The bacteriophage (λ)'s cI mRNA was used to examine the importance of the 5′-terminal phosphate on expression of leadered and leaderless mRNA in Escherichia coli. A hammerhead ribozyme was used to produce leadered and leaderless mRNAs, in vivo and in vitro, that contain a 5′-hydroxyl. Although these mRNAs may not occur naturally in the bacterial cell, they allow for the study of the importance of the 5′-phosphorylation state in ribosome binding and translation of leadered and leaderless mRNAs. Analyses with mRNAs containing either a 5′-phosphate or a 5′-hydroxyl indicate that leaderless cI mRNA requires a 5′-phosphate for stable ribosome binding in vitro as well as expression in vivo.
Keywords: translation initiation, leaderless mRNA, ribosome binding, 5′-phosphate
Abstract
The bacteriophage λ's cI mRNA was utilized to examine the importance of the 5′-terminal phosphate on expression of leadered and leaderless mRNA in Escherichia coli. A hammerhead ribozyme was used to produce leadered and leaderless mRNAs, in vivo and in vitro, that contain a 5′-hydroxyl. Although these mRNAs may not occur naturally in the bacterial cell, they allow for the study of the importance of the 5′-phosphorylation state in ribosome binding and translation of leadered and leaderless mRNAs. Analyses with mRNAs containing either a 5′-phosphate or a 5′-hydroxyl indicate that leaderless cI mRNA requires a 5′-phosphate for stable ribosome binding in vitro as well as expression in vivo. Ribosome-binding assays show that 30S subunits and 70S ribosomes do not bind as strongly to 5′-hydroxyl as they do to 5′-phosphate containing leaderless mRNA and the tRNA-dependent ternary complex is less stable. Additionally, filter-binding assays revealed that the 70S ternary complex formed with a leaderless mRNA containing a 5′-hydroxyl has a dissociation rate (koff) that is 4.5-fold higher compared with the complex formed with a 5′-phosphate leaderless mRNA. Fusion to a lacZ reporter gene revealed that leaderless cI mRNA expression with a 5′-hydroxyl was >100-fold lower than the equivalent mRNA with a 5′-phosphate. These data indicate that a 5′-phosphate is an important feature of leaderless mRNA for stable ribosome binding and expression.
INTRODUCTION
Bacterial mRNA's contain a 5′-triphosphate, and the majority of them contain untranslated leader sequences upstream of their coding regions. In the conventional translation pathway, initiation begins with binding of initiation factors (IFs) 1 and 2, initiator tRNA, and an mRNA's translation initiation region by a 30S subunit that is prebound with IF3 (Schmeing and Ramakrishnan 2009). The translation initiation region of leadered mRNAs contains features and signals, including a canonical Shine-Dalgarno (SD) sequence (Shine and Dalgarno 1974), which are important for the formation of the 30S initiation complex. The SD sequence, contained within the untranslated leader, base pairs with the anti-SD (ASD) sequence located near the 3′end of the 16S rRNA (Steitz and Jakes 1975; Hui and de Boer 1987; Jacob et al. 1987) and contributes to formation of the 30S preinitiation complex, while the mRNA is docked on the platform of the 30S subunit (Marzi et al. 2007; Simonetti et al. 2009). IFs and initiator tRNA aid in mRNA accommodation into the ribosome's mRNA channel to form the 30S intiation complex (Simonetti et al. 2009). This complex is joined by the 50S subunit to form the 70S initiation complex.
Recently, a large number of non-SD-led genes, including those that encode leaderless mRNAs (LL mRNA), have been identified through a computational search of 162 prokaryotic genomes (Chang et al. 2006). LL mRNAs, which lack a 5′-untranslated region, are present in a number of prokaryotic species and are especially common in Archaea (Nakagawa et al. 2010). Although sequences downstream from the 5′AUG start codon can greatly increase translational efficiency (Martin-Farmer and Janssen 1999), conserved features downstream from the start codon of LL mRNAs that enhance translation have not been identified. Furthermore, a 5′-terminal AUG start codon is the only known translation signal common to leaderless mRNAs and is required for recognition and efficient translation of LL mRNAs in E. coli (O'Donnell and Janssen 2001; Brock et al. 2008). Also, the presence of a 5′AUG, not codon–anticodon pairing alone, is required for the efficient translation of LL mRNAs in E. coli (Van Etten and Janssen 1998).
The translation initiation of LL mRNAs is thought to occur through a novel pathway involving 70S ribosomes (Balakin et al. 1992; O'Donnell and Janssen 2002; Moll et al. 2004; Udagawa et al. 2004); however, a mechanism involving initiation with 30S subunits has not been ruled out. The role of IFs in LL mRNA translation is not clearly defined, but IF2 enhances LL mRNA translation (Grill et al. 2001) and IF3 discriminates against ribosome binding and translation of LL mRNA (Tedin et al. 1999; O'Donnell and Janssen 2002; Udagawa et al. 2004). Recent evidence reveals that “61S” ribosomes, resulting from kasugamycin treatment of cells, lack several 30S subunit proteins and are capable of selectively translating leaderless mRNAs (Kaberdina et al. 2009). It is evident that the translation initiation pathway of LL mRNA differs from that of canonical mRNA. Recent cross-linking studies suggest that specific ribosomal proteins might contribute to the recognition of a LL mRNA's 5′-terminal AUG and may facilitate initial mRNA recognition and binding in the absence of IFs and initiator tRNA (Brock et al. 2008). Our focus is to identify the features of LL mRNA that interact with the ribosome to facilitate recognition, binding, and translation.
In this study, we investigate the importance of an mRNA's 5′-terminal phosphate for ribosome binding and translation. Like SD mRNA's, LL mRNAs contain a 5′-triphosphate as a consequence of transcription initiation at the A of the AUG start codon. In order to study the effect of the 5′-phosphate, we used a hammerhead ribozyme to synthesize mRNAs, in vivo and in vitro, that contain a 5′-hydroxyl after self-cleavage. Since initiation is the rate-limiting step of translation, we examined the binding of these mRNAs to 30S subunits and 70S ribosomes. While 30S subunits bound similarly to leadered mRNAs regardless of a 5′-terminal phosphate or hydroxyl, neither 30S subunits nor 70S ribosomes bound as strongly to LL mRNAs containing a 5′-hydroxyl as they did to LL mRNA with a 5′-phosphate. The decrease in ribosome binding to 5′-hydroxyl LL mRNA indicates a variation from leadered mRNA in ternary complex formation and/or stability. Thus, we utilized toeprint and filter-binding assays to determine the stability of ternary complexes containing LL mRNA with either a 5′-phosphate or a 5′-hydroxyl. The results from these assays revealed that the 70S ternary complex with LL mRNA containing a 5′-hydroxyl is unstable with a koff that is 4.5-fold higher relative to the complex containing 5′-phosphate LL mRNA.
The expression from translational fusions shows that leadered mRNAs are expressed well with either a 5′-terminal phosphate or hydroxyl. Conversely, expression from LL mRNAs containing a 5′-hydroxyl are dramatically reduced when compared with expression from LL mRNA with a 5′-phosphate. Taken together, our results suggest that 5′-terminal phosphate is important for ribosome binding and expression of LL mRNA.
RESULTS
Gene fusions for production of mRNAs with a 5′-hydroxyl
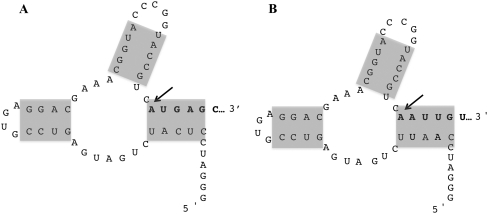
A hammerhead ribozyme, adapted and modified from the sequence of Price et al. (1995), was added to the 5′ end of leaderless and lac-leadered cI-lacZ fusions (Fig. 1, A and B, respectively) in order to investigate the contribution of a mRNA's 5′-terminal phosphate to ribosome binding and translation. Transcription in vivo or in vitro is predicted to produce self-cleaving RNAs encoding leaderless (expressed from plasmid pOH-LLcI) or lac-leadered (expressed from plasmid pOH-SDcI) cI-lacZ mRNAs with a hydroxyl group at the 5′ position of the cleavage site (5′OHApUpGpApGpCp… or 5′OHApApUpUpGpUp…, respectively) (Fig. 1A,B). For comparison, gene fusions were constructed that lacked the ribozyme sequence; transcription from these constructs produced leaderless (expressed from plasmid pLLcI) and lac-leadered (expressed from plasmid pSDcI) cI-lacZ fusion mRNAs that contain a 5′-triphosphate.
FIGURE 1.
Hammerhead ribozyme sequence, proposed structure, and site of cleavage (arrow). Shaded regions represent areas of base pairing. (A) Structure resulting in leaderless cI-lacZ mRNA with a 5′-OH after cleavage. (B) Structure resulting in lac-leadered cI-lacZ mRNA with a 5′-OH after cleavage. Sequence in bold identifies the beginning of the cI coding sequence (A) or the lac-untranslated leader sequence (B).
A hammerhead ribozyme sequence was also added upstream of the cI coding sequence, which included an additional A or G nucleotide upstream of the start codon (expressed from plasmids pOH-A-LLcI and pOH-G-LLcI, respectively); after ribozyme cleavage, the mRNA 5′ end is predicted to be 5′OHApApUpGp… or 5′OHGpApUpGp…, respectively, with the start codon underlined. These fusions were constructed to examine the expression of “leaderless” mRNAs containing a 5′-hydroxyl as well as an extra phosphodiester-linked nucleotide immediately upstream of the start codon. Similar cI-lacZ fusions lacking the ribozyme sequence were constructed; transcription from these constructs yield mRNA containing a 5′-triphosphate as well as the additional A or G nucleotide upstream of the start codon (expressed from plasmids pA-LLcI and pG-LLcI, respectively).
Leaderless mRNA with a 5′-hydroxyl does not support translation in vivo
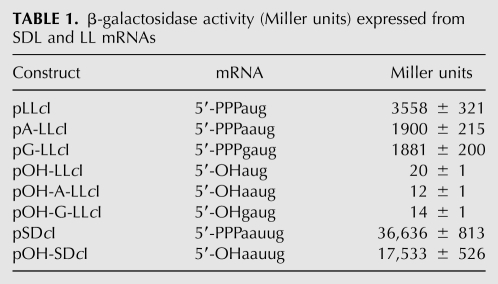
β-galactosidase assays performed on cells expressing the leaderless cI-lacZ mRNA containing a 5′-hydroxyl (expressed from pOH-LLcI) revealed that expression was <1% relative to leaderless mRNA with a 5′-triphosphate (expressed from pLLcI), whereas expression from lac-leadered cI-lacZ mRNA containing a 5′-hydroxyl (expressed from pOH-SDcI) was reduced only twofold relative to leadered mRNA with a 5′-triphosphate (expressed from pSDcI) (Table 1).
TABLE 1.
β-galactosidase activity (Miller units) expressed from SDL and LL mRNAs
To test the possibility that the absence of a phosphodiester bond upstream of the 5′-AUG initiation codon was responsible for the decrease in translation from leaderless mRNA with a 5′-hydroxyl, leaderless cI-lacZ mRNAs containing either an A or a G nucleotide upstream of the start codon were assayed for expression. Expression from leaderless cI-lacZ mRNA containing a 5′-hydroxyl and an additional G or A nucleotide upstream of the start codon (expressed from pOH-G-LLcI or pOH-A-LLcI, respectively) was reduced more than 100-fold relative to the equivalent mRNA with a 5′-triphosphate (expressed from pG-LLcI or pA-LLcI, respectively) (Table 1). Thus, a phosphodiester bond immediately upstream of the start codon did not restore expression for a leaderless cI-lacZ mRNA containing a 5′-hydroxyl, suggesting that a 5′-terminal triphosphate is required for efficient expression of leaderless mRNA in vivo. However, the 5′-terminal triphosphate is not necessary for translation of leadered cI-lacZ mRNA.
Mapping 5′ ends of mRNA resulting from transcription initiation or ribozyme cleavage
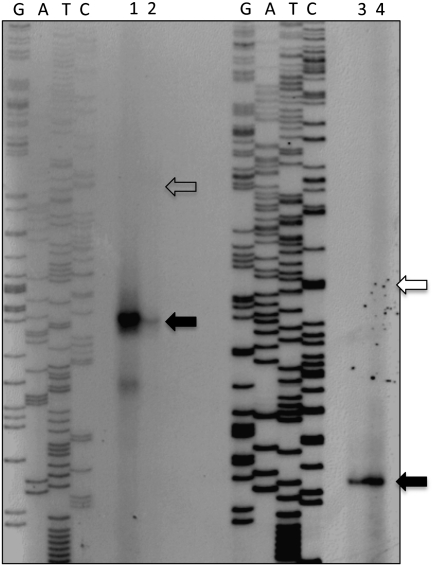
The 5′ ends of mRNA expressed from leaderless cI-lacZ fusions, with or without the ribozyme sequence, were identified by primer extension analysis (Fig. 2). In vivo (Fig. 2, lane 3) or in vitro (data not shown) transcription of the ribozyme-containing leaderless cI-lacZ mRNA, after self-cleavage, resulted in mRNA containing the cI AUG start codon at the 5′ end (the sequence at the 5′ end of leaderless cI mRNA is shown in Fig. 1A). Similarly, transcription of ribozyme-containing cI-lacZ fusions with a single nucleotide leader, after self-cleavage, resulted in mRNA containing an additional A or G nucleotide upstream of the AUG (data not shown). Signals corresponding to uncleaved, ribozyme-containing in vitro or in vivo leaderless mRNA were not detected by this primer extension analysis. Analysis of in vivo-synthesized leaderless cI-lacZ mRNA that lacked the ribozyme sequence revealed transcription initiating at the A of the AUG start codon (Fig. 2, lane 4).
FIGURE 2.
Mapping of mRNA 5′-ends by primer extension analysis of leadered (lanes 1,2) and leaderless (lanes 3,4) cI-lacZ fusions with a 5′-terminal hydroxyl or triphosphate. The closed arrows identify the 5′-end of the lac-untranslated leader (lanes 1,2) or the 5′-end (i.e., the translational start site) of LL mRNAs (lanes 3,4). The open arrows identify the expected migratory positions of uncleaved, hammerhead-containing mRNA. Lanes 2 and 3 show in vivo mRNA containing a 5′-hydroxyl, and lanes 1 and 4 show in vivo mRNA containing a 5′-triphosphate. Lanes labeled G, A, T, and C indicate the dideoxy termination sequencing reactions used to map the 5′ ends of transcripts.
In vivo (Fig. 2, lane 2) or in vitro (data not shown) transcription of the ribozyme-containing lac-leadered cI-lacZ mRNA, after self-cleavage, resulted in mRNA containing the 38-nucleotide lac untranslated leader upstream of the cI coding sequence (the sequence at the 5′ end of leadered cI mRNA is shown in Fig. 1B). Upon overexposure of the primer extension autoradiogram of in vivo mRNA (Fig. 2, lane 2), two signals were observed. The shorter extension product maps to the ribozyme cleavage site, and the larger extension product corresponds, in size, to the 5′ end of the ribozyme sequence. The intensity of the larger signal, corresponding in size to uncleaved mRNA, was minimal. Analysis of in vivo-synthesized lac-leadered cI-lacZ mRNA that lacked the ribozyme sequence revealed transcription initiating at the first nucleotide of the 38-nucleotide lac leader (Fig. 2, lane 1).
Conversion of mRNA's 5′-triphosphate to 5′-hydroxyl decreases the in vivo abundance of full-length mRNA
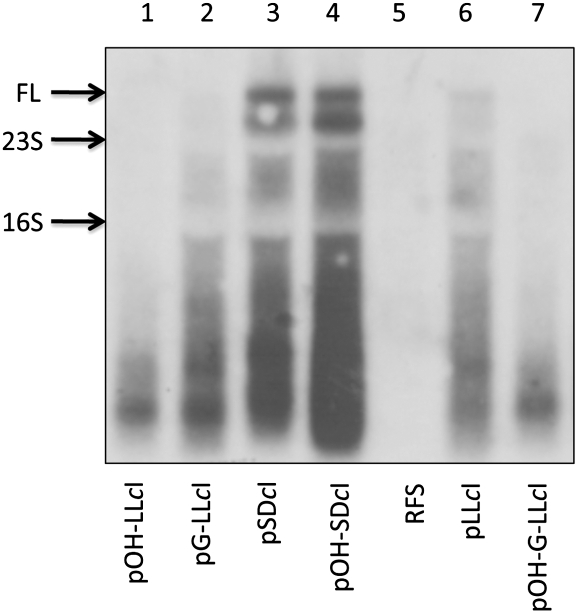
The relative abundance of in vivo leadered and leaderless cI-lacZ mRNAs was analyzed by Northern hybridizations (Fig. 3). The largest hybridizing signal, ∼4200 nucleotides (indicated by arrow), is the predicted size of full-length cI-lacZ mRNA. The high amount of lacZ mRNA fragmentation has been reported and is consistent with the mRNA's synthesis time being similar to the mRNA's half-life (Murakawa et al. 1991).
FIGURE 3.
Analysis of cI-lacZ mRNA by Northern hybridization. Autoradiogram of size-fractionated in vivo RNA probed for cI-lacZ mRNA. Arrows indicate predicted positions of full-length mRNA (FL), 23S, and 16S rRNA. Lanes are labeled according to total in vivo RNA extracted from the E. coli RFS859 host strain (RFS) or the host strain containing plasmids expressing cI-lacZ fusions described in Table 1.
Leadered cI-lacZ mRNAs containing a 5′-hydroxyl or a 5′-triphosphate revealed similar amounts of full-length mRNA (Fig. 3, lanes 3,4). Although expressed from the same lac promoter, the leaderless mRNAs were significantly reduced in abundance compared with SD-leadered mRNAs. Leaderless mRNAs containing a 5′-triphosphate, expressed from pLLcI or pG-LLcI (Fig. 3, lanes 6 and 2, respectively), revealed more large molecular weight cI-lacZ mRNA relative to leaderless mRNAs containing a 5′-hydroxyl expressed from pOH-LLcI or pOH-G-LLcI (Fig. 3, lanes 1 and 7, respectively). These results indicate that cells expressing leaderless mRNAs with a 5′-triphosphate contain more large molecular weight mRNA than cells expressing mRNAs with a 5′-hydroxyl, and suggest that the lack of expression observed for the 5′-hydroxyl containing LL mRNA relates to the low abundance of in vivo mRNA.
Absence of the 5′-terminal phosphate affects ribosome binding to LL mRNA, but not to leadered mRNA in vitro
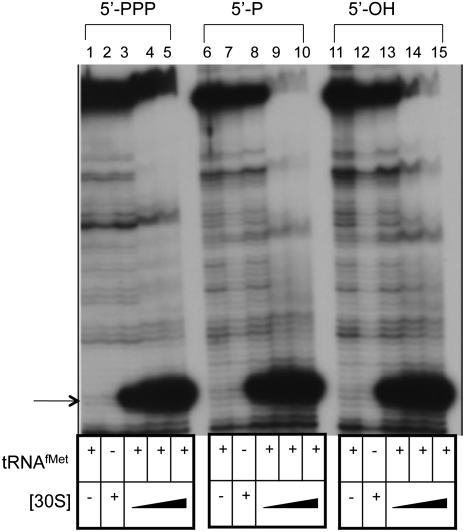
Primer extension inhibition (toeprint) assays were used to compare ribosome binding of leadered (expressed from pSDcI or pOH-SDcI) and LL (expressed from pLLcI or pOH-LLcI) cI-lacZ mRNAs with a 5′-phosphate or a 5′-hydroxyl. Messenger RNAs with a 5′-phosphate contained either a triphosphate, resulting from transcription initiation, or a monophosphate that resulted from the treatment of the 5′-hydroxyl mRNA with T4 polynucleotide kinase and ATP. SD-leadered (SDL) cI-lacZ mRNA with a 5′-phosphate or a 5′-hydroxyl was used to form ternary complexes with 30S subunits and tRNAfMet (Fig. 4). Both 5′-triphosphate and 5′-monophosphate (Fig. 4, lanes 3–5, and lanes 8–10, respectively) and 5′-hydroxyl (Fig. 4, lanes 13–15) SDL mRNAs show a tRNAfMet-dependent toeprint signal. Additionally, toeprint signals with 30S subunits at 2X (Fig. 4, lanes 3,8,13), 6X (Fig. 4, lanes 4,9,14), and 12X (Fig. 4, lanes 5,10,15) are similar for 5′-hydroxyl or 5′-triphosphate and 5′-monophosphate containing SDL RNAs (1X). These results suggest that 30S subunit binding to SDL cI-lacZ mRNA is not greatly affected by the presence of a phosphate or hydroxyl at the 5′-terminus.
FIGURE 4.
Primer extension inhibition (toeprint) analysis of 30S subunit binding to SDL cI-lacZ mRNA with a 5′-triphosphate (lanes 1–5), a 5′-monophosphate (lanes 6–10), or a 5′-hydroxyl (lanes 11–15). Lanes 1, 6, and 11 omit 30S subunits, and lanes 2, 7, and 12 omit tRNAfMet (1 μM when included). (Lanes 3,8,13) 30S subunit concentration (88 nM) is twofold higher than mRNA (44 nM) concentration. (Lanes 4,9,14) 30S subunit concentration (264 nM) is sixfold higher than mRNA concentration. (Lanes 2,5,7,10,12,15) 30S subunit concentration (528 nM) is 12-fold higher than mRNA concentration. The position of the toeprint signal is identified by an arrow.
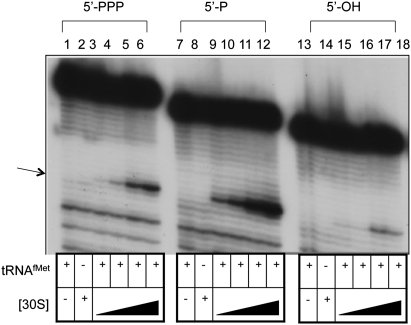
Leaderless mRNAs with a 5′-phosphate (mono- or triphosphate, as described above for leadered mRNAs) or a 5′-hydroxyl were also used to form ternary complexes with 30S subunits (Fig. 5). The LL mRNAs all produce tRNAfMet -dependent toeprint signals when bound to 30S subunits. However, subunit binding to mRNA (1X) containing a 5′-hydroxyl is decreased when compared with the 5′-triphosphate and 5′-monophosphate mRNA with a 30S subunit excess of 2X (Fig. 5, cf. lane 15 and lanes 3 and 9, respectively), 6X (Fig. 5, cf. lane 16 and lanes 4 and 10, respectively), 12X (Fig. 5, cf. lane 17 and lanes 5 and 11, respectively) and 18X (Fig. 5, cf. lane 18 and lanes 6 and 12, respectively). The results indicate that 30S subunits produce the strongest toeprint signal with LL mRNA containing a 5′-monophosphate, followed by a 5′-triphosphate, with the weakest signal observed with 5′-hydroxyl mRNA.
FIGURE 5.
Primer extension inhibition (toeprint) analysis of 30S subunit binding to LL cI-lacZ mRNA with a 5′-triphosphate (lanes 1–6), a 5′-monophosphate (lanes 7–12), or a 5′-hydroxyl (lanes 13–18). Lanes 1, 7, and 13 exclude 30S subunits, and lanes 2, 8, and 14 exclude tRNAfMet (1 μM when included). (Lanes 3,9,15) 30S subunit concentration (88 nM) is twofold higher than mRNA (44 nM) concentration. (Lanes 4,10,16) 30S subunit concentration (264 nM) is sixfold higher than mRNA concentration. (Lanes 5,11,17) 30S subunit concentration (528 nM) is 12-fold higher than RNA concentration. (Lanes 2,6,8,12,14,18) 30S subunit concentration (792 nM) is 18-fold higher than mRNA concentration. The position of the toeprint signal is identified by an arrow.
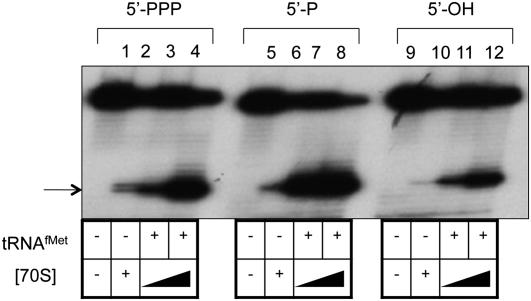
Since leaderless mRNAs bind more strongly to 70S ribosomes than they do to 30S subunits (O'Donnell and Janssen 2002), toeprint assays with ternary complexes of 70S ribosomes, tRNAfMet, and LL mRNA containing a 5′-phosphate (mono- or triphosphate) or a 5′-hydroxyl were performed (Fig. 6). The toeprint signal with 5′-hydroxyl LL mRNA (1X) is reduced compared with the signal with 5′-triphosphate and 5′-monophosphate LL mRNA (1X) when the ribosome concentration is 2X (Fig. 6, cf. decreased intensity in lane 11 and lanes 3 and 7, respectively) or 6X (Fig. 6, cf. lane 12 and lanes 4 and 8, respectively). As with 30S subunits, toeprint signals with 70S ribosomes were strongest with leaderless mRNAs containing a 5′-monophosphate, followed by a 5′-triphosphate, and weakest with a 5′-hydroxyl mRNA. Binding to LL mRNA increased as the concentration of 70S ribosomes increased, and binding to 5′-hydroxyl LL mRNA is reduced relative to 5′-phosphate LL mRNAs. Occasionally, at high ribosome concentrations, we observe signal in the absence of tRNA (Fig. 6, lanes 2, 6, 10).
FIGURE 6.
Primer extension inhibition (toeprint) analysis of 70S ribosome binding to LL cI-lacZ mRNA with a 5′-triphosphate (lanes 1–4), a 5′-monophosphate (lanes 5–8), or a 5′-hydroxyl (lanes 9–12). Lanes 1, 5, and 9 exclude 70S ribosomes, and lanes 2, 6, and 10 exclude tRNAfMet (1 μM when included). (Lanes 3,7,11) 30S subunit concentration (88 nM) is twofold higher than mRNA (44 nM) concentration. (Lanes 2,4,6,8,10,12) 70S ribosome concentration (264 nM) is sixfold higher than mRNA concentration. The position of the toeprint signal is identified by an arrow.
From these results, we conclude that both 30S subunit and 70S ribosome binding to LL mRNA is reduced when the 5′-phosphate is changed to a 5′-hydroxyl. In addition, 30S subunit binding to SDL mRNA is not significantly altered when the 5′-phosphate is changed to a 5′-hydroxyl.
The 5′-terminal phosphate of a LL mRNA influences ternary complex stability in vitro
Competitive toeprint assays were used to investigate whether the decrease in toeprint signal observed with the 5′-hydroxyl LL mRNA resulted from a decrease in ternary complex stability. In these assays, ternary complexes are preformed with LL mRNA, 70S ribosomes, and tRNAfMet, followed by addition of excess competitor LL mRNA containing a 5′-monophosphate or 5′-hydroxyl, and stability of the preformed complex is followed over time.
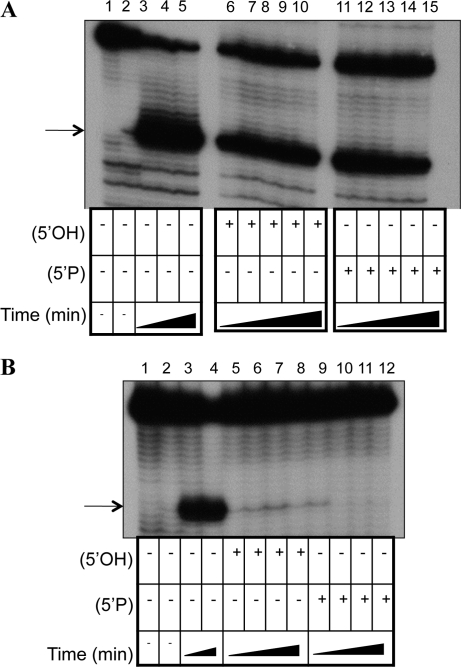
The stability of a preformed ternary complex containing 5′-monophosphate LL mRNA was examined (Fig. 7A). In the absence of competitor mRNA, the toeprint signal remains stable for the 30-min time course used in the assay (Fig. 7A, lanes 3–5). After the addition of 5′-hydroxyl competitor mRNA (Fig. 7A, lanes 6–10), there is an initial decrease in the toeprint signal (compared with the no competitor control), but the toeprint signal remains relatively stable as the time course proceeds from 0 to 30 min (Fig. 7A, cf. signal intensity in lanes 6–10). Similarly, after the addition of 5′-monophosphate competitor mRNA, the signal intensity shows a slight decrease over time from 0 to 30 min (Fig. 7A, lanes 11–15). These results suggest that the ternary complex formed with 5′-phosphate LL mRNA remains relatively stable in the presence of competitor LL mRNA containing a 5′-monophosphate or a 5′-hydroxyl.
FIGURE 7.
Competition primer extension inhibition (toeprint) analysis of LL cI-lacZ mRNA bound to 70S ribosomes with or without tRNAfMet. Prebound ternary complexes containing LL cI-lacZ mRNA with a 5′-monophosphate (A) or 5′-hydroxyl (B) are challenged with excess competitor mRNA containing a 5′-hydroxyl or a 5′-monophosphate. (A) Stability of ternary complexes containing 5′-phosphate LL mRNA. Lane 1 excludes ribosomes, and lane 2 excludes tRNAfMet. (Lanes 3–5) The ternary complex incubated for 0, 20, and 30 min after the initial binding, respectively, with no addition of competitor mRNA. (Lanes 6–10) A 5′-hydroxyl LL mRNA competitor (20X) was added 15 min after initial ternary complex formation with primer-annealed 5′phosphate mRNA (1X). (Lanes 11–15) A 5′-phosphate LL competitor mRNA (20X) was added 15 min after initial ternary complex formation with primer-annealed 5′phosphate mRNA (1X). Reverse transcription was initiated at 0, 5, 10, 20, or 30 min after the competitor addition. (B) Stability of ternary complexes containing 5′-hydroxyl LL mRNA. Lane 1 excludes ribosomes, and lane 2 excludes tRNAfMet. (Lanes 3,4) The ternary complex incubated for 0 and 20 min, respectively, after the complex formation, with no addition of competitor mRNA. (Lanes 5–8) A 5′-hydroxyl LL mRNA competitor (20X) was added to the reactions; (lanes 9–12) a 5′-phosphate LL mRNA competitor (20X) was added 15 min after initial ternary complex formation with primer-annealed 5′hydroxyl mRNA (1X). Reverse transcription was initiated at 0, 5, 10, and 20 min after the competitor addition. The position of the toeprint signals are identified by arrows.
Next, the stability of the ternary complex formed with 5′-hydroxyl LL RNA, 70S ribosomes, and tRNAfMet was tested (Fig. 7B). In the presence of excess ribosomes and the absence of competitor mRNA (Fig. 7B, lanes 3,4), a ternary complex is formed and the toeprint signal remains constant for the 30-min time course used in this assay. However, after the addition of 5′-hydroxyl competitor (Fig. 7B, lanes 5–8) or 5′-monophosphate competitor (Fig. 7B, lanes 9–12), the toeprint signal is nearly undetectable at the first time-point tested and remains negligible throughout the time course examined. These results indicate that the ternary complex containing a LL mRNA with a 5′-hydroxyl is unstable in the presence of excess LL mRNAs containing either a 5′-monophosphate or 5′-hydroxyl.
Taken together, these toeprint assays suggest that LL cI mRNA containing either a 5′-phosphate or 5′-hydroxyl are able to form ternary complexes with 70S ribosomes (Figs. 6, 7). The ternary complex remains stable if the LL mRNA contains a 5′-phosphate (Fig. 7A), but is unstable if the LL mRNA has a 5′-hydroxyl (Fig. 7B).
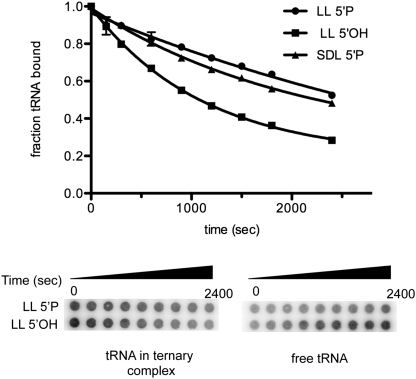
As another measure of ternary complex stability, filter-binding assays were used to compare the dissociation rates of tRNAfMet from 70S ternary complexes (Shoji et al. 2009) with leaderless mRNA (Fig. 8). The amount of radiolabeled tRNAfMet that remains bound to the 70S ribosomes was measured over time, following a 100-fold dilution into a solution containing 400-fold excess unlabled tRNAfMet; thus, as ternary complexes dissociate, reformation with radiolabeled tRNAfMet is minimized by both dilution and the excess unlabeled competitor tRNAfMet.
FIGURE 8.
Filter binding analysis of tRNAfMet dissociation from 70S ternary complexes with 5′-phosphate or 5′-hydroxyl LL cI-lacZ mRNA. Ternary complexes are formed with mRNA (0.5 μM), 30S subunits or 70S ribosomes (0.7 μM), and 32P-labeled tRNAfMet (≤0.05 μM). Reactions were diluted 100-fold into buffer containing 400-fold excess unlabeled tRNAfMet. Samples were spotted onto a double membrane system over time (bottom). The nitrocellulose upper membrane shows tRNAfMet bound to ribosomes (i.e., tRNA in ternary complex) and the positively charged lower membrane shows tRNAfMet not in ternary complex (i.e., free tRNA). The graph shows the proportion of tRNAfMet bound to ribosomes after phosphorimaging quantification of radiolabeled tRNA bound to each membrane and is used to derive the complex stabilities for radiolabeled tRNA in ternary complexes containing LL cI-lacZ mRNA with a 5′-phosphate (LL 5′P) or a 5′-hydroxyl (LL 5′OH), or ternary complexes containing SDL cI-lacZ mRNA with a 5′-phosphate (SDL 5′P).
The dissociation rate (koff) of tRNAfMet from complexes formed with 5′-hydroxyl mRNA is estimated at 8.94 x 10−4 sec−1, whereas the dissociation rate of tRNAfMet from complexes formed with 5′-monophosphate LL mRNA is estimated at 1.99 x 10−4 sec−1 (Fig. 8). These results indicate that initiator tRNAfMet is more stably bound to complexes containing 5′-phosphate LL mRNA than to complexes containing 5′-hydroxyl LL mRNA. The fact that tRNAfMet is more stably bound to ternary complexes with 5′-phosphate mRNA is consistent with the increased toeprint stability (Fig. 7) and the increased in vivo expression (Table 1) from LL mRNA with a 5′-phosphate. Thus, the 5′-terminus of LL cI-lacZ mRNA affects 70S ternary complex stability. Additionally, the koff of tRNAfMet from complexes containing SDL mRNA and 30S subunits is estimated at 4.75 × 10−4 sec−1 (Fig. 8). By this assay, the stability of LL mRNA (5′-monophosphate) on 70S ribosomes is slightly more stable than SDL mRNA on 30S subunits (without initiation factors).
DISCUSSION
The 5′-phosphorylation of leaderless cI-lacZ mRNA is important for ribosome binding and translation. A hammerhead ribozyme was used to produce Shine–Dalgarno leadered (SDL) and leaderless (LL) mRNAs that contained a 5′-terminal hydroxyl group, thereby allowing for the in vitro and in vivo comparisons of ribosome binding and expression from mRNAs containing phosphate or hydroxyl groups at their 5′-termini. Our results show that a 5′-terminal phosphate is required for efficient ribosome binding, ternary complex stability, and translation of LL, but not SDL, mRNA. For SDL mRNAs, the presence of signals within the untranslated leader (e.g., SD sequence, S1-binding pyrimidine-rich tracks) presumably direct ribosome binding to the internal initiation codon, with little influence by the 5′ phosphorylation state on ribosome binding and expression.
Leaderless mRNA with a 5′-hydroxyl does not support translation in vivo
Our in vivo data comparing expression of both SDL and LL cI-lacZ mRNAs suggest that the 5′-phosphorylation state greatly affects expression only of LL mRNAs. When the 5′-triphosphate, which is present on nascent bacterial LL mRNAs, was altered to a 5′-hydroxyl, the in vivo expression was essentially abolished. Interestingly, expression was not rescued by addition of a 5′-A or 5′-G nucleotide, indicating that phosphorylation of the 5′-terminus itself rather than an upstream phosphodiester bond plays a role in the translational efficiency of LL mRNAs. Northern blotting revealed that the abundance of 5′-hydroxyl LL mRNA was decreased, suggesting that the 5′-hydroxyl mRNA might be turned over more rapidly than 5′-triphosphate LL mRNA. RNase E, a major endoribonuclease responsible for mRNA turnover in E. coli, discriminates between mRNAs based on their 5′-phosphorylation state but does not inherently prefer a 5′-hydroxyl containing mRNA as substrate (Jiang and Belasco 2004). Thus, our results are consistent with the interpretation that the LL mRNA's 5′-hydroxyl leads to a decrease in ribosome binding, which can result in decreased expression by leaving the unbound mRNA more susceptible to degradation (Dreyfus 2009).
The 5′-terminal phosphate of a LL mRNA affects in vitro ribosome binding through ternary complex destabilization
The toeprint competition assays showed that 70S ternary complexes containing 5′-hydroxyl LL mRNA were unstable compared with complexes with 5′-monophosphate LL mRNA. However, the 5′-hydroxyl LL mRNA toeprint signal, in the presence of excess ribosomes and tRNA, but no competitor, remained constant over time, suggesting a ribosome association/disassociation equilibrium. The reduced binding of 5′-hydroxyl LL mRNA, compared with 5′-monophosphate LL mRNA, was observed at lower ribosome concentrations, and the reduced stability was readily apparent when competitor mRNAs were added to prebound ternary complexes. In support of the instability observed with toeprint assays, filter-binding assays with 70S ternary complexes also showed instability and revealed that initiator tRNA dissociated nearly five times faster from complexes containing 5′-hydroxyl than 5′-monophosphate LL mRNAs.
We observed that the 5′-monophosphate LL mRNA produced stronger toeprint signals than the 5′-triphosphate LL mRNA. Krishnan et al. (2010) have shown that AUG proximity to the 5′ end is an important determinant of ribosome binding and expression for LL mRNA; therefore, the difference we observed could relate to the extra nucleotide present on the mRNA synthesized in vitro with T7 RNA polymerase, which is not present on the HH-cleaved and phosphorylated mRNA. It is also possible that increased ribosome binding results from a decreased charge or steric hindrance associated with a 5′-terminal monophosphate compared with a triphosphate.
The 5′-terminal phosphate of LL mRNA affects P-site stability
Our toeprint data show that 5′-hydroxyl LL mRNA can form a tRNA-dependent ternary complex, indicating that the 5′AUG is able to reach the P-site. The instability of this ternary complex, measured by both toeprint and filter-binding assays, reflects an inability of the 5′-hydroxyl LL mRNA to stabilize in the P-site and suggests that the 5′-terminal phosphate may be involved in start codon stabilization at the P-site. We hypothesize that the 5′-terminal phosphate interacts with a component of the ribosome (rRNA or ribosomal proteins) or initiator tRNA that contributes to ternary complex stability.
Cross-linking studies (Brock et al. 2008) show that the “U” of a LL mRNA's 5′AUG start codon contacts S7, suggesting that the 5′-phosphate could contact this protein during ribosome binding. A portion of the C-terminal α-helix of S7 reaches the tunnel between the head and the platform near the E site (Yusupova et al. 2001; Simonetti et al. 2009) and may provide a site for start codon contact. It seems unlikely that additional ribosomal proteins could involve the 5′-phosphate in P-site stabilizing interactions, because the decoding site is an rRNA-rich environment that is relatively devoid of protein.
In both the E- and P-sites, high-resolution structural studies reveal that mRNA/ribosome interactions occur mainly through 16S rRNA contacts (Berk et al. 2006; Korostelev et al. 2006; Selmer et al. 2006). The “U” of the AUG start codon for SDL (La Teana et al. 1995) and LL (Brock et al. 2008) mRNAs both contact 16S rRNA near position 1530, suggesting that this contact could be important for the start codon of all mRNAs to reach the P-site. Also, structural data indicate that the conserved 16S rRNA nucleotide G926 is positioned properly to interact (hydrogen bond) with the phosphate 5′ of a start codon's +1 nucleotide when it is positioned in the P-site (Berk et al. 2006). However, because 5′-hydroxyl LL mRNA expression was not rescued by addition of a phosphodiester-linked upstream A or G nucleotide (i.e., 5′ of the AUG start codon), thereby restoring the hydrogen bonding opportunity for interaction with G926, it seems unlikely that G926 is responsible for the ternary complex instability observed with 5′-hydroxyl LL mRNA. While we cannot rule out the possibility that the single A or G nucleotide addition is sterically destabilizing, recent data show that leaderless mRNAs with a 5′-triphosphate and two or three additional nucleotides at the 5′ end bind ribosomes and are expressed (Krishnan et al. 2010), thereby indicating that ribosome binding and initiation can tolerate upstream nucleotides as long as the 5′ end is phosphorylated. It remains unclear why leaderless mRNA with a 5′ phosphate is expressed, while a similar mRNA containing an additional phosphodiester-linked 5′ nucleotide, but lacking a 5′ phosphate, is not expressed (Table 1).
Interactions of E-site rRNA residues with a leaderless mRNA's 5′-phosphate might be necessary for ternary complex stabilization. The antibiotic kasugamycin (ksg) inhibits translation initiation by interfering with contacts between the mRNA channel and nucleotides immediately upstream of the start codon (Schuwirth et al. 2006). Ksg is held in place and stabilized by 16S rRNA nucleotides A792, A794, G926, A1499, G1505, and U1506 that reside within the E- and P-sites of the mRNA channel, suggesting that one or more of these nucleotides could possibly contribute to start codon stabilization. However, expression of 5′-triphosphate containing LL cI-lacZ mRNA is not affected by ksg (Schuwirth et al. 2006), suggesting that these 16S rRNA positions are not needed for LL mRNA ternary complex stability and are unlikely to be responsible for the instability observed with 5′-hydroxyl LL mRNA.
A LL mRNA's 5′-phosphate may directly or indirectly affect the stability of initiator tRNA; in the absence of a 5′-phosphate, initiator tRNA may more readily dissociate from a ternary complex. Transfer RNA's affinity to and stability in the P-site depends on anticodon stem–loop interactions with mRNA and the 30S subunit (Shoji et al. 2009); possibly, binding or P-site placement of LL mRNA with a 5′-hydroxyl disturbs or does not allow an interaction that is needed for tRNA stability.
While our ribosome binding data show that 5′-hydroxyl containing LL mRNA can form tRNA-dependent ternary complexes, these complexes are unstable. We hypothesize that this inability to form a stable ternary complex results in the complete loss of in vivo expression. This loss of expression may result from the failure to compete with other cellular mRNAs for ribosome binding or, possibly, a defect in moving to the elongation phase of translation. Additionally, the decreased ribosome binding likely leads to more rapid mRNA degradation and a lower mRNA abundance, as observed by Northern blotting. Interestingly, recent work by Mitchell et al. (2010) suggests that the 5′-5′ triphosphate of the 5′-7-methylguanosine cap of eukaryotic mRNA inhibits the inappropriate binding of mRNA to preinitiation complexes. While uncapped mRNAs with a 5′-hydroxyl were still recruited to a preinitiation complex, the complex appeared unable to scan for, or unable to stably interact with the start codon. The importance of the mRNA's 5′ phosphorylation state in establishing a preinitiation complex at the 5′ end of leaderless mRNA and at the 5′-end of capped eukaryotic mRNA is noteworthy and may reflect an mRNA 5′-end feature that was conserved during the evolution of the eukaryotic cap structure.
Final remarks
The results presented here and elsewhere (Van Etten and Janssen 1998; O'Donnell and Janssen 2001; Brock et al. 2008; Krishnan et al. 2010) suggest that the combination of a 5′-phosphate and 5′-terminal AUG triplet represent a novel translation signal that uniquely identifies the 5′ end of a leaderless mRNA for ribosome binding and translation initiation. Leaderless mRNAs provide a model system for studies of novel mRNA–ribosome interactions that may be different from those that occur with SDL mRNAs. Additionally, ribosome contacts that identify the start codon for LL mRNA may also be important for SDL mRNAs, but their importance has been overshadowed by the SD–ASD interaction. Leaderless mRNA studies are likely to be relevant to all translation systems, since all domains (Bacteria, Eukarya, and Archaea) contain and are able to express leaderless mRNAs (Janssen 1993; Malys and McCarthy 2010). Additional analysis of the mechanisms by which leaderless mRNAs recognize and bind ribosomes will contribute to our understanding of mRNA–ribosome interactions, ribosome structure–function, and possibly reveal novel mechanisms of translational control.
MATERIALS AND METHODS
Bacterial strains
E. coli DH5α (New England Biolabs [NEB]) was used as the host for all plasmid DNA manipulations. E. coli RFS859 (F-, thr-1, araC859, leuB6, Δlac74, tsx-274, λ-, gyrA111, recA11, relA1, thi-1) (Schleif 1972) was used as host for the expression and assay of lacZ mRNA and enzyme activity for cI-lacZ constructs. E. coli MRE600 was used as the host strain for ribosome isolation.
Reagents and recombinant DNA procedures
Radio-labeled [γ-32P]ATP (6000 Ci/mmol, 150 mCi/mL) was purchased from Perkin Elmer. Restriction endonucleases, T4 DNA ligase, T4 polynucleotide kinase, and T7 RNA polymerase were purchased from NEB and used according to manufacturer's directions. RNase-free DNase I (Roche), AMV reverse transcriptase (Life Sciences), and Pfu DNA polymerase (Stratagene) were used according to the manufacturer's specifications. DNA manipulations, plasmid isolations, and preparation and transformation of competent cells were performed according to Sambrook et al. (1989). The lacZ-specific oligonucleotide 5′-GTTTTCCCAGTCACGACGTTG-3′ was used in DNA sequencing reactions, primer extensions, and toeprint assays, and anneals to positions +99 to +78 of the lacZ coding sequences in both leadered and leaderless cI-lacZ fusions.
Construction of leadered and leaderless cI-lacZ fusions with and without a hammerhead ribozyme sequence
Plasmids encoding leaderless (pLLcI) or lac-leadered (pSDcI) cI-lacZ fusions (O'Donnell and Janssen 2001) containing cI codons 1–16 fused to the fifth codon of a lacZ reporter gene were used to generate in vivo mRNAs with a 5′-triphosphate. These plasmids were also used as templates in PCR amplifications to produce DNA fragments containing a T7 promoter sequence, thereby allowing for in vitro transcription with T7 RNA polymerase and production of RNA used in toeprint reactions. Oligonucleotides were used to assemble DNA fragments encoding the hammerhead ribozyme sequences (Fig. 1), with subsequent cloning into plasmids containing upstream lac or T7 promoters; transcription in vivo from an E. coli lac promoter or in vitro from a T7 promoter was used to produce leaderless (pOH-LLcI) and lac-leadered (pOH-SDcI) cI-lacZ mRNA containing, after ribozyme cleavage, a 5′-hydroxyl. Other ribozyme-containing cI-lacZ fusions were constructed that encoded mRNA containing, after self-cleavage, a 5′-OH and an additional nucleotide (G or A) upstream of the AUG initiation codon.
β-galactosidase activity measurements
β-galactosidase assays were performed as described (Miller 1992).
Radiolabeling of oligonucleotides
Oligoucleotides were end-labeled as described previously (Van Etten and Janssen 1998).
RNA isolation and primer extension analysis
Total RNA was isolated as described previously (Wu and Janssen 1996). Primer extension reactions containing 60 μg of in vivo RNA or 1 μg of in vitro synthesized RNA and 2 pmol of an end-labeled lacZ-specific oligoucleotide were performed as described previously (Brown et al. 1988). The primer extension reactions were subjected to electrophoresis against appropriate dideoxy sequencing reactions and visualized by autoradiography.
Determination of phosphorylation efficiency by T4 polynuceotide kinase
The efficiency of 5′-OH phosphorylation by T4 polynucleotide kinase was determined by following the transfer of 32P from [γ-32P]ATP (6000 Ci/mmol, 150 mCi/mL; Perkin Elmer) to the 5′ end of pOH-AUGcI mRNA. The reaction contained 20 mM Tris-HCl (pH 8), 10 mM MgOAc, 10 mM DTT, 2 mM ATP, 225 μCi [γ-32P]ATP, 10 μg of RNA, and 20 U of T4 polynucleotide kinase, and was incubated at 37°C for 30 min. The enzyme was inactivated at 65°C for 15 min. Dilutions of the radiolabeled mRNA were precipitated with 5% trichloroacetic acid and collected on glass fiber filters (Reeve Angel). Filters were washed with 0.1% HCl, followed by ethanol, and air-dried. Incorporated 32P was determined by comparing the amount of 32P incorporated to the total ratio of labeled and unlabeled ATP in the reaction. The efficiency of the phosphorylation reaction was determined to be 53%.
Preparation of in vitro synthesized transcripts
RNAs were synthesized and purified as described (Fredrick and Noller 2002). RNAs used in toeprint and filter-binding assays were synthesized by combining DNA templates (hammerhead SDL or LL cI-lacZ fusions containing a T7 promoter) and T7 RNAP in 1X buffer (40 mM Tris at pH 7.8, 25 mM MgCl2, 1 mM spermidine, 0.01% Triton X-100, 5 mM each NTP, and 30 mM DTT). Transcription reactions were incubated for 4 h at 37°C, and 40 mM EDTA was added. Samples were treated with DNase (Roche) for 15 min at 37°C. RNA was EtOH precipitated and suspended in RNA loading dye (50% formamide, 0.05% bromophenol blue, 0.05% xylene cyanol). Samples were subjected to PAGE (6% acrylamide, 7 M urea) and full-length products were excised using UV shadowing. Gel slices were rocked overnight at room temperature in elution buffer (300 mM NaOAc at pH 5.2, 0.1% SDS, 1 mM EDTA). The supernatant was phenol extracted and EtOH precipitated. To phosphorylate the 5′-hydroxyl termini when needed, RNA was incubated with T4 PNK and 1 mM ATP in 1X kinase buffer (0.7 M Tris-HCl at pH 7.6, 100 mM MgCl2, 50 mM DTT) for 30 min, phenol extracted, and NH4OAc/EtOH precipitated. Samples were passed through IllustraTM MicroSpinTM G-25 columns (GE Healthcare) to remove excess ATP.
Primer extension inhibition (toeprint) assays
DNA oligonucleotides were phosphorylated at the 5′-terminus using [γ-32P]ATP (6000 Ci/mmol, 150 mCi/mL; Perkin Elmer) and T4 PNK in 1X kinase buffer for 30 min at 37°C and annealed to 3′-termini of RNA as described (Martin-Farmer and Janssen 1999).
Annealed RNA is mixed with 30S subunits or 70S ribosomes with or without tRNAfMet for 15 min at 37°C. Reactions were transferred to ice, and reverse transcriptase was added. The reactions were incubated for 15 min at 37°C and stopped by the addition of NaOAc and EtOH. Precipitated complexes were collected by centrifugation and dissolved in loading dye (80% deionized formamide, 10 mM NaOH, 1 mM EDTA, and 0.5% bromophenol blue and xylene cyanol), followed by heat treatment (95°C, 5 min) and PAGE (6% acrylamide, 7 M urea) in 1X TBE.
In competition (time course) toeprint assays, primer-annealed RNA (44 nM) was prebound to 70S ribosomes (264 nM) with or without tRNAfMet (1 μM) for 15 min at 37°C. Competitor RNA (lacking the 3′ DNA oligonucleotide annealing site; at 880 nM) is added to the reactions and incubated for various lengths of time, followed by the addition of reverse transcriptase.
Filter binding
The filter-binding apparatus was prepared as described (Fahlman et al. 2004). In short, a positively charged nylon membrane was placed on the manifold and covered with a reinforced nitrocellulose membrane.
For tRNAfMet dissociation assays, tRNAfMet 5′ ends were dephosphorylated by treatment with CIP (NEB) for 30 min at 37°C. Following phenol extraction and precipitation, dephosphorylated tRNAfMet was phosphorylated with [γ-32P]ATP (6000 Ci/mmol, 150 mCi/mL; Perkin Elmer) and T4 PNK in 1X imidazole buffer (50 mM imidazole-Cl at pH 6.4, 18 mM MgCl2, 5 mM DTT) with 5% PEG 8000. Radiolabeled tRNAfMet was passed through MicroSpinTM G-25 columns (IllustraTM GE Healthcare) to remove excess ATP.
To measure tRNAfMet dissociation from ternary complexes (koff rates), assays were performed as described (Shoji et al. 2009). Labeled tRNAfMet (≤0.05 μM), 70S ribosomes (0.7 μM), and mRNA (0.5 μM) were incubated for 20 min at 37C in 1x SB to allow for ternary complex formation. At t = 0, 2 μL of this mixture was diluted 100-fold in buffer containing 0.2 μM unlabeled tRNAfMet, and incubation at 37°C is continued. At times (indicated in Fig. 7 legend), 20 μL samples were filtered through the double-membrane apparatus (described above). Phosphorimage analysis was used for pixel quantification and conversion of pixels to amount of tRNAfMet in ternary complex. Nonlinear regression analysis software (GraphPad Prism) was used to determine koff values.
ACKNOWLEDGMENTS
We thank Jay Brock, Karthik Krishnan, Racheal Desmone, and John Hawes for intellectual contributions to this work. We also thank John P. Richardson for discussions that led to use of the hammerhead ribozyme approach in this work. This work was supported by grant GM065120 (G.R.J.) from the National Institutes of Health.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.027698.111.
REFERENCES
- Balakin AG, Skripkin EA, Shatsky IN, Bogdanov AA 1992. Unusual ribosome binding properties of mRNA encoding bacteriophage λ repressor. Nucleic Acids Res 20: 563–571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berk V, Zhang W, Pai RD, Cate JHD 2006. Structural basis for mRNA and tRNA positioning on the ribosome. Proc Natl Acad Sci 103: 15830–15834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brock JE, Pourshahian S, Giliberti J, Limbach PA, Janssen GR 2008. Ribosomes bind leaderless mRNA in Escherichia coli through recognition of their 5′-terminal AUG. RNA 14: 2159–2169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown JW, Thomm M, Beckler GS, Frey G, Stetter KO, Reeve JN 1988. An archaebacterial RNA polymerase binding site and transcription initiation of the hisA gene in Methanococcus vannielii. Nucleic Acids Res 16: 135–150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang B, Halgamuge S, Tang S 2006. Analysis of SD sequences in completed microbial genomes: Non-SD-led genes are as common as SD-led genes. Gene 373: 90–99 [DOI] [PubMed] [Google Scholar]
- Dreyfus M 2009. Killer and protective ribosomes. Prog Mol Biol Transl Sci 85: 423–466 [DOI] [PubMed] [Google Scholar]
- Fahlman RP, Dale T, Uhlenbeck OC 2004. Uniform binding of aminoacylated transfer RNAs to the ribosomal A and P sites. Mol Cell 16: 799–805 [DOI] [PubMed] [Google Scholar]
- Fredrick K, Noller HF 2002. Accurate translocation of mRNA by the ribosome requires a peptidyl group or its analog on the tRNA moving into the 30S P site. Mol Cell 9: 1125–1131 [DOI] [PubMed] [Google Scholar]
- Grill S, Moll I, Hasenohrl D, Gualerzi CO, Blasi U 2001. Modulation of ribosomal recruitment to 5′-terminal start codons by translation initiation factors IF2 and IF3. FEBS Lett 495: 167–171 [DOI] [PubMed] [Google Scholar]
- Hui A, de Boer HA 1987. Specialized ribosome system: preferential translation of a single mRNA species by a subpopulation of mutated ribosomes in Escherichia coli. Proc Natl Acad Sci 84: 4762–4766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob WF, Santer M, Dahlberg AE 1987. A single base change in the Shine-Dalgarno region of 16S rRNA of Escherichia coli affects translation of many proteins. Proc Natl Acad Sci 84: 4757–4761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssen GR. 1993. Eubacterial, archaeabacterial, and eukaryotic genes that encode leaderless mRNA. In Industrial microorganisms: basic and applied molecular genetics. (ed. R Baltz et al.), pp. 59–67. ASM Press, Washington, D.C. [Google Scholar]
- Jiang X, Belasco JG 2004. Catalytic activation of multimeric RNase E and RNase G by 5′-monophosphorylated RNA. Proc Natl Acad Sci 101: 9211–9216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaberdina AC, Szaflarski W, Nierhaus KH, Moll I 2009. An unexpected type of ribosomes induced by kasugamycin: a look into ancestral times of protein synthesis? Mol Cell 33: 227–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korostelev A, Trakhanov S, Laurberg M, Noller HF 2006. Crystal structure of a 70S ribosome-tRNA complex reveals functional interactions and rearrangements. Cell 126: 1065–1077 [DOI] [PubMed] [Google Scholar]
- Krishnan KM, Van Etten WJ 3rd, Janssen GR 2010. Proximity of the start codon to a leaderless mRNA's 5′-terminus is a strong positive determinant of ribosome binding and expression in Escherichia coli. J Bacteriol 192: 6482–6485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Teana A, Gualerzi CO, Brimacombe R 1995. From stand-by to decoding site. Adjustment of the mRNA on the 30S ribosomal subunit under the influence of the initiation factors. RNA 1: 772–782 [PMC free article] [PubMed] [Google Scholar]
- Malys N, McCarthy JE 2010. Translation initiation: variations in the mechanism can be anticipated. Cell Mol Life Sci 68: 991–1003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Farmer JA, Janssen GR 1999. A downstream CA repeat sequence increases translation from leadered and unleadered mRNA in Escherichia coli. Mol Microbiol 31: 1025–1038 [DOI] [PubMed] [Google Scholar]
- Marzi S, Myasnikov AG, Serganov A, Ehresmann C, Romby P, Yusupov M, Klaholz BP 2007. Structured mRNAs regulate translation initiation by binding to the platform of the ribosome. Cell 130: 1019–1031 [DOI] [PubMed] [Google Scholar]
- Miller J. 1992. A short course in bacterial genetics. In A laboratory manual for Escherichia coli and related bacteria. Vol 1, pp. 72–74. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Mitchell SF, Walker SE, Algire MA, Park E, Hinnebusch AG, Lorsch JR 2010. The 5′-7-methylguanosine cap on eukaryotic mRNAs serves both to stimulate canonical translation initiation and to block an alternative pathway. Mol Cell 39: 950–962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moll I, Hirokawa G, Kiel MC, Kaji A, Blasi U 2004. Translation initiation with 70S ribosomes: an alternative pathway for leaderless mRNAs. Nucleic Acids Res 32: 3354–3363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakawa GJ, Kwan C, Yamashita J, Nierlich DP 1991. Transcription and decay of the lac messenger: role of an intergenic terminator. J Bacteriol 173: 28–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa S, Niimura Y, Miura K, Gojobori T 2010. Dynamic evolution of translation initiation mechanisms in prokaryotes. Proc Natl Acad Sci 107: 6382–6387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Donnell SM, Janssen GR 2001. The initiation codon effects ribosome binding and translational efficiency in Escherichia coli of cI mRNA with or without the 5′ untranslated leader. J Bacteriol 183: 1277–1283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Donnell SM, Janssen GR 2002. Leaderless mRNAs bind 70S ribosomes more strongly than 30S ribosomal subunits in Escherichia coli. J Bacteriol 184: 6730–6733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price SR, Ito N, Oubridge C, Avis JM, Nagai K 1995. Crystallization of RNA-protein complexes I. Methods for the large-scale preparation of RNA suitable for crystallographic studies. J Mol Biol 249: 398–408 [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T 1989. Molecular cloning: A laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- Schleif R 1972. Fine-structure deletion map of the Escherichia coli L-arabinose operon. Proc Natl Acad Sci 11: 3479–3484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmeing TM, Ramakrishnan V 2009. What recent ribosome structures have revealed about the mechanism of translation. Nature 461: 1234–1242 [DOI] [PubMed] [Google Scholar]
- Schuwirth BS, Day JM, Hau CW, Janssen GR, Dahlberg AE, Cate JH, Vila-Sanjurjo A 2006. Structural analysis of kasugamycin inhibition of translation. Nat Struct Mol Biol 10: 879–886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selmer M, Dunham CM, Murphy FV, Weixlbaumer A, Petry S, Kelley AC, Weir JR, Ramakrishnan V 2006. Structure of the 70S ribosome complexed with mRNA and tRNA. Science 313: 1935–1942 [DOI] [PubMed] [Google Scholar]
- Shine J, Dalgarno L 1974. The 3′-terminal sequence of Escherichia coli 16S ribosomal RNA: complementarity to nonsense triplets and ribosome binding sites. Proc Natl Acad Sci 71: 1342–1346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoji S, Abdi NM, Bundschuh R, Fredrick K 2009. Contribution of ribosomal residues to P-site tRNA binding. Nucleic Acids Res 37: 4033–4042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonetti A, Marzi S, Jenner L, Myasnikov A, Romby P, Yusupova G, Klaholz BP, Yusupov M 2009. A structural view of translation initiation in bacteria. Cell Mol Life Sci 66: 423–436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steitz JA, Jakes K 1975. How ribosomes select initiator regions in mRNA: base pair formation between the 3′ terminus of 16S rRNA and the mRNA during initiation of protein synthesis in Escherichia coli. Proc Natl Acad Sci 72: 4734–4738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tedin K, Moll I, Grill S, Resch A, Gualerzi CO, Blasi U 1999. Translation initiation factor 3 antagonizes authentic start codon selection on leaderless mRNAs. Mol Microbiol 31: 67–77 [DOI] [PubMed] [Google Scholar]
- Udagawa T, Shimizu Y, Ueda T 2004. Evidence for the translation initiation of leaderless mRNAs by intact 70S ribosomes without its dissociation into subunits in eubacteria. J Biol Chem 279: 8539–8546 [DOI] [PubMed] [Google Scholar]
- Van Etten W, Janssen GR 1998. An AUG initiation codon, not codon-anticodon complementarity, is required for the translation of unleadered mRNA in Escherichia coli. Mol Microbiol 27: 987–1001 [DOI] [PubMed] [Google Scholar]
- Wu CJ, Janssen GR 1996. Translation of vph mRNA in Streptomyces lividans and Escherichia coli after removal of the 5′ untranslated leader. Mol Microbiol 22: 339–355 [DOI] [PubMed] [Google Scholar]
- Yusupova GZ, Yusupov MM, Cate JH, Noller HF 2001. The path of messenger RNA through the ribosome. Cell 106: 233–241 [DOI] [PubMed] [Google Scholar]