemental Fig. S4B). Interestingly, for 33 miRNA species, the most abundant form did not correspond to the annotated sequence in miRBase (Supplemental Fig. S5A,B). Further investigation of the deep-sequencing data of miRBase revealed that for 16 miRNAs, the same isomiR variation was identified as the highest expressed sequence also by others (Supplemental Fig. S5A). For the remaining 17, the most abundant sequence shown in the deep-sequencing data was either the annotated sequence or a different isomiR variation (Supplemental Fig. S5B).
Within these 33 miRNAs, the most observed variations were single-base mutations in the tail (nucleotides 10–20) or small (1- to 2-nt) 3′ additions or deletions. However, 5-nt or even 6-nt deletions were not uncommon. Occasionally we identified sequence variants corresponding to no annotated SNP and A-to-G transitions, potentially attributable to adenosine-to-inosine editing events when the SOLiD platform is used (Schulte et al. 2010).
Discovery of novel miRNAs
To identify novel miRNAs, we devised an analysis pipeline even more stringent than the one used to analyze annotated miRNAs (Fig. 4). Quality filtered reads of all four libraries were mapped against the human genome (UCSC, assembly hg19), resulting in about 140 million hits to the genome per library. We found that many reads did not align to previously annotated RNAs such as miRBase miRNAs, coding and noncoding mRNAs, transfer RNAs, ribosomal RNAs, and other small RNAs (e.g., small nuclear RNAs, Piwi-interacting RNAs [piRNAs], small nucleolar RNAs), indicating that many small RNAs remain to be discovered (Supplemental Fig. S6; Yang et al. 2010). To identify novel miRNAs among these hits, we applied a series of tight selection criteria, aimed at minimizing the number of false discoveries: Hit number was reduced by applying a high-stringency quality filter (23.6 million), requiring a minimum alignment of 19 nt and allowing only one mismatch and one sequencing error. Then, we removed reads already annotated in miRBase and other ncRNA databases (19.9 million) and all reads mapping to repetitive sequences (14.1 million). We also speculated that sequencing artifacts and random Drosha/Dicer processing products may yield less abundant hits. Thus, we introduced an abundance filter, accepting only genomic locations harboring 100 or more alignments with an identical 5′ end start (6.6 million hits). One prominent feature of miRNAs is their biogenesis from a RNA hairpin structure (Winter et al. 2009; Krol et al. 2010). Thus, we intersected the remaining hits with a predicted short RNA hairpin database. We found that more than 270,000 counts, representing 511 genomic locations, mapped in the stem (but not in the loop) of one of the predicted hairpins (Supplemental Table S3).
FIGURE 4.
Pipeline of genome mapping. Flowchart describing data analysis pipeline designed for the identification of novel potential miRNA candidates. Numbers in italics indicate the amount of remaining reads/hits/miRNAs after each filtering step, averaged across all four libraries. Filtering threshold for alignment length was set at ≥19 nt, allowing not more than one mismatch and one sequencing error. After final overlap with a hairpin prediction database, more than 270,000 hits remained, representing 511 candidates for putative novel miRNA locations.
In order to validate our analysis criteria, reads corresponding to the mature miRNA species identified with the miRBase mapping were passed through the same pipeline. Interestingly, in spite of the higher stringency and the increased abundance threshold, we found that 126 annotated miRNAs were identified (data not shown).
The 511 miRNA candidates were further examined, and remapping against fRNAdb and miRBase version 18.0 confirmed that none of the selected sequences were previously annotated as small noncoding RNA. This candidate miRNAs data set was considered as a subset of expressed RNAs potentially enriched for novel miRNAs species. To confirm this interpretation, 28 of the newly identified candidate miRNAs were randomly chosen for further experimental validation.
Novel miRNAs are associated to the RISC
In order to validate the candidate miRNAs, qPCR assays were developed. We found that 23 out of 28 miRNA candidates tested were readily detectable in both HUVECs and HEK-293 cells. However, a caveat applied to novel-735 since its sequence was similar to that of hsa-miR-151, and the adopted assay may not discriminate between these two miRNAs.
In keeping with qPCR data, the expression of novel-181 and novel-1322 was also confirmed by Northern blotting (Supplemental Fig. S7).
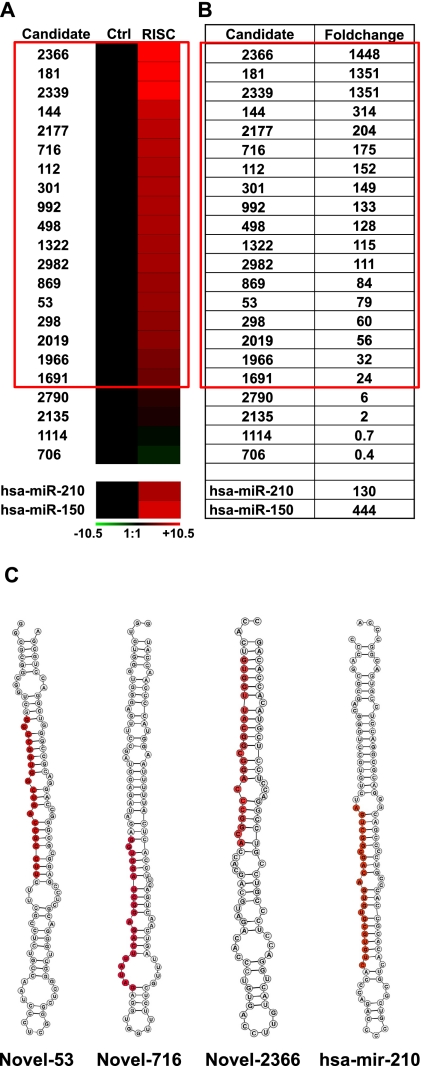
One prominent biological feature of miRNAs is their association to the RISC effector complex (Winter et al. 2009; Krol et al. 2010). To assay whether the candidates miRNAs identified as expressed in HUVECs were associated to the RISC, we tested if they were detectable in immunoprecipitates of Ago2, a crucial RISC component. HEK-293 cells were transfected with an expression vector for a c-Myc–tagged allele of Ago2, resulting in cells enriched with c-Myc-Ago2–containing RISCs. Then, cell lysates were derived and immunopreciptated using an antibody against c-Myc. The levels of coimmunoprecipitated miRNAs were measured by qPCR and compared with the background control, represented by c-Myc immunoprecipitates derived from backbone vector transfected cells. Figure 5, A and B, shows that 18 out of 22 candidate miRNAs displayed a more than 20-fold enrichment in RISCs and were thereby confirmed as genuine novel miRNAs. Two annotated miRNAs adopted as positive controls, miR-150 and miR-210, displayed similar enrichment levels. Predicted hairpin folding of the novel miRNAs is shown in Figure 5C and Supplemental Figure S8.
FIGURE 5.
Novel miRNAs are associated to RISCs. (A) Newly identified miRNAs were measured in Myc-Ago2–containing RISC immunocomplexes, and their levels are expressed as average fold change compared with control (n = 3). Red and green colors indicate up- and down-regulation, respectively, and modulations are indicated using a log2 scale (−ΔΔCt). (B) Table showing the same values displayed in A in a linear scale. Candidates highlighted in the red square display a statistically significant, greater than 20-fold enrichment. (C) Secondary structures, predicted using RNAfold software (Vienna RNA package), of three representative novel miRNA stem–loops and of one annotated miRNA (miR-210). Mature miRNAs are highlighted in red.
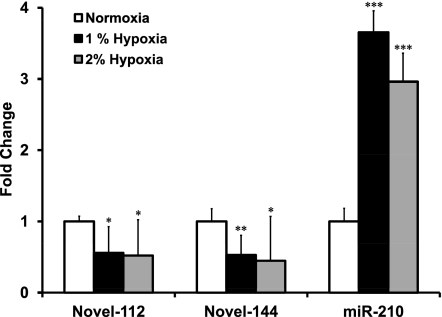
Novel-112 and novel-144 miRNAs are down-modulated by hypoxia
We assayed whether experimentally validated novel miRNAs were modulated by hypoxia. Given the low numerosity of the samples analyzed by deep-sequencing, we considered for further validation only those miRNAs displaying a minimum fold change of three and a P-value <0.005 in at least one of the two sequencing data sets. As shown in Figure 6, qPCR experiments confirmed that novel-112 and novel-144 were significantly down-modulated in HUVECs exposed to both 1% and 2% oxygen for 24 h. Furthermore, qPCR measurements showed that novel-112 and novel-144 down-modulation was maintained when 1% oxygen exposure was extended to 48 h (Supplemental Fig. S9). As expected, miR-210 was induced at both time-points and under both hypoxic conditions (Fig. 6; Supplemental Fig. S9; Fasanaro et al. 2008, 2009; Chan et al. 2009).
FIGURE 6.
Differential miRNA expression upon hypoxia. Levels of novel-112, novel-144, and hsa-miR-210 were measured by qPCR in HUVECs exposed to 1% or 2% oxygen for 24 h. At both oxygen concentrations, novel-112 and novel-144 miRNAs were down-modulated and miR-210 was induced (n = 7; *P < 0.05; **P < 0.001; ***P < 0.001).
Target prediction and pathway analysis
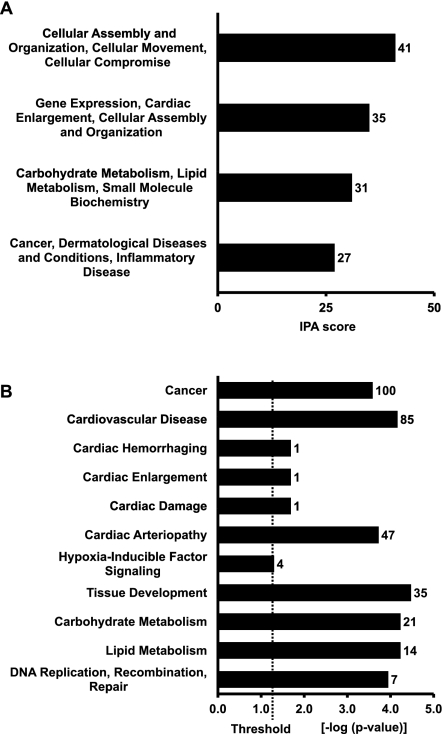
To investigate the biological relevance of the miRNAs significantly modulated by hypoxia in HUVECs, functional pathway analysis (IPA) was performed of the target genes predicted for novel-112, novel-144, and miR-210. To this aim, TargetScan and DIANA–microT algorithms were used, which allow target prediction of nonpreviously annotated miRNAs. Experimentally confirmed miR-210 targets were also used (Devlin et al. 2011). Intriguingly, predicted targets of hypoxia-modulated miRNAs were significantly associated to both cardiovascular and neoplastic diseases and, more specifically, to hypoxia-inducible factor signaling (Fig. 7).
FIGURE 7.
Ingenuity pathway analysis (IPA) of targets predicted for miRNAs modulated in HUVECs upon hypoxia exposure. (A) Top network functions significantly associated to targets predicted for novel-112, novel-144, and miR-210. Bars, scores computed by IPA. (B) Pathways and biological functions significantly associated to targets predicted for novel-112, novel-144, and miR-210. Bars, P-values evaluated by exact Fisher test expressed in logarithmic scale. Threshold indicates the minimally accepted significance level of P < 0.05. Numbers beside the bars indicate number of predicted targets involved significantly in the indicated pathways.
We also searched novel-112, novel-144, and miR-210 predicted targets among hypoxia-modulated genes. We found that novel-112, novel-144 and miR-210 predicted targets displayed a fivefold enrichment (P < 0.00001) in HUVECs exposed to hypoxia conditions identical to these adopted in our study.
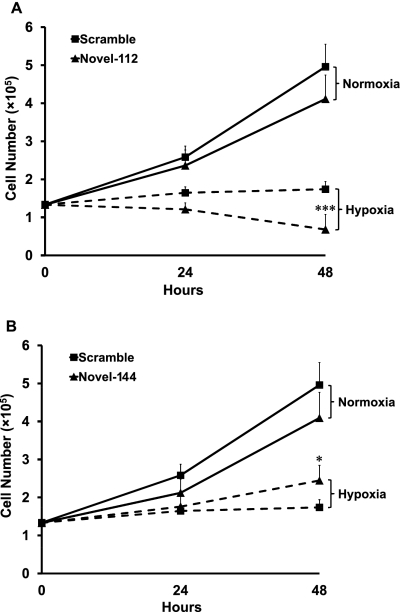
Novel-112 and novel-144 miRNAs affect endothelial cell growth in hypoxia
To gain further insight into the biological function of hypoxia-repressed miRNAs, HUVECs were transfected with mimics of novel-112 and novel-144 or with a negative control sequence and were exposed to 1% oxygen for 24 and 48 h.
As assessed by growth curves, hypoxia inhibited HUVEC proliferation, as expected. The overexpression of the two novel miRNAs had a significant impact on hypoxic cells: novel-112 and novel-144 overexpression resulted in lower and higher cell numbers, respectively, compared with scramble controls (Fig. 8).
FIGURE 8.
Novel-112 and novel-144 affect hypoxia-induced growth arrest. HUVECs were transfected with novel-112 or novel-144 mimics or with a control scramble sequence. The next day (T0), cells were exposed to 1% hypoxia for the indicated time and counted. Following hypoxia, novel-112 expression significantly decreased (A) and novel-144 expression significantly increased (B) cell number compared with that of the control (n = 16; *P < 0.003; ***P < 0.0005).
DISCUSSION
Deep-sequencing is a powerful technique, allowing not only absolute quantification of millions of sequences in an unbiased manner but also the identification of novel miRNAs. In the present study, the deep-sequencing approach was used to delineate the complete repertoire of miRNA species expressed in endothelial cells and to identify miRNA modulations upon exposure to hypoxic stress.
Very stringent filtering steps were introduced in the custom-designed pipelines, preferring the loss of possibly interesting sequences over contamination with false positives. Thus, the fraction of annotated miRNAs among the sequenced small RNAs was lower than that reported by others (Kawaji et al. 2008). For the identification of novel miRNAs, exclusion criteria were even tighter: along with very stringent alignment quality cutoffs, sequences mapping to other small noncoding RNAs, like snoRNAs, rRNAs, and tRNAs, were also excluded from the analysis. While this avoided the contamination of random fragments, it may also lead to the exclusion of genuine miRNA species. Indeed, very recent studies identified snoRNA-derived molecules with miRNA-like functions (Ender et al. 2008; Brameier et al. 2011; Ono et al. 2011).
The expression levels of annotated miRNA measured by deep-sequencing were similar to those previously obtained by our group with independent profiling techniques, i.e., qPCR and microarrays (Fasanaro et al. 2008). Albeit the analyzed HUVEC populations were not the same, a high correlation degree was found between the read counts and both Ct or fluorescence intensity. We also found that a small number of miRNA species largely dominated the miRNA population: 32% of all annotated miRNAs detectable in HUVECs were represented by miR-21; 7%, by miR-126. However, much less abundant miRNAs, such as miR-210 (<0.02% of the miRNA population in normoxia), also have a proven biological role in endothelial cells (Fasanaro et al. 2008; Chan et al. 2009). The biological significance of these wide abundance differences is currently unknown but may relate to the abundance of the corresponding target mRNAs (Bartel 2009; Matkovich et al. 2010). Investigation of differential expression among annotated miRNAs by deep-sequencing analysis revealed the expected significant induction of miR-210 in both libraries. Two other miRNAs, miR-150 and miR-328, that we previously identified as significantly up-regulated by hypoxia in endothelial cells (Fasanaro et al. 2008) were significantly induced only in one of the two libraries. This might be due to the slow kinetics of their up-modulation, which reach a more than fivefold induction only after 48 h exposure to hypoxic conditions (Fasanaro et al. 2008).
Deep-sequencing analysis also allowed appreciation of the high degree of variability of miRNAs (Landgraf et al. 2007). Indeed, the occurrence of editing events or the presence of unknown variations in sequence length are not quantified with traditional profiling platforms and might interfere with the precision of differential expression analysis, especially when looped reverse transcription primers are used (Benes and Castoldi 2010).
Differences in sequence length are likely due to variability in either Dicer1 or Drosha cleavage positions (Morin et al. 2008; Wu et al. 2009), whereas single base changes can be attributed to SNPs or editing events, reported as frequently occurring in miRNAs (Kawahara et al. 2007; Landgraf et al. 2007; Morin et al. 2008; Dupuis and Maas 2010; Marti et al. 2010). We cannot exclude that differences in sequence length could be due to aspecific degradation, but we would expect these events to affect all miRNA species equally. Conversely, we found that certain miRNAs showed many more 3′/5′ deletions than did others, suggesting that most of them likely are genuine isomiRs. Similarly, although the SOLiD 2-base system interrogates each base twice during the sequencing process, we cannot rule out that sequencing errors occur. Nevertheless, isoforms found at high levels may be accepted with a good degree of confidence.
Given the importance of the seed region (nucleotides 2–8 from the 5′) of the miRNA for recognition of the target mRNA (Lewis et al. 2005; Bartel 2009), variations at the 5′ end are particularly interesting: It has been suggested that multiple mature miRNAs, descending from one miRNA precursor, might have different seed sequences, thus recognizing a different set of targets (Azuma-Mukai et al. 2008; Chiang et al. 2010).
Interestingly, the annotated mature sequence was not always the most abundant form: For 33 miRNAs species, an isomiR variant was expressed at higher levels. For instance, a longer miR-21 isomiR was seven times more abundant than the form annotated in miRBase. By making use of the miRBase deep-sequencing data, we found that for 16 of these miRNAs, others had the same results. For the remaining 17, the observed sequence was found as the most abundant in endothelium only and may be possibly attributed to the cell type of origin.
Finally, further studies are warranted to determine whether stress stimuli such as hypoxia might affect miRNA post-transcriptional regulation and the differential accumulation of isomiRs (Fernandez-Valverde et al. 2010).
Strand selection seems to be based on the stability of 5′ base-paired ends in the miRNA/miRNA* duplex (Winter et al. 2009; Krol et al. 2010). However, much remains to be discovered about this process. In contrast to the general conception that miRNA* sequences are not functionally active, recent studies revealed that the abundance, possible function, and physiological relevance of miRNA*s have been underestimated (Mah et al. 2010). Arm switching by alternative strand selection seems to provide a fundamental mechanism to evolve the function of a miRNA locus and target gene network (Ruby et al. 2007; de Wit et al. 2009; Griffiths-Jones et al. 2011). Moreover, strand selection is highly regulated since it shows tissue-, cell-, and condition-specific modulation. Indeed, miRNA* species seem to have an impact on gene expression and regulatory networks, suggesting their contribution to disease states (Ro et al. 2007; Chiang et al. 2010; Biasiolo et al. 2011; Yang et al. 2011).
In keeping with these observations, in endothelial cells several annotated miRNA* forms were also much more abundant than their mature miRNA siblings (miR-33a, -214, and -616). This suggests that in endothelial cells, these annotated star sequences do not constitute the passenger strand, fated to degradation, but rather the guide strand, having regulatory activity. Further investigations are necessary to verify these findings. Whether these miRNAs also display alternative duplex strand selection in other cell types remains to be determined. Furthermore, by detailed analysis of sequences mapping to both arms of annotated stem–loops, we discovered nine novel, not yet annotated miRNA* sequences.
For the discovery of novel miRNAs, highly stringent exclusion criteria based on the abundance, quality of alignment, and localization on the stem of predicted hairpins were applied. By these means, 511 candidates for potential novel miRNAs were identified.
Given the ongoing miRNA identification techniques, the miRNAs discovered so far are expected to be the most abundantly and ubiquitously expressed. Other frequently used identification parameters include evolutionary conservation (Mendes et al. 2009; Yousef et al. 2009) and clusterization with other miRNAs (Sewer et al. 2005). Thus, we were not surprised that the average expression of miRNA candidates was lower than that of annotated miRNAs. However, several novel miRNAs displayed expression levels similar to these of many annotated ones. Moreover, most novel miRNAs displayed very low conservation among vertebrates, with some exceptions, such as novel-53.
A recent article indicated that convincing evidence was lacking about the ability of nearly one-third of the mouse miRNA genes annotated in miRBase version 14.0 to generate authentic miRNAs (Chiang et al. 2010). While further studies are needed, we addressed the concern that many nonconserved candidate miRNAs, often expressed at low levels, could be sequencing artifacts or random products of Drosha/Dicer processing instead of genuine miRNAs. To this purpose, we selected 28 miRNA candidates and assayed their levels by qPCR and found that 23 candidates were readily detectable. Thus, most small RNAs were validated with an independent technique. Moreover, the candidate miRNAs that we failed to detect are not necessarily artifacts. Indeed, we did not troubleshoot the custom qPCR assays extensively, and these tests may lack the necessary sensitivity. For the same reason, we did not further validate novel-735, since it is very similar to miR-151 and we had no conclusive evidence of that our qPCR assay could discriminate between these miRNA species, likely belonging to the same family. Furthermore, two of the newly identified miRNAs could be detected by Northern blotting. Then, we speculated that association to the RISC is the most stringent assay to determine whether a candidate miRNA is indeed functional (Karginov et al. 2007; Fasanaro et al. 2009). We found that 18 out of 22 candidate miRNAs were indeed associated to RISC immunoprecipitates, and we considered these as genuine novel miRNAs. It is highly likely that additional novel miRNAs are present in the candidate data set. However, this requires individual experimental testing. Thus, we intend the candidate miRNAs data set to be a subset of expressed RNAs highly enriched for miRNAs species.
To gain further insight into the possible biological significance of newly identified and annotated miRNAs modulated in hypoxic conditions, a pathway-based analytical tool was used (IPA). Targets predicted for novel-112, novel-144, and miR-210 were analyzed for a significant association to biological pathways and networks. Most interestingly, IPA analysis revealed a significant association to the hypoxia-inducible factor signaling pathway. Encouragingly, a significant relation of 85 and 100 predicted target genes to cardiovascular diseases and cancer, respectively, were also identified. Indeed, in both these diseases, hypoxia plays a critical pathogenetic role (Semenza 2010a,b). Moreover, novel-112, novel-144, and miR-210 predicted targets were enriched among the hypoxia-modulated genes in HUVECs exposed to hypoxia, further confirming the likely biological relevance of the regulation of these miRNAs. In keeping with this interpretation, overexpression of novel-112 and novel-144 in HUVECs under hypoxic conditions had a significant impact on the endothelial cell growth.
CONCLUSIONS
In conclusion, deep-sequencing is confirmed as a valid technological platform to investigate the complete repertoire of miRNA and miRNA variants expressed in vascular cells. A first description of the miRNA population characterizing human endothelial cells was delineated. It appears clear that in spite of the close attention of the scientific community in the past few years, the effort toward the identification of human miRNAs is far from over. Many new miRNA genes remain to be discovered, likely displaying low conservation and cell lineage–restricted expression. In this study only, we report the discovery of 18 new miRNAs and nine miRNA* species and pave the way for the discovery of many more. Furthermore, we also highlighted the high degree of variability of the miRNAs expressed in endothelial cells.
Hypoxia is a crucial pathogenetic component of ischemia characterizing very widespread diseases such as cancer, myocardial infarction, stroke, and ischemic ulcers. We think that this study may further improve our understanding of the molecular mechanisms underpinning these diseases. In this respect, further studies are needed to explore whether the two novel miRNAs that we found to be modulated by hypoxia in vitro are involved in tissue response to ischemia.
MATERIALS AND METHODS
Cell culture
Primary HUVECs (Lonza) were grown in EGM-2 medium (Lonza) containing 2% FBS. The human embryonic kidney 293 cell line (HEK-293, ATCC) was cultured in Dulbecco's modified Eagle's medium (DMEM) containing 10% fetal bovine serum (Fasanaro et al. 2008). HUVECs were kept under hypoxic conditions (1% and 2% oxygen) using a hypoxic incubator (Forma Series II 3110 Water Jacketed CO2 Incubator, Thermo Scientific) where the O2 concentration is continuously monitored and adjusted to the desired level by N2 injections (Fasanaro et al. 2008).
RNA purification and small RNA library generation and sequencing
Total RNA was extracted using TRIzol (Invitrogen). For library preparation, total RNA was extracted from two independent sample pairs of HUVECs, exposed to 1% oxygen or normoxic conditions for 24 h. In addition, total RNA of one sample pair was further enriched for small RNAs up to 60 bp by size-selection using a flashPAGE fractionator following the Ambion protocol (Ambion). RNA purity, integrity, and size distribution were assessed using an Agilent 2100 Bioanalyzer (Agilent Technologies). Then, four sequencing libraries were generated using the Small RNA Expression Kit according to the manufacturer's protocol (SREK Rev. C, Applied Biosystems). Briefly, RNA was first hybridized and ligated with the adapter mix B and subsequently reverse transcribed and treated with RNase H. The obtained cDNA libraries were PCR amplified, purified, and size-selected by PAGE, resulting in libraries containing inserted small RNA sequences of 20–40 bp in length. Size, integrity, and purity of the libraries were verified by the Agilent 2100 Bioanalyzer (Agilent Technologies). cDNA libraries were amplified onto beads using emulsion PCR; templated beads were deposited on slides and analyzed using the Applied Biosystems SOLiD-3 Sequencer. The obtained sequence reads and corresponding quality values were first filtered with an in-house generated perl script in order to eliminate any read with one or more “noncalled” bases. The remaining sequences were then filtered for a minimal quality value of 9 with the SOLiD Software Development Community csfasta_quality_filter.pl script (http://solidsoftwaretools.com/gf/). Since SOLiD reads (35 nt) were longer than the expected length of mature miRNAs (20–23 nt), the miRNA reads apparently contained part of the adapter sequence. To avoid high rates of sequencing errors or mismatches due to the adapter, the reads were trimmed to 25 nt in length after removing all reads that map to the genome over larger intervals. The GEO accession number for this study is GSE26497 and can be accessed at http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?token=rnkdxeuaiyquqdm&acc=GSE26497.
Annotated miRNA analysis and mapping
The quality filtered reads were mapped against all annotated human mature and precursor miRNA sequences (miRBase version 14.0) (Griffiths-Jones et al. 2008) using a specifically designed set of SHRiMP-parameters (Rumble et al. 2009) for optimal miRNA sequence detection, as displayed in Figure 9. All reads were also mapped against a nonredundant compilation of other small noncoding RNA sequences retrieved from GenBank (Benson et al. 2009), RNAdb (version 2.0) (Pang et al. 2007), and NONCODE (He et al. 2008), in addition to ribosomal RNA and transfer RNA sequences downloaded from GenBank and coding and noncoding RefSeq sequences from the UCSC corresponding to the hg19 assembly version. Any sequence mapping with a maximum one mismatch in color space against one of these collections was subsequently excluded from the miRNA alignment data sets in order to avoid false-positive counts. The miRNA mappings were passed through an alignment quality filter, requiring a minimum alignment length of 17 nt between the read and reference, allowing not more than one mismatch and not more than three sequencing errors. Reads with sequencing errors in the first 15 nt were excluded, and no mismatches were allowed in the seed region of the miRNA. Finally, each read was assigned to its corresponding miRNA, and the number of sequences detected per miRNA (counts) were calculated. Then, the number of counts for each of the miRBase annotated miRNAs was normalized by expressing the counts in percentages of the total library size (percentage counts). After applying an abundance threshold of 0.001% of total counts per library, analysis of the expression levels and mature sequence variations (isomiRs), together with identification of novel miRNA star sequences, was carried out.
FIGURE 9.
SHRiMP mask. Illustration of the set of masks (5 × 20 bases) used for local alignment of the reads against the human miRBase and the human genome by SHRiMP. The symbol 1 indicates positions that must be identical to the corresponding nucleotide in the reference sequence, while the symbol 0 indicates positions that may mismatch (with a penalty to final score). Only reads with perfect alignment of the seed region (location 3–8) and with a maximum of two mismatches in the following bases at any position (location 9–18) were accepted. At the first and last two positions, mismatches were allowed to identify also miRNA length variants. Further selection criteria were adopted at later analysis stages.
Discovery of novel miRNA* sequences
To identify novel miRNA* sequences, the output of the SHRiMP-mapping versus human stem–loop sequences of miRBase was screened for miRNAs with annotation only for the mature sequence, harboring five or more reads mapping to an identical position on the opposite arm in all four libraries and showing an average abundance of at least 0.001% percentage counts. Subsequently, only the putative novel star sequences lying within ±3 nt opposite of, and sharing at least 50% sequence complementarity with, their respective annotated mature sequence were accepted as novel star candidates.
miRNA IsomiR analysis
For isomiR analysis (additions, deletions, and single-base variations in the annotated mature miRNA sequence), counts were divided between the sequence variations identified for each mature miRNA. IsomiRs with average percentage counts <0.001% were removed, and corresponding statistical analyses were computed. miRNA species showing a higher abundance for at least one isomiR compared with the annotated sequence were analyzed singularly and mapped against the human genome using BLAT (Kent 2002) to confirm genomic alignment.
Alternative arm selection of annotated miRNA/miRNA* pairs
The ratios of the miRNA/miRNA* species percentage counts were calculated. Any annotated miRNA* sequence with at least 0.001 percentage counts (averaged over all four libraries) and at least a five times higher expression level compared with the relevant mature sequence was further investigated.
Small RNA genomic mapping
Small RNA SOLiD sequences were mapped against the human hg19 genome sequences with parallel instances of SHRiMP (Rumble et al. 2009), with the same original parameter set used in the annotated miRNAs (see Fig. 9). After filtering the sequences that mapped against the human genome for alignment quality (minimum alignment of 19 nt and only one mismatch/sequencing error allowed), all reads mapping to the genome coordinates corresponding to any annotated small noncoding RNA comprised in miRBase (Griffiths-Jones et al. 2008), GenBank (Benson et al. 2009), RNAdb (version 2.0) (Pang et al. 2007) or NONCODE (He et al. 2008) were excluded. Additionally, reads mapping more than three times to the human genome (nonspecific) or overlapping with simple repeats (http://hgdownload.cse.ucsc.edu/goldenPath/hg18/database/) were removed from the data set. Subsequently, all reads with alignments starting at an identical 5′ position were merged, and the genomic positions harboring 100 or more counts in at least one of the four libraries were retrieved. Finally, these coordinates were intersected with a custom database containing genome-wide short RNA hairpin structure predictions. Predictions of plausible miRNA precursor-like structures were made by scanning across the entire human genome, including repetitive regions. RNALfold and a perl script were used to identify miRNA precursor-like structures, using a sliding window size of 110 nt, which allows a maximum stem length of ∼45–50 nt. Predicted single unbranched stems were selected with folding energies of −30kcal/mol or better, terminal loops ≤20 nt, and bulges <5 nt on one side of the stem. We identified 2,443,320 pre-miRNA–like structures, of which 1,564,161 did not contain repetitive SINE elements. This set predicts 71% of known human miRNAs (miRBase version 9.0) (Griffiths-Jones et al. 2006). In the UCSC browser, most predictions are located in introns and intergenic sequences, with the prediction set showing enrichment for 5′ UTRs, knownGene exons, and splice sites, which may be a consequence of the higher GC content of these regions. Afterward, only the genomic locations positioned on the stem of a predicted hairpin (not in a side bulge or loop) were accepted for further analysis. Reads identified as mature human miRNA species during the miRBase mapping were used as positive controls by running them through the same pipeline.
To exclude reads aligning >25 nt, results of genome mappings obtained with the alignment tool Bowtie (Langmead et al. 2009) were used to intersect the reads of normoxic and hypoxic library 2, now trimmed to three different lengths (i.e., 23, 28, and 33 nt) with the coordinates of the predicted precursors. Candidate precursors overlapping with reads aligning for >23 nt were excluded if the counts of these aligned reads trimmed to 28 and/or 33 nt in any of the two libraries represented >5% of the counts of the 23-nt trimmed reads.
An essential requirement for a genuine mature miRNA is the consistent processing of its 5′ end. For the identification of short RNA sequences due to degradation processes, the flanking regions of the putative 5′ end of all candidates were investigated. According to criteria described before (Chiang et al. 2010), we determined the total number of reads mapping within ±30 nt from the candidate 5′ end. Candidates were excluded if <50% of all reads mapping within the investigated region were present within ±2 nt or if <50% of the reads within the region of ±2 nt were mapping to the precise 5′ end.
Each candidate miRNA, translated in sequence space from color space, was mapped against human genome (hg19, UCSC) using BLAT (Kent 2002) to confirm genomic location and aligned against the fRNAdb (Pang et al. 2007) and miRBase (version 18.0) (Griffiths-Jones et al. 2008) to confirm their absence in these data sets.
Comparison of deep-sequencing with other miRNA profiling techniques
Annotated miRNA data obtained by deep-sequencing were compared to HUVEC profiles obtained with different techniques. For this purpose, the averaged, Log10 transformed counts of the miRNAs expressed by the two normoxic HUVEC populations were compared with the profiles previously obtained by either qPCR or microarrays in independent HUVEC populations. In this deep-sequencing data analysis, miRNAs were considered as expressed if at least one read was measured in each normoxic HUVEC library. For qPCR/deep-sequencing comparison, 159 miRNA/miRNAs* were tested in four different HUVEC samples by TaqMan qPCR (Fasanaro et al. 2008). For this analysis, only miRNAs showing average Ct values <38 were used. After normality assessment using the Kolmogorov-Smirnov test, Pearson correlation was calculated for 114 miRNA species. For array/deep-sequencing analysis, seven independent miRNA profiles were obtained using the miRCURY LNA microRNA Array version 11.0 (Exiqon) platform. Average Log10 transformed Hy3 intensities of the miRNAs displaying signal to noise ratio >3 were used. After normality assessment using the Kolmogorov-Smirnov test, Spearmann correlation was calculated for 269 miRNA/miRNA* species.
qPCR
Annotated and candidate miRNAs were assayed by real-time reverse transcriptase (RT) qPCR using the miScript PCR System (Qiagen) according to the manufacturer's protocols. Total RNAs (25 ng) of the HUVECs were first reverse transcribed and then amplified by SYBR Green PCR, using the miScript Universal Primer and the miRNA-specific forward primer, custom-designed for the newly discovered sequences. When applicable, data were normalized to hsa-miR-16 levels, and the relative expression was calculated using the comparative Ct method (2–ΔΔCt) (Livak and Schmittgen 2001). Reactions run without either RT or the template were used as negative controls. Amplification reactions were performed using the 7900HT Fast Real-Time PCR System (Applied Biosystems), and a melting curve analysis was carried out to verify specificity of the amplification products.
RNA/RISC coimmunoprecipitation
HEK-293 cells were transfected with either an expression vector for c-Myc-Ago2 (Karginov et al. 2007) or vector alone using Fugene 6 (Roche) according to the manufacturer instructions. RISC immunoprecipitation was performed as previously described (Fasanaro et al. 2009). Briefly, transfected cells were harvested in 1 mL/p15 dish of cold lysis buffer (25 mmol/L Tris-HCl at pH 7.4, 150 mmol/L KCl, 0.5% [w/v] NP40, 5 mmol/L EDTA) supplemented with 5 mmol/L dithiothreitol, protease inhibitors mixture tablets (Roche), 1 mmol/L phenylmethylsulfonyl fluoride, and 100 units/mL of RNasin Plus (Promega). After 30 min at 4°C, samples were precleared for 10 min with 75 μL/mL of A/G-agarose beads (Santa Cruz) and spun at 4°C at 20.000g in a microcentrifuge for 30 min. Next, lysates were incubated with 2.5 μg/mL of anti-c-myc antibody (9E10, Santa Cruz) for 3 h at 4°C, and then 50 μL/mL of A/G-agarose beads were added to each sample. After 1 h, immunocomplexes were washed twice with lysis buffer and resuspended in 200 μL of TRIzol (Invitrogen). RNA was purified, and miRNAs were measured by qPCR. The enrichment of candidate and annotated miRNAs was determined by calculating the ΔCT between immunoprecipitates derived from c-Myc-Ago2–transfected HEK-293 cells and their respective controls.
miRNA overexpression and growth curves
Customer-designed miScript miRNA mimics (Qiagen) or negative control siRNA sequence (Qiagen) were transfected by small, interfering RNA transfection reagent (Santa Cruz Biotechnology) in 40% confluent HUVECs (4 × 103/cm2) at the final concentration of 40 nM. After 16 h, cells were incubated with fresh medium for 4 h and exposed to 1% oxygen for 24 and 48 h. The cell number was measured by the Countess cell counter (Invitrogen).
Northern blot
Total RNA was separated on a 15% polyacrylamide/7 M urea gel (10 μg/lane) by electrophoresis and transferred onto Amersham Hybond-N+ membrane (GE Healthcare) by electroblotting. Customer-designed miRCURY probes (Locked Nucleic Acid oligonucleotides, Exiqon) for novel-181 and novel-1322 were terminally labeled with γ-32P-ATP, and hybridization was performed overnight at 55°C, as previously described (Fasanaro et al. 2008). MicroRNA Marker (New England BioLabs) was used as a size marker.
Target prediction and pathway analysis
Bioinformatic prediction of target gene sites for miRNAs significantly modulated by hypoxia (novel-112, novel-144, and hsa-miR-210) was performed using TargetScan Human 5.1 (Lewis et al. 2005) and DIANA-microT version 3.0 (Maragkakis et al. 2009); experimentally confirmed targets of miR-210 were also included (Devlin et al. 2011). Enriched pathways among miRNA targets were generated by Ingenuity Pathways Analysis (version 8.8, Ingenuity Systems), using the Ingenuity Pathways Knowledge Base as reference set and assuming direct and indirect relationships. Fischer's exact test was used for statistical analysis and a P-value ≤0.05 was considered significant.
The targets predicted for novel-112, novel-144, and hsa-miR-210 were intersected with mRNA expression data derived from HUVECs exposed to hypoxia conditions identical to these adopted in our study (P Fasanaro and F Martelli, unpubl.). Genes displaying a significant (P < 0.01) expression difference between normoxic and hypoxic cells were identified by the class comparison function of BRB-ArrayTools software (version 4.1.0, Beta_3 Release, October 2010). These genes were then intersected with the computationally predicted miRNA targets, and only opposite modulated miRNA/mRNA couples were considered. Significance of predicted target-gene enrichment for novel-112, novel-144 and hsa-miR-210, was calculated with a χ2 test.
Statistical analysis
Variables were analyzed by two-tailed Student t-test unless indicated differently, using GraphPad Prism version 4.00 (GraphPad Software). A value of P ≤ 0.05 was deemed statistically significant. Results are reported as mean ± SE.
DATA DEPOSITION
The identified 18 novel miRNAs were submitted to miRBase and the officially assigned names are as follows: novel-144=hsa-mir-6068, novel-2019=hsa-mir-6069, novel-1966=hsa-mir-6070, novel-1691=hsa-mir-6071, novel-298=hsa-mir-6072, novel-498=hsa-mir-6073, novel-716=hsa-mir-6074, novel-2366=hsa-mir-6075, novel-869=hsa-mir-6076, novel-181=hsa-mir-6077, novel-301=hsa-mir-6078, novel-112=hsa-mir-6079, novel-1322=hsa-mir-6080, novel-2982=hsa-mir-6081, novel-2339=hsa-mir-6082, novel-2177=hsa-mir-6083, novel-53=hsa-mir-6084, novel-992=hsa-mir-6085.
Furthermore, the nine novel miRNA* species will be added to miRBase as: hsa-miR-216a-3p, hsa-miR-376a-2-5p, hsa-miR-376b-5p, hsa-miR-376c-5p, hsa-miR-381-5p, hsa-miR-495-5p, hsa-miR-503-3p, hsa-miR-758-5p, hsa-miR-98-3p.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
ACKNOWLEDGMENTS
This work was partly supported by the Ministero della Salute (Italian Ministry of Public Health, RF07Onc-26/1, RC11, RFS07).
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.027615.111.
REFERENCES
- Audic S, Claverie JM 1997. The significance of digital gene expression profiles. Genome Res 7: 986–995 [DOI] [PubMed] [Google Scholar]
- Azuma-Mukai A, Oguri H, Mituyama T, Qian ZR, Asai K, Siomi H, Siomi MC 2008. Characterization of endogenous human Argonautes and their miRNA partners in RNA silencing. Proc Natl Acad Sci 105: 7964–7969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP 2009. MicroRNAs: target recognition and regulatory functions. Cell 136: 215–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benes V, Castoldi M 2010. Expression profiling of microRNA using real-time quantitative PCR, how to use it and what is available. Methods 50: 244–249 [DOI] [PubMed] [Google Scholar]
- Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, Sayers EW 2009. GenBank. Nucleic Acids Res 37: D26–D31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biasiolo M, Sales G, Lionetti M, Agnelli L, Todoerti K, Bisognin A, Coppe A, Romualdi C, Neri A, Bortoluzzi S 2011. Impact of host genes and strand selection on miRNA and miRNA* expression. PLoS ONE 6: e23854 doi: 10.1371/journal.pone.0023854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brameier M, Herwig A, Reinhardt R, Walter L, Gruber J 2011. Human box C/D snoRNAs with miRNA like functions: expanding the range of regulatory RNAs. Nucleic Acids Res 39: 675–686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan SY, Zhang YY, Hemann C, Mahoney CE, Zweier JL, Loscalzo J 2009. MicroRNA-210 controls mitochondrial metabolism during hypoxia by repressing the iron-sulfur cluster assembly proteins ISCU1/2. Cell Metab 10: 273–284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang HR, Schoenfeld LW, Ruby JG, Auyeung VC, Spies N, Baek D, Johnston WK, Russ C, Luo S, Babiarz JE, et al. 2010. Mammalian microRNAs: experimental evaluation of novel and previously annotated genes. Genes Dev 24: 992–1009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Wit E, Linsen SE, Cuppen E, Berezikov E 2009. Repertoire and evolution of miRNA genes in four divergent nematode species. Genome Res 19: 2064–2074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devlin C, Greco S, Martelli F, Ivan M 2011. miR-210: More than a silent player in hypoxia. IUBMB Life 63: 94–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorn GW 2nd 2010. Therapeutic potential of microRNAs in heart failure. Curr Cardiol Rep 12: 209–215 [DOI] [PubMed] [Google Scholar]
- Dupuis DE, Maas S 2010. MiRNA editing. Methods Mol Biol 667: 267–279 [DOI] [PubMed] [Google Scholar]
- Ender C, Krek A, Friedlander MR, Beitzinger M, Weinmann L, Chen W, Pfeffer S, Rajewsky N, Meister G 2008. A human snoRNA with microRNA-like functions. Mol Cell 32: 519–528 [DOI] [PubMed] [Google Scholar]
- Fasanaro P, D'Alessandra Y, Di Stefano V, Melchionna R, Romani S, Pompilio G, Capogrossi MC, Martelli F 2008. MicroRNA-210 modulates endothelial cell response to hypoxia and inhibits the receptor tyrosine kinase ligand Ephrin-A3. J Biol Chem 283: 15878–15883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fasanaro P, Greco S, Lorenzi M, Pescatori M, Brioschi M, Kulshreshtha R, Banfi C, Stubbs A, Calin GA, Ivan M, et al. 2009. An integrated approach for experimental target identification of hypoxia-induced miR-210. J Biol Chem 284: 35134–35143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fasanaro P, Greco S, Ivan M, Capogrossi MC, Martelli F 2010. microRNA: emerging therapeutic targets in acute ischemic diseases. Pharmacol Ther 125: 92–104 [DOI] [PubMed] [Google Scholar]
- Fernandez-Valverde SL, Taft RJ, Mattick JS 2010. Dynamic isomiR regulation in Drosophila development. RNA 16: 1881–1888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garzon R, Calin GA, Croce CM 2009. MicroRNAs in cancer. Annu Rev Med 60: 167–179 [DOI] [PubMed] [Google Scholar]
- Griffiths-Jones S, Grocock RJ, van Dongen S, Bateman A, Enright AJ 2006. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res 34: D140–D144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ 2008. miRBase: tools for microRNA genomics. Nucleic Acids Res 36: D154–D158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths-Jones S, Hui JH, Marco A, Ronshaugen M 2011. MicroRNA evolution by arm switching. EMBO Rep 12: 172–177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He S, Liu C, Skogerbo G, Zhao H, Wang J, Liu T, Bai B, Zhao Y, Chen R 2008. NONCODE v2.0: decoding the non-coding. Nucleic Acids Res 36: D170–D172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivan M, Harris AL, Martelli F, Kulshreshtha R 2008. Hypoxia response and microRNAs: no longer two separate worlds. J Cell Mol Med 12: 1426–1431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karginov FV, Conaco C, Xuan Z, Schmidt BH, Parker JS, Mandel G, Hannon GJ 2007. A biochemical approach to identifying microRNA targets. Proc Natl Acad Sci 104: 19291–19296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawahara Y, Zinshteyn B, Chendrimada TP, Shiekhattar R, Nishikura K 2007. RNA editing of the microRNA-151 precursor blocks cleavage by the Dicer-TRBP complex. EMBO Rep 8: 763–769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaji H, Nakamura M, Takahashi Y, Sandelin A, Katayama S, Fukuda S, Daub CO, Kai C, Kawai J, Yasuda J, et al. 2008. Hidden layers of human small RNAs. BMC Genomics 9: 157 doi: 10.1186/1471-2164-9-157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ 2002. BLAT—the BLAST-like alignment tool. Genome Res 12: 656–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krol J, Loedige I, Filipowicz W 2010. The widespread regulation of microRNA biogenesis, function and decay. Nat Rev Genet 11: 597–610 [DOI] [PubMed] [Google Scholar]
- Landgraf P, Rusu M, Sheridan R, Sewer A, Iovino N, Aravin A, Pfeffer S, Rice A, Kamphorst AO, Landthaler M, et al. 2007. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell 129: 1401–1414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10: R25 doi: 10.1186/gb-2009-10-3-r25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis BP, Burge CB, Bartel DP 2005. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 120: 15–20 [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- Mah S, Buske C, Humphries R, Kuchenbauer F 2010. miRNA*: A passenger stranded in RNA-induced silencing complex? Crit Rev Eukaryot Gene Expr 20: 141–148 [DOI] [PubMed] [Google Scholar]
- Maragkakis M, Reczko M, Simossis VA, Alexiou P, Papadopoulos GL, Dalamagas T, Giannopoulos G, Goumas G, Koukis E, Kourtis K, et al. 2009. DIANA-microT web server: elucidating microRNA functions through target prediction. Nucleic Acids Res 37: W273–W276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marti E, Pantano L, Banez-Coronel M, Llorens F, Minones-Moyano E, Porta S, Sumoy L, Ferrer I, Estivill X 2010. A myriad of miRNA variants in control and Huntington's disease brain regions detected by massively parallel sequencing. Nucleic Acids Res 38: 7219–7235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matkovich SJ, Zhang Y, Van Booven DJ, Dorn GW 2nd 2010. Deep mRNA sequencing for in vivo functional analysis of cardiac transcriptional regulators: application to Gαq. Circ Res 106: 1459–1467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormick R, Buffa FM, Ragoussis J, Harris AL 2010. The role of hypoxia regulated microRNAs in cancer. Curr Top Microbiol Immunol 345: 47–70 [DOI] [PubMed] [Google Scholar]
- Mendes ND, Freitas AT, Sagot MF 2009. Current tools for the identification of miRNA genes and their targets. Nucleic Acids Res 37: 2419–2433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morin RD, O'Connor MD, Griffith M, Kuchenbauer F, Delaney A, Prabhu AL, Zhao Y, McDonald H, Zeng T, Hirst M, et al. 2008. Application of massively parallel sequencing to microRNA profiling and discovery in human embryonic stem cells. Genome Res 18: 610–621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono M, Scott MS, Yamada K, Avolio F, Barton GJ, Lamond AI 2011. Identification of human miRNA precursors that resemble box C/D snoRNAs. Nucleic Acids Res 39: 3879–3891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang KC, Stephen S, Dinger ME, Engstrom PG, Lenhard B, Mattick JS 2007. RNAdb 2.0–an expanded database of mammalian non-coding RNAs. Nucleic Acids Res 35: D178–D182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ro S, Park C, Young D, Sanders KM, Yan W 2007. Tissue-dependent paired expression of miRNAs. Nucleic Acids Res 35: 5944–5953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruby JG, Stark A, Johnston WK, Kellis M, Bartel DP, Lai EC 2007. Evolution, biogenesis, expression, and target predictions of a substantially expanded set of Drosophila microRNAs. Genome Res 17: 1850–1864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rumble SM, Lacroute P, Dalca AV, Fiume M, Sidow A, Brudno M 2009. SHRiMP: accurate mapping of short color-space reads. PLoS Comput Biol 5: e1000386 doi: 10.1371/journal.pcbi.1000386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulte JH, Marschall T, Martin M, Rosenstiel P, Mestdagh P, Schlierf S, Thor T, Vandesompele J, Eggert A, Schreiber S, et al. 2010. Deep sequencing reveals differential expression of microRNAs in favorable versus unfavorable neuroblastoma. Nucleic Acids Res 38: 5919–5928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semenza GL 2010a. Defining the role of hypoxia-inducible factor 1 in cancer biology and therapeutics. Oncogene 29: 625–634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semenza GL 2010b. Vascular responses to hypoxia and ischemia. Arterioscler Thromb Vasc Biol 30: 648–652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sewer A, Paul N, Landgraf P, Aravin A, Pfeffer S, Brownstein MJ, Tuschl T, van Nimwegen E, Zavolan M 2005. Identification of clustered microRNAs using an ab initio prediction method. BMC Bioinformatics 6: 267 doi: 10.1186/1471-2105-6-267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shendure J 2008. The beginning of the end for microarrays? Nat Methods 5: 585–587 [DOI] [PubMed] [Google Scholar]
- Urbich C, Kuehbacher A, Dimmeler S 2008. Role of microRNAs in vascular diseases, inflammation, and angiogenesis. Cardiovasc Res 79: 581–588 [DOI] [PubMed] [Google Scholar]
- Winter J, Jung S, Keller S, Gregory RI, Diederichs S 2009. Many roads to maturity: microRNA biogenesis pathways and their regulation. Nat Cell Biol 11: 228–234 [DOI] [PubMed] [Google Scholar]
- Wu H, Ye C, Ramirez D, Manjunath N 2009. Alternative processing of primary microRNA transcripts by Drosha generates 5′ end variation of mature microRNA. PLoS ONE 4: e7566 doi: 10.1371/journal.pone.0007566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang JH, Shao P, Zhou H, Chen YQ, Qu LH 2010. deepBase: a database for deeply annotating and mining deep sequencing data. Nucleic Acids Res 38: D123–D130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang JS, Phillips MD, Betel D, Mu P, Ventura A, Siepel AC, Chen KC, Lai EC 2011. Widespread regulatory activity of vertebrate microRNA* species. RNA 17: 312–326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yousef M, Showe L, Showe M 2009. A study of microRNAs in silico and in vivo: bioinformatics approaches to microRNA discovery and target identification. FEBS J 276: 2150–2156 [DOI] [PubMed] [Google Scholar]