Translation of the full-length messenger RNA (mRNA) of the human immunodeficiency virus type 1 (HIV-1) generates the precursor of the viral enzymes via a programmed −1 ribosomal frameshift. Here the authors investigated whether the highly structured 5′ untranslated region (UTR) of this mRNA, which interferes with translation initiation, can modulate HIV-1 frameshift efficiency and showed that, when the 5′ UTR of HIV-1 mRNA occupies the 5′ end of the reporter mRNA, HIV-1 frameshift efficiency is increased about fourfold, compared with a control dual-luciferase reporter with a short unstructured 5′ UTR. This increase was related to an interference with cap-dependent translation initiation by the TAR-Poly(A) region at the 5′ end of the messenger. A model is presented that accounts for the variations in frameshift efficiency depending on the 5′ UTR and the presence of Tat, and it is proposed that a range of frameshift efficiencies is compatible with the virus replication.
Keywords: HIV-1 programmed −1 ribosomal frameshift, translation initiation, TAR, Tat, IRES
Abstract
Translation of the full-length messenger RNA (mRNA) of the human immunodeficiency virus type 1 (HIV-1) generates the precursor of the viral enzymes via a programmed −1 ribosomal frameshift. Here, using dual-luciferase reporters, we investigated whether the highly structured 5′ untranslated region (UTR) of this mRNA, which interferes with translation initiation, can modulate HIV-1 frameshift efficiency. We showed that, when the 5′ UTR of HIV-1 mRNA occupies the 5′ end of the reporter mRNA, HIV-1 frameshift efficiency is increased about fourfold in Jurkat T-cells, compared with a control dual-luciferase reporter with a short unstructured 5′ UTR. This increase was related to an interference with cap-dependent translation initiation by the TAR-Poly(A) region at the 5′ end of the messenger. HIV-1 mRNA 5′ UTR also contains an internal ribosome entry site (IRES), but we showed that, when the cap-dependent initiation mode is available, the IRES is not used or is weakly used. However, when the ribosomes have to use the IRES to translate the dual-luciferase reporter, the frameshift efficiency is comparable to that of the control dual-luciferase reporter. The decrease in cap-dependent initiation and the accompanying increase in frameshift efficiency caused by the 5′ UTR of HIV-1 mRNA is antagonized, in a dose-dependent way, by the Tat viral protein. Tat also stimulates the IRES-dependent initiation and decreases the corresponding frameshift efficiency. A model is presented that accounts for the variations in frameshift efficiency depending on the 5′ UTR and the presence of Tat, and it is proposed that a range of frameshift efficiencies is compatible with the virus replication.
INTRODUCTION
Conventional translation of the human immunodeficiency virus type 1 (HIV-1) full-length messenger RNA (mRNA) produces Gag, the precursor of its structural proteins. It also produces Gag-Pol, the precursor of its enzymes, via a programmed −1 ribosomal frameshift (Jacks et al. 1988; for review, see Brierley and Dos Ramos 2006; Brakier-Gingras and Dulude 2010). This frameshift occurs in a specific region of HIV-1 full-length mRNA (Fig. 1), the slippery sequence, which is followed by an irregular stem–loop, the frameshift stimulatory signal (FSS) that controls frameshift efficiency (Dulude et al. 2002; Gaudin et al. 2005; Staple and Butcher 2005). Current models propose that the encounter between ribosomes and the FSS forces them to make a pause. This pause would be responsible for an incomplete translocation for a small proportion of the ribosomes, which triggers the −1 frameshift (Namy et al. 2006; Leger et al. 2007; Liao et al. 2011). Whether they frameshift or not, ribosomes continue translation by unfolding the FSS with their helicase-associated activity (Takyar et al. 2005). We previously showed (Gendron et al. 2008) that the frameshift event is affected by changes in the rate of translation initiation, although it occurs during the elongation step of translation. A ribosome must encounter the folded FSS to frameshift. A high initiation rate results in the ribosomes being close to each other, reducing the time available for the FSS to refold between the passage of the ribosomes, whereas a low initiation rate has the converse effect. HIV-1 frameshift efficiency is thus inversely proportional to the translation initiation rate. This was demonstrated by using a control dual-luciferase reporter containing the HIV-1 frameshift region inserted between the Renilla (Rluc) and the firefly (Fluc) luciferase genes, such that a −1 frameshift is required to produce Fluc (expressed as a Rluc-Fluc fusion protein), whereas Rluc is produced following the conventional rules of translation. The control dual-luciferase mRNA reporter has a short (∼90 nt long) and unstructured 5′ untranslated region (UTR). This contrasts with the ∼300-nt-long 5′ UTR of HIV-1 full-length mRNA, which is highly structured. This 5′ UTR contains several extremely conserved signals (Fig. 1) that are required for several steps in virus replication (for review, see Berkhout 1996). HIV-1 full-length mRNA is capped and polyadenylated and can therefore be translated, like the majority of cellular mRNAs, via a cap-dependent translation initiation (for reviews on translation, see Gebauer and Hentze 2004; Pestova et al. 2007; Sonenberg and Hinnebusch 2009; Jackson et al. 2010). This mode of initiation involves loading of the 40S ribosomal subunit and associated factors at the 5′ end of the mRNA and subsequent scanning in the 3′ direction until an AUG codon in an appropriate context is encountered. An alternative translation initiation way is via an internal ribosome entry site (IRES) to which the 40S subunit binds within the mRNA, proximal to the AUG start codon (for reviews, see Lopez-Lastra et al. 2005; Doudna and Sarnow 2007; Hellen 2009; Pacheco and Martinez-Salas 2010). Such an entry site was detected in the 5′ UTR of HIV-1 full-length mRNA (Fig. 1; Brasey et al. 2003). Therefore, the translation of this mRNA can be initiated in a cap-dependent and also in an IRES-dependent way (for reviews, see Yilmaz et al. 2006; Ricci et al. 2008; Balvay et al. 2009; Chamond et al. 2010).
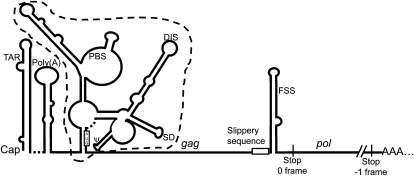
FIGURE 1.
Schematic representation of the 5′ UTR and frameshifting region of HIV-1 full-length mRNA. This mRNA is capped and polyadenylated. Its 5′ UTR is shown under the Weeks conformation, as adapted from Wilkinson et al. (2008). The regulatory motifs present in this region are indicated: the TAR stem–loop, the poly(A) stem–loop containing the polyadenylation signal, the primer binding site (PBS), the dimerization initiation site (DIS), the major splice donor site (SD), and the packaging signal (ψ). The IRES encompasses the region delineated by the dashed line. The AUG initiator codon of gag is boxed. The rest of the mRNA is represented by a thin line except for the HIV-1 frameshift region. The slippery sequence, UUUUUUA, where the frameshift takes place, is indicated, followed by the frameshift stimulatory signal (FSS). The gag coding region is delineated by the AUG initiator codon and the stop codon in the zero frame. The pol coding sequence is delineated by the slippery sequence and the stop codon in the −1 frame.
The 5′ UTR of HIV-1 full-length mRNA is known to slow down mRNA translation (SenGupta et al. 1990; Geballe and Gray 1992; Miele et al. 1996), an effect mostly due to the TAR stem–loop at the 5′ end (Cullen 1986), which is enhanced by the additional presence of the sequence encoding the neighbor poly(A) hairpin (Parkin et al. 1988). It likely results from an interference of TAR with the binding of the 40S subunit to the mRNA. In addition, depending on its concentration, TAR can activate or inhibit the double-stranded RNA-dependent kinase PKR (for reviews, see Bannwarth and Gatignol 2005; Clerzius et al. 2011). This effect, which has an influence on global protein synthesis, is much weaker than the blockade of accessibility of the 5′ end of the mRNA (SenGupta et al. 1990; Geballe and Gray 1992). The activation of PKR causes translation inhibition by phosphorylation of the α-subunit of eIF2, an initiation factor essential for cap-dependent translation initiation, whereas the inhibition of PKR, by decreasing the amount of phosphorylated eIF2, has the opposite effect (for review, see Clerzius et al. 2011).
Several cellular proteins, such as La autoantigen (Chang et al. 1994; Svitkin et al. 1994), Staufen (Dugre-Brisson et al. 2005), and the TAR RNA-binding protein (TRBP) (Dorin et al. 2003), bind the TAR stem–loop and alleviate the blockade of translation initiation caused by TAR, probably by destabilizing this structure. Some cellular RNA helicases also contribute to antagonize the effect of TAR on translation such as RHA, which binds to TAR (Fujii et al. 2001) and enhances the translation of HIV-1 mRNAs (Bolinger et al. 2010). DDX3 also enhances the translation of HIV-1 mRNAs (Liu et al. 2011), although it is not known so far whether it binds to TAR. Among HIV-1 proteins, it was observed more than two decades ago that the Tat protein can counteract the effect of TAR and stimulates translation of the viral mRNAs, but other studies contradicted these observations (see below). The major role of Tat is the transactivation of transcription of viral transcripts by binding to TAR (for reviews, see Brady and Kashanchi 2005; Gatignol 2007), but, by interacting with a variety of cellular proteins, it has other pleiotropic effects (Pugliese et al. 2005; Gautier et al. 2009; Romani et al. 2009; Van Duyne et al. 2009; Johri et al. 2011). Interestingly, Tat up-regulates several cellular RNA helicases among which are DDX3 (Yedavalli et al. 2004) and probably RHA, with which it directly interacts (Van Duyne et al. 2009). As to the effect of Tat on the translation initiation of TAR-containing RNA transcripts, it was shown that in cultured cells, in reticulocyte lysates, or in Xenopus oocytes (Cullen 1986; SenGupta et al. 1990; Braddock et al. 1993), Tat counteracted the inhibitory effect of TAR, but other researchers did not see this effect of Tat either in cultured cells (Chin et al. 1991), in reticulocyte lysates, or in Xenopus oocytes (Parkin et al. 1988; Svitkin et al. 1994).
Tat also has a stimulatory effect on global protein synthesis by competing with eIF2 for phosphorylation by PKR or by inhibiting PKR activity, independently of the presence of TAR (for review, see Clerzius et al. 2011). Because of the conflicting results in the literature, the role of Tat in the translation of mRNAs containing TAR at their 5′ ends remained controversial. In the present study, we investigated, in the absence and in the presence of Tat, the effect of the 5′ UTR of HIV-1 full-length mRNA on the efficiency of the programmed −1 frameshift occurring upon translation of this messenger. We constructed dual-luciferase reporters in which the HIV-1 frameshift region is inserted between the two luciferases, with the complete HIV-1 full-length mRNA 5′ UTR, or a part of it, at the 5′ end of the reporter mRNA. Our results show that, when CD4+ Jurkat T-cells or HEK 293T cells were transfected with these different plasmids, the frameshift efficiency with the constructs containing the TAR-Poly(A) region significantly increased compared with the frameshift efficiency obtained with the control dual-luciferase reporter with a short and unstructured 5′ UTR. With a long structured 5′ UTR devoid of the TAR-Poly(A) region, the frameshift efficiency was comparable to or slightly greater than that of the control dual-luciferase reporter. We constructed another reporter in which the translation initiation for Rluc and Rluc-Fluc was exclusively IRES-dependent and found that its frameshift efficiency was comparable to that of the control dual-luciferase. With all of the reporters, the addition of Tat decreased the frameshift efficiency, the effect being more pronounced with the constructs containing the TAR-Poly(A) region at their 5′ ends or using exclusively an IRES-dependent mode of translation initiation.
RESULTS
Description of dual-luciferase constructs containing the 5′ UTR of HIV-1 full-length mRNA or portions of this 5′ UTR
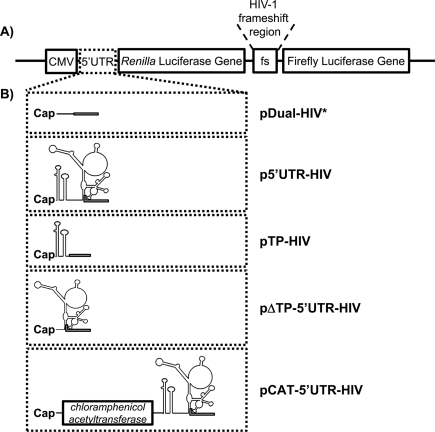
Our aim was to investigate the impact of the structured 5′ UTR of HIV-1 full-length mRNA on HIV-1 frameshift efficiency. To this end, we used dual-luciferase constructs coding for Rluc and Fluc separated by the HIV-1 frameshift region (Fig. 2A,B). The pDual-HIV* mRNA reporter contains a short and relatively unstructured 5′ UTR and corresponds to reporters commonly used to study HIV-1 frameshift so far. We first studied the influence of the complete 5′ UTR of HIV-1 full-length mRNA on frameshift efficiency, using p5′UTR-HIV. Secondly, we determined the impact of different parts of this 5′ UTR by studying the influence of the TAR-Poly(A) structures at the 5′ end of the mRNA reporter, with pTP-HIV, and the influence of the portion of the 5′ UTR of HIV-1 full-length mRNA encompassing an IRES (as determined by Brasey et al. 2003) and without TAR-Poly(A), with pΔTP-5′UTR-HIV. Plasmids pDual-HIV* and pTP-HIV use a cap-dependent mode of initiation, whereas pΔTP-5′UTR-HIV and p5′UTR-HIV could use both the cap-dependent and the IRES-dependent initiation mode. A last reporter, pCAT-5′UTR-HIV, was made that forced the ribosomes to use the IRES-dependent mode to translate the luciferases. The chloramphenicol acetyltransferase (CAT) coding sequence was added upstream of HIV-1 5′ UTR in p5′UTR-HIV, such that the CAT coding sequence is translated via a cap-dependent initiation mode, whereas Rluc and Rluc-Fluc are produced via an IRES-dependent initiation mode.
FIGURE 2.
Scheme of the vectors used to assess HIV-1 frameshift efficiency. (A) Representation of a dual-luciferase reporter. All of the reporters used in this study are derivatives of this dual-luciferase reporter, which differ in their 5′ UTR. They contain the Renilla (Rluc) and the firefly luciferase (Fluc) coding sequences, under control of a CMV promoter, and separated by the HIV-1 frameshift region (nucleotides 1608–1685 in pLAI). Rluc is synthesized by all of the ribosomes translating the mRNA, whereas Fluc is synthesized, as a fusion to Rluc, only by ribosomes that make a −1 frameshift in the frameshift region. (B) Details on the 5′ UTR of the different reporter mRNAs used in this study. Plasmid pDual-HIV* has a short and unstructured 5′ UTR. The plasmid p5′UTR-HIV contains the complete 5′ UTR of HIV-1 full-length mRNA (nucleotides 1–335 in pLAI). The plasmid pTP-HIV contains the beginning of the 5′ UTR of HIV-1 full-length mRNA encompassing the TAR and poly(A) structures (nucleotides 1–106 in pLAI). The plasmid pΔTP-5′UTR-HIV contains a part of the HIV-1 full-length mRNA 5′ UTR (nucleotides 95–335 in pLAI), which encompasses HIV-1 IRES but not TAR and poly(A). Finally, the plasmid pCAT-5′UTR-HIV was designed such that translation of the CAT sequence results from a cap-dependent initiation, whereas the production of the luciferases, preceded by the 5′ UTR of HIV-1 full-length mRNA, results from an IRES-dependent initiation. In pDual-HIV* and pTP-HIV, only the cap-dependent initiation is available. In pΔTP-5′UTR-HIV and p5′UTR-HIV, both cap- and IRES-dependent initiations could be possible. In all of the plasmids, the AUG initiator codon for Rluc is followed by the first 30 nt from the gag coding region. Also, an oligonucleotide coding for a peptide linker was inserted between the nucleotide from gag and the beginning of the Rluc coding sequence. The AUG codon and the 30 nt from gag plus the linker are represented by an open box.
The 5′ UTR of HIV-1 full-length mRNA modulates HIV-1 frameshift efficiency
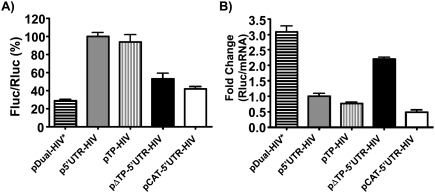
We assessed how the different 5′ UTRs of the mRNA reporters influence HIV-1 frameshift efficiency when the reporters were transfected in cultured cells. Since HIV-1 infects T-lymphocyte cells, we first chose CD4+ Jurkat T-cells (Fig. 3A). Using a reporter containing the HIV-1 full-length mRNA 5′ UTR (p5′UTR-HIV), we observed a fourfold increase in frameshift efficiency compared with a short and unstructured 5′ UTR (pDual-HIV*). This increase mostly results from the presence of the TAR-Poly(A) region, since this region alone (pTP-HIV) causes an increase in frameshift efficiency comparable to that of the complete 5′ UTR. The remaining structured region of the 5′ UTR, devoid of the TAR-Poly(A) region (pΔTP-5′UTR-HIV), causes about a twofold increase in frameshift efficiency when compared with pDual-HIV* but does not contribute to further increase the frameshift efficiency obtained when TAR-Poly(A) is present. HIV-1 IRES is contained within this remaining structured region of the 5′ UTR. We also observed that mutations in the IRES known to increase or decrease by twofold the IRES activity (Gendron et al. 2011) did not affect frameshift efficiency when introduced in p5′UTR-HIV (data not shown). As to the frameshift efficiency of the pCAT-5′UTR-HIV reporter, for which translation of the luciferases is IRES-dependent, it was slightly higher than that of pDual-HIV* and about one-third of the frameshift efficiency observed with the p5′UTR-HIV reporter. Thus, the use of an IRES-dependent translation initiation mode when HIV-1 full-length mRNA is translated results in a frameshift efficiency that strongly differs from that observed with a cap-dependent initiation mode. Note that the changes in frameshift efficiency were assessed by comparing the Fluc/Rluc ratios in the constructs where Fluc expression depends on a −1 frameshift (−1 constructs). Such a direct comparison of these ratios is valid to assess changes in frameshift efficiency because the frameshift site and its local context are identical in the different constructs. In some assays, the frameshift efficiencies were also assessed by dividing the Fluc/Rluc ratio in the −1 constructs by that of the Fluc/Rluc ratio in constructs where Rluc and Fluc are in-frame (0 constructs) (see Grentzmann et al. 1998). The frameshift efficiency was found to be 9.1% ± 0.8% in pDual-HIV* and the variations of frameshift efficiency with pTP-HIV, p5′UTR-HIV, and pΔTP-5′UTR-HIV, when compared with pDual-HIV*, reproduced the changes observed when comparing the Fluc/Rluc ratios in the −1 constructs.
FIGURE 3.
Influence of the 5′ UTR of the reporter mRNA on frameshift efficiency with a dual-luciferase system in Jurkat T-cells. (A) The Fluc/Rluc ratio represents the relative frameshift efficiency corresponding to the different 5′ UTRs inserted upstream of the dual-luciferase coding sequences. The Fluc/Rluc ratios are presented relative to the p5′UTR-HIV construct, with the complete 5′ UTR of HIV-1 full-length mRNA, which is arbitrarily set at 100%. Each value represents the mean ± SEM of at least three independent triplicate experiments. (B) Effect of the 5′ UTR on translation efficiency. The Rluc activities were divided by the corresponding mRNA levels that were quantified by qPCR. The Rluc/mRNA ratios are presented relative to the p5′UTR-HIV construct, which is arbitrarily set at 1. Rluc/mRNA ratios were measured in triplicate, and one representative experiment is shown.
We next investigated whether the changes seen in frameshift efficiency were directly related to translation efficiency by measuring the ratio of Rluc per mRNA produced, using real-time quantitative polymerase chain reaction (qPCR) (Fig. 3B). This experiment showed that the level of intact reporter mRNA produced in transfected cells was similar for each construct, eliminating the possibility that premature termination or aberrant splicing could have occurred in some of the constructs (data not shown). It also showed changes in translational efficiency depending on the 5′ UTR of the construct. The presence of a structured 5′ UTR, devoid of TAR-Poly(A) as in pΔTP-5′UTR-HIV, causes a 1.5-fold decrease in translation efficiency when compared with the short and unstructured 5′ UTR of pDual-HIV*. The TAR-Poly(A) region, as seen with pTP-HIV, or the complete 5′ UTR, as seen in p5′UTR-HIV, reduces translation efficiency about fourfold when compared with pDual-HIV*. With these constructs, a high frameshift efficiency parallels a low translation efficiency. In contrast, the translation efficiency of pCAT-5′UTR-HIV (Fig. 3B) is lower than that of pTP-HIV and p5′UTR-HIV, whereas its frameshift efficiency (Fig. 3A) is much lower than that of those two plasmids. There is thus no apparent inverse correlation between translation rate and frameshift efficiency when the IRES-dependent mode is used.
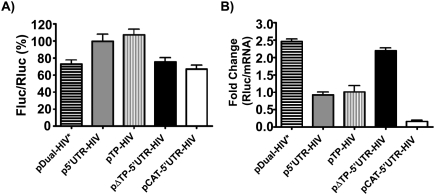
We also verified whether and how the 5′ UTR of HIV-1 full-length mRNA influences HIV-1 frameshift efficiency in HEK 293T, another cell line that is commonly used to study HIV-1 replication (Fig. 4A). We observed that the presence of the complete 5′ UTR of HIV-1 full-length mRNA (p5′UTR-HIV) increases frameshift efficiency by ∼1.5-fold compared with the control pDual-HIV*. The TAR-Poly(A) region (pTP-HIV) causes the same increase of the frameshift efficiency as the complete 5′ UTR, while the HIV-1 5′ UTR devoid of TAR-Poly(A) (pΔTP-5′UTR-HIV) does not affect the frameshift efficiency compared with pDual-HIV*. With pCAT-5′UTR-HIV, the frameshift efficiency is comparable to that of pDual-HIV*. Therefore, the general trend for changes in frameshift efficiency is the same for Jurkat T-cells and HEK 293T cells, although the latter cell line appears less sensitive than the Jurkat T-cells to variations in the 5′ UTR of the reporter mRNA. As for the Jurkat cells, the changes in the frameshift efficiency in HEK 293T were assessed by measuring the changes in the Fluc/Rluc ratios in the −1 constructs. When the frameshift efficiencies were measured by dividing the Fluc/Rluc ratios in the −1 constructs by those in the corresponding in-frame constructs, the frameshift efficiency was found to be 11.0% ± 1.0% in pDual-HIV* and was increased ∼1.5-fold in pTP-HIV or p5′UTR-HIV but did not change in pΔTP-5′UTR-HIV, in agreement with the results obtained when comparing the Fluc/Rluc ratios in the −1 constructs.
FIGURE 4.
HIV-1 frameshifting relative to the type of 5′ UTR of the dual-luciferase reporter mRNA in HEK 293T cells. (A) The Fluc/Rluc ratio represents the relative frameshift efficiency, which was arbitrarily set at 100% for p5′UTR-HIV. Each value represents the mean ± SEM of at least three independent triplicate experiments. (B) The Rluc activities were divided by the level of mRNA quantified by qPCR. The Rluc/mRNA ratio is shown relative to p5′UTR-HIV, which was arbitrarily set at 1. Rluc/mRNA ratios were measured in triplicate, and one representative experiment is shown.
The different 5′ UTRs were also seen to affect translation efficiency of Rluc in HEK 293T cells, as examined by the same procedure described above for Jurkat T-cells (Fig. 4B). TAR-Poly(A), as seen with pTP-HIV and p5′UTR-HIV, reduces translation efficiency by ∼2.5-fold compared with pDual-HIV*. A structured region lacking TAR-Poly(A), as in pΔTP-5′UTR-HIV, has a translation rate comparable to pDual-HIV*. The translation rate of Rluc from pCAT-5′UTR-HIV is smaller (about one-fifth) than that of the reporter mRNAs containing TAR-Poly(A) (pTP-HIV and p5′UTR-HIV). Again, the general trend for the changes in translation efficiency was comparable to what was observed in Jurkat T-cells, although there are quantitative differences between the two types of cells.
Impact of the Tat viral protein on frameshift efficiency
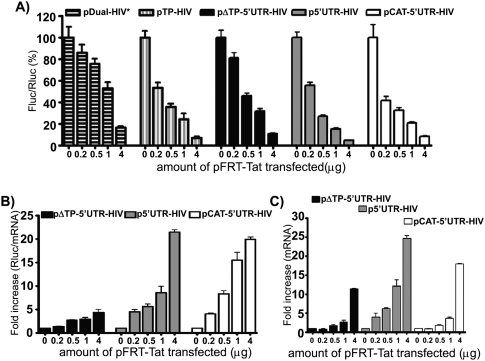
As explained in the Introduction, the presence of the Tat viral protein counteracts the blockade imposed by TAR on translation initiation according to several researchers. However, this observation was contradicted by several studies, casting a doubt on a role of Tat in the control of translation of HIV-1 transcripts. In the present study, Tat was transiently expressed in Jurkat T-cells, as assessed by Western blotting (data not shown). We investigated the effect of Tat on frameshift efficiency with luciferase reporters either containing the TAR-Poly(A) structures or not containing them (Fig. 5A). Our results show a strong dose-dependent decrease in frameshift efficiency with increasing quantities of Tat when TAR is present at the 5′ end of the mRNA (pTP-HIV and p5′UTR-HIV). For the lowest quantity of Tat expressed, the frameshift efficiency amounts to 50% of its value in the absence of Tat and the decline in frameshift efficiency continues, although less abruptly, for further increases in Tat expression. As to pΔTP-5′UTR-HIV, with the 5′ UTR of HIV-1 mRNA devoid of TAR-Poly(A), there is a slow decrease in frameshift efficiency in the presence of increasing quantities of Tat, this efficiency amounting to 80% of its value in absence of Tat for the lowest quantity of Tat expressed. A similar pattern was seen with the control plasmid, pDual-HIV*. When an IRES-dependent mode of initiation was used, with pCAT-5′UTR-HIV, the frameshift efficiency strongly and rapidly decreased in the presence of Tat, as for pTP-HIV and p5′UTR-HIV.
FIGURE 5.
Tat decreases HIV-1 frameshift efficiency in a dose-dependent manner. (A) The Fluc/Rluc ratio was assessed in lysates from Jurkat T-cells cotransfected with pFRT-Tat and pDual-HIV*, pTP-HIV, pΔTP-5′UTR-HIV, p5′UTR-HIV, or pCAT-5′UTR-HIV. The Fluc/Rluc ratio is presented relative to its value in absence of Tat, which is set at 100% for each construct. Results are the mean ± SEM of at least three independent triplicate experiments. (B,C) Cotransfection with pFRT-Tat and pΔTP-5′UTR-HIV, p5′UTR-HIV, or pCAT-5′UTR-HIV. (B) Impact of Tat on translation efficiency. The Rluc activity was divided by the level of Rluc mRNA quantified by qPCR. The Rluc/mRNA ratio is presented relative to the value in the absence of Tat, which is set arbitrarily at 1. Rluc/mRNA ratios were measured in triplicate, and one representative experiment is shown. (C) Relative expression of the dual-luciferase mRNA in presence of Tat. The value in the absence of Tat was arbitrarily set at 1. The mRNA levels were quantified in cell lysates by qPCR. The analysis was performed in triplicate, and one representative experiment is shown.
Next, we examined the translation efficiency in the presence of Tat by measuring the Rluc/mRNA ratio (Fig. 5B). Three constructs were selected for these assays: pΔTP-5′UTR-HIV, p5′UTR-HIV, and pCAT-5′UTR-HIV. In all three cases, translation efficiency increases when Tat expression increases, likely accounting for the decrease in frameshift efficiency. The effect of Tat is stronger in the presence of TAR-Poly(A) (with p5′UTR-HIV), the stimulation of translation being twofold to fourfold that observed in absence of TAR-Poly(A) (with pΔTP-5′UTR-HIV). The effect of Tat is also stronger with the reporter using the IRES mode of initiation for the luciferases (pCAT-5′UTR-HIV). Therefore, our results clearly demonstrate that, in cultured cells, Tat stimulates translation of the different reporters, whether translation is cap-dependent or IRES-dependent, and also decreases the frameshift efficiency.
It is observed that Tat slightly increases the amount of reporter mRNA in the absence of TAR at its 5′ end, whereas the effect of Tat is stronger in its presence (Fig. 5C). This transcriptional effect of Tat with heterologous promoters has already been described (Dingwall et al. 1990; Kim and Risser 1993). An increased transcription could increase the frameshift efficiency by decreasing the ribosome load, thus antagonizing the effect of Tat. The inhibitory effect of Tat on frameshifting could thus have been even greater if Tat was not also stimulating the production of mRNA.
Finally, we also investigated the effect of a mutant of Tat, TatK41T (Kuppuswamy et al. 1989), on frameshift efficiency and on translation efficiency in Jurkat T-cells. This mutant is transcriptionally inactive, and the mutation is located in a region (amino acids 40–58) involved in the interaction of Tat with various cellular proteins, among which is PKR (Cai et al. 2000; Bres et al. 2002; Brady and Kashanchi 2005). The expression of TatK41T in Jurkat T-cells was verified by Western blot (data not shown) and was similar to that of wild-type Tat. We did not observe any significant effect on frameshift efficiency and on translation efficiency with this mutant, whether TAR was present or not (data not shown), suggesting that the effect of Tat on translation and, hence, on frameshift could be related to its capacity to interact with some cellular proteins.
DISCUSSION
Using a dual-luciferase reporter containing either the complete 5′ UTR from HIV-1 full-length mRNA or portions of this UTR, we showed that mostly the TAR-Poly(A) structures, at the 5′ end of this mRNA, affect the programmed −1 ribosomal frameshift that generates HIV-1 enzymes upon translation of this messenger. As explained in the Introduction, a ribosome that encounters an FSS downstream from a slippery sequence makes a pause that can result in a −1 frameshift. Whether it makes a −1 frameshift or not, the ribosome then unfolds the FSS when continuing translation. The spacing between ribosomes on the mRNA is determined by the rate of translation initiation and, depending on the distance between the elongating ribosomes, the FSS will refold or not between the passage of the ribosomes. The results analyzed below were obtained with Jurkat T-cells, but comparable results were obtained with HEK 293T cells. A strong increase in the frameshift efficiency is observed in presence of the complete 5′ UTR, or of only the TAR-Poly(A) stem–loops, whereas a smaller increase is seen with the HIV-1 5′ UTR devoid of TAR-Poly(A). As mentioned in the Introduction, it has been known for a long time that the 5′ UTR of HIV-1 full-length mRNA decreases translation initiation, and we show here that an increase in HIV-1 frameshift efficiency can be linked to that effect. When translation is cap-dependent, the presence of the TAR-Poly(A) region, by interfering with the binding of the 40S subunit to the mRNA, slows down translation initiation such that the distance between the ribosomes provides more time for the FSS to refold between their passage. It is likely that each ribosome encounters a folded FSS, and an interference with the movement of the ribosomes such as that provoked by the portion of the 5′ UTR following TAR-Poly(A) does not further affect the probability that a ribosome encounters a folded FSS. Consequently, a comparable frameshift efficiency is observed in the presence of TAR-Poly(A), with or without the rest of the HIV-1 full-length mRNA 5′ UTR. Most studies on programmed ribosomal frameshifts were carried out with cassettes encompassing the slippery site and flanking sequence in the RNA whose translation can involve a frameshift. No attention was paid to the possible effect of distant sequence features of this mRNA. There are, however, notable exceptions such as the work of the group of Miller (Barry and Miller 2002), demonstrating that an interaction between a structure 4 kb distant from the shift site and a structure downstream from this shift site influences the frameshift in Barley yellow dwarf virus RNA. Also, a recent study by Kurian et al. (2011) investigated the synthesis of Saccharomyces cerevisiae ornithine decarboxylase antizyme, a protein that controls the level of polyamines and whose synthesis depends on a ribosomal frameshift. They demonstrated that the N-terminal part of the antizyme controls the completion of the synthesis of this protein, thus indirectly involving the portion of the antizyme mRNA coding for this N-terminal part in this control. The present study emphasizes the importance of the 5′ UTR of HIV-1 full-length mRNA for an event that occurs at a large distance but is nevertheless influenced by this region via an effect on the translation rate, which determines whether a ribosome will encounter or not encounter a folded FSS.
In the absence of TAR, the Tat viral protein increases the rate of translation and decreases the frameshift efficiency with our dual-luciferase reporters. The same type of response is seen in the presence of TAR, but it is more pronounced. The absence of TAR corresponds to the situation observed with the control plasmid and plasmid pΔTP-5′UTR-HIV, and the effect of Tat can be explained by the fact that it competes with eIF2 for phosphorylation or inhibits PKR (for review, see Clerzius et al. 2011). With TAR-containing reporter mRNAs (p5′UTR-HIV and pTP-HIV), the competition with eIF2 for phosphorylation or the inhibition of PKR by Tat likely contributes to its stimulation of translation. However, this stimulation is stronger than in the absence of TAR, implying that another effect of Tat is involved. As mentioned in the Introduction, TAR interferes with the accessibility of the 5′ end of the mRNA to the ribosomes, and Tat was shown to antagonize this interference. An attractive suggestion is that Tat destabilizes the TAR structure, which would also contribute to its stimulation of translation with TAR-containing constructs. The mechanism for this destabilization is still hypothetical. It does not seem that Tat interacts with TAR in the cytoplasm since its tight specific binding to TAR in the nucleus requires the presence of a nuclear protein, cyclin T (Luo and Peterlin 1993; Stauber and Pavlakis 1998; Wei et al. 1998). A simple model that accounts for the effect of Tat on the translation of TAR-containing mRNAs would be that it is mostly exerted in the nucleus by stimulating the expression of specific proteins, which then counteract the blockade exerted by TAR on translation in the cytoplasm. This requires the capacity of Tat to interact with various proteins such as transcription factors, a property that is likely missing in the TatK41T mutant, which does not affect translation and frameshift efficiency and is altered in a domain of interaction with several cellular proteins. We propose that Tat indirectly destabilizes TAR by increasing the amount of RNA helicases, such as DDX3 and RHA, which stimulate HIV-1 mRNA translation. Other cellular proteins that destabilize the TAR stem–loop could also participate in this effect. We cannot explain the discrepancy in the literature concerning the antagonist effect of Tat on TAR in translation in cultured cells. That the effect of Tat was not seen in reticulocyte lysates makes sense since such a system is inappropriate if the effect of Tat depends on up-regulation of DDX3 and RHA. The same comment can apply to the Xenopus oocytes. Interestingly, Tat was recently found to increase translation of its own mRNA, which possesses a truncated version of the HIV-1 full-length mRNA 5′ UTR including the TAR stem–loop (Charnay et al. 2009). This increase was not seen in a reticulocyte lysate but in cultured HeLa cells.
HIV-1 full-length mRNA possesses the characteristics allowing two modes of initiation to be used: a cap-dependent and an IRES-dependent mode. However, when both initiation modes are available, the cap-dependent mode seems to be highly favored. Indeed, the frameshift efficiency corresponding to the IRES-dependent initiation mode is much smaller than that observed with the cap-dependent initiation mode. However, the frameshift efficiency for the construct containing the complete 5′ UTR with the IRES is comparable to that observed for the construct lacking the IRES region and containing only the TAR-Poly(A) region at its 5′ end. Also, introduction of mutations in the complete HIV-1 full-length mRNA 5′ UTR that increase or decrease the activity of HIV-1 IRES by twofold did not affect the frameshift efficiency. These observations strongly support the notion that the IRES is not used or is weakly used under standard conditions of cellular growth. A similar conclusion was reached by Berkhout et al. (2011), using a mutagenesis approach. However, the IRES could become important under conditions triggered by the viral infection, in line with the findings of Brasey et al. (2003) and Vallejos et al. (2011), showing that HIV-1 IRES is active under specific conditions, such as when the cell cycle is blocked in the G2/M phase, and requires stimulatory factors present during this phase of the cell cycle. We found that, when the Rluc-Fluc reporter was translated exclusively via the IRES contained within the 5′ UTR of HIV-1 full-length mRNA, its translation efficiency was low, being smaller than that of a reporter containing the TAR-Poly(A) region or the complete 5′ UTR of HIV-1 mRNA. This observation contrasts with the fact that the frameshift efficiency corresponding to the use of the IRES was comparable to that obtained with the control pDual-HIV* plasmid and, therefore, much lower than that of the reporters containing TAR-Poly(A). This suggests that only a fraction of the ribosomes encounter the folded FSS when the IRES is used, in spite of their low translation efficiency. The IRES-dependent initiation depends on the conformation of the IRES (Fitzgerald and Semler 2009; Hellen 2009). One can hypothesize that the IRES oscillates between various conformations, but in the period of time during which it adopts an active conformation, a bunch of ribosomes bind quickly. These few ribosomes that succeeded in initiating are close together, and the FSS does not have time to refold between their passage, accounting for the low frameshift efficiency. When a cap-dependent initiation is possible, the adoption of an active IRES conformation must be much rarer due to the movement of the scanning ribosomes across the IRES sequence. Interestingly, Tat also increases the translation efficiency when IRES-dependent initiation is used. This increase could result in part from the up-regulation by Tat of proteins optimizing the conformation of the IRES. It is worth mentioning that up-regulation of the RNA helicase DDX3 by Tat was recently shown to stimulate HIV-1 IRES activity (Liu et al. 2011).
The frameshift event constitutes an interesting target for the development of novel anti-HIV drugs. We observed in this study that the frameshift efficiency is extremely sensitive to the concentration of Tat. It is, however, unlikely that the concentration of Tat remains invariable during the viral replication cycle. This raises the point that there should be a range of frameshift efficiencies that are compatible with the virus replication. It has been shown that a decrease in frameshift efficiency handicaps HIV-1 replication (Telenti et al. 2002; Dulude et al. 2006), and it had been assumed that either decreasing or increasing the frameshift efficiency would interfere with HIV-1 replication (see Dinman and Wickner 1992). We propose that there is a minimal value providing a Gag-Pol/Gag ratio appropriate for virus replication. Decreasing this frameshift efficiency below the minimal value would interfere with the virus replication, but increasing this frameshift efficiency should be tolerated to a given extent. This is in agreement with studies from the group of Yamamoto (Miyauchi et al. 2006) and from our group (D Dulude, A-S Guenier, J Duchaine, A Iannello, A Ahmad, D Lamarre, L Brakier-Gingras, and N Heveker, in prep.) showing that doubling HIV-1 frameshift efficiency does not handicap viral replication. Under these conditions, Dulude et al. also showed that the incorporation of Gag-Pol within the virions was unchanged. However, Shehu-Xhilaga et al. (2001) had shown that a greater increase in frameshift efficiency (about fourfold) decreases the infectivity of the virions, and this decrease was related to a perturbation in the dimerization of HIV-1 genomic RNA. All of these observations combined with our results have implications for the development of drugs targeting the HIV-1 frameshift, since it is not sufficient to alter the frameshift efficiency with a given drug to handicap the virus replication, but it is important to bring this efficiency outside values acceptable for the viral replication.
MATERIALS AND METHODS
Plasmids
Frameshift efficiency was measured using plasmids that contain a dual-luciferase reporter based on a system pioneered by Atkins and coworkers (Grentzmann et al. 1998). Briefly, the HIV-1 frameshift region (nucleotides 1608–1685 according to pLAI, a vector expressing a molecular clone of HIV-1 group M subtype B proviral DNA) (Peden et al. 1991) is inserted between the Rluc and the Fluc such that every ribosome that initiates translation of the reporter mRNA produces Rluc, but only those that make a −1 frameshift in the frameshift region produce Fluc (as a Rluc-Fluc fusion protein). Derivatives of pDual-HIV(−1) (Dulude et al. 2006) with a modified 5′ UTR upstream of the Rluc were created. Briefly, p5′UTR-HIV contains the complete 5′ UTR of the HIV-1 full-length mRNA (nucleotides 1–335 from pLAI, where nucleotide 1 corresponds to the transcription start point). pΔTP-5′UTR-HIV contains a portion of the 5′-UTR region of HIV-1 full-length mRNA lacking the TAR and poly(A) structures but containing the HIV 1 IRES region (nucleotides 95–335 in pLAI). Plasmid pTP-HIV contains the HIV-1 TAR-Poly(A) region (nucleotides 1–106 according to pLAI). In pCAT-5′UTR-HIV, the translation initiation of the luciferases was made IRES-dependent by adding the bacterial chloramphenicol acetyltransferase (CAT) coding sequence upstream of the 5′ UTR in p5′UTR-HIV. Finally, the control pDual-HIV* contains a short and unstructured 5′ UTR. In all derivatives, the first 30 nt of gag after the initiator codon and a 30-nt glycine-rich linker were added between the AUG initiator codon and the Rluc coding sequence (for all the details and the primers used for these constructs, see the Supplemental Material). In-frame derivatives (0 constructs) of pDual-HIV*, pTP-HIV, p5′UTR-HIV, and pΔTP-5′UTR-HIV, where the coding sequences of Rluc and Fluc are in-frame, were also created. This was done by using pDual-HIV(0), which contains the HIV-1 frameshift region (Dulude et al. 2006). In this plasmid, an adenine was added after the slippery sequence of the HIV-1 frameshift region, and the slippery sequence was inactivated by mutagenesis to CUUCCUC. The frameshift region of pDual-HIV(0) was extracted with BsaBI and BamHI and inserted into pDual-HIV*, pTP-HIV, p5′UTR-HIV, and pΔTP-5′UTR-HIV digested with these enzymes.
The plasmid pFRT-Tat was derived from pCMV1-Tat (Gatignol et al. 1991). It expresses a Tat protein of 86 amino acids, which contains an HA tag. The Tat-coding fragment was amplified by PCR, using primers KK and LL (see the Supplemental Table), and the fragment was inserted into the KpnI and the BamHI sites of pcDNA5/FRT (Invitrogen). The 86-amino-acid form of Tat, which is frequently used in studies on Tat, corresponds to a C-terminal truncated form of the natural full-length 101-amino-acid Tat form and was obtained with a number of laboratory-passaged viral strains such as pLAI, pHXB2, and pN4-L3 (Jeang et al. 1999).
Transfection of CD4+ Jurkat T-cells
CD4+ Jurkat T-cells were maintained in RPMI 1640 medium (GIBCO) supplemented with 10% (v/v) FBS (Wisent). Transfection of these cells was made by electroporation, using the Neon transfection system (Invitrogen) according to the manufacturer's instructions. Briefly, 1.25 μg of DNA was mixed with 2 × 106 cells and subjected to one pulse of 1150 V for 40 msec. Two transfections were mixed per well of a six-well plate. For assays with pFRT-Tat, cotransfections were performed with 1 μg of dual-luciferase reporters and increasing amounts of pFRT-Tat ranging from 0 to 4 μg. An empty plasmid, pcDNA5/FRT was added, when required, to maintain an equivalent DNA input. The cells were harvested 24 h post-transfection.
Transfection of HEK 293T cells
HEK 293T cells (human embryonic kidney cells transformed with adenovirus and simian virus 40 large T) were maintained in DMEM (Wisent) supplemented with 10% (v/v) FBS (Wisent). Transfections were performed with 1 μg of dual-luciferase reporters per well, in six-well plates containing HEK 293T cells (3 × 105) prepared 1 d before transfection, using polyethylenimine (PEI; Polysciences, Inc.) as described before (Gendron et al. 2008). The cells were harvested 48 h post-transfection.
Luciferase assays
The Fluc and the Rluc activities of the constructs were measured as relative light units with a Hidex Triathler multilabel tester, as previously described (Gendron et al. 2011). A Dual-Luciferase Reporter Assay System kit (Promega) was used, following the manufacturer's protocol. The relative frameshift efficiency is assessed by the ratio of the Fluc activity to the Rluc activity (Fluc/Rluc).
Real-time quantitative PCR (qPCR)
The Rluc mRNA expression was measured relative to that of GAPDH mRNA. To this end, total RNA was isolated, using the TRIzol reagent (Invitrogen), from CD4+ Jurkat T-cells or HEK 293T cells transfected with the different plasmids, following the manufacturer's protocol. DNAse I (Fermentas) was then used to remove DNA from the extract, and cDNA was generated, using the High Capacity cDNA Reverse Transcription Kit (Applied Biosystems) with 1 μg of total RNA. Real-time analyses were performed on a Light Cycler 480 multi-well plate 96 (Roche Diagnostics) in a final volume of 10 μL. The reaction samples contained 2× SYBR Green Supermix (Bio-Rad) , 0.125 μM each of the gene-specific primers (Rluc-fwd: 5′-GCTGCCCAGCAAGATCATGAGAAA-3′ and Rluc-rev: 5′-GCTCTCAGGTAGGCGTTGTAGTTT-3′) or GAPDH (GAPDH-fwd: 5′-ACATCGCTCAGACACCATG-3′ and GAPDH-rev: 5′-TGTAGTTGAGGTCAATGAAGGG-3′), and 2.5 μL of a 1:5 dilution of cDNA as template. All primer sets were tested before use in this work to ensure that only a single product was amplified. The optimal thermal cycling parameters included a pre-incubation for 5 min at 95°C; 50 cycles of amplification for 10 sec at 95°C, for 15 sec at 65°C, and for 20 sec at 72°C; melting curve analysis for 5 sec at 95°C (segment 1), 65°C/1 min (segment 2), and 95°C/0 sec (segment 3). Controls lacking reverse transcriptase showed no significant amplification.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
ACKNOWLEDGMENTS
This work was supported by a grant (HOP-86864) from the Canadian Institutes of Health Research (CIHR). J.C. and K.G. acknowledge a studentship from CIHR, and G.F. acknowledges a senior fellowship from FRSQ (Fonds de la Recherche en Santé du Québec). We thank Luc Desgroseillers and Dominic Dulude for critical reading of this manuscript and for stimulating discussions and numerous suggestions, and we thank Nikolaus Heveker for his interest in this work. We also thank Anne Gatignol (Lady Davis Institute, Montreal) and Martin Holcik (University of Ottawa) for their generous gift of pCMV1-Tat and pβgal/CAT, respectively.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.030346.111.
REFERENCES
- Balvay L, Soto Rifo R, Ricci EP, Decimo D, Ohlmann T 2009. Structural and functional diversity of viral IRESes. Biochim Biophys Acta 1789: 542–557 [DOI] [PubMed] [Google Scholar]
- Bannwarth S, Gatignol A 2005. HIV-1 TAR RNA: The target of molecular interactions between the virus and its host. Curr HIV Res 3: 61–71 [DOI] [PubMed] [Google Scholar]
- Barry JK, Miller WA 2002. A −1 ribosomal frameshift element that requires base pairing across four kilobases suggests a mechanism of regulating ribosome and replicase traffic on a viral RNA. Proc Natl Acad Sci 99: 11133–11138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berkhout B 1996. Structure and function of the human immunodeficiency virus leader RNA. Prog Nucleic Acid Res Mol Biol 54: 1–34 [DOI] [PubMed] [Google Scholar]
- Berkhout B, Arts K, Abbink TE 2011. Ribosomal scanning on the 5′-untranslated region of the human immunodeficiency virus RNA genome. Nucleic Acids Res 39: 5232–5244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolinger C, Sharma A, Singh D, Yu L, Boris-Lawrie K 2010. RNA helicase A modulates translation of HIV-1 and infectivity of progeny virions. Nucleic Acids Res 38: 1686–1696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braddock M, Powell R, Blanchard AD, Kingsman AJ, Kingsman SM 1993. HIV-1 TAR RNA-binding proteins control TAT activation of translation in Xenopus oocytes. FASEB J 7: 214–222 [DOI] [PubMed] [Google Scholar]
- Brady J, Kashanchi F 2005. Tat gets the “green” light on transcription initiation. Retrovirology 2: 69 doi: 10.1186/1742-4690-2-69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brakier-Gingras L, Dulude D 2010. Programmed −1 ribosomal frameshift in the human immunodeficiency virus of type 1. In Recoding: Expansion of decoding rules enriches gene expression (ed. J Atkins, R Gesteland), pp. 175–192. Springer, New York. [Google Scholar]
- Brasey A, Lopez-Lastra M, Ohlmann T, Beerens N, Berkhout B, Darlix JL, Sonenberg N 2003. The leader of human immunodeficiency virus type 1 genomic RNA harbors an internal ribosome entry segment that is active during the G2/M phase of the cell cycle. J Virol 77: 3939–3949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bres V, Kiernan R, Emiliani S, Benkirane M 2002. Tat acetyl-acceptor lysines are important for human immunodeficiency virus type-1 replication. J Biol Chem 277: 22215–22221 [DOI] [PubMed] [Google Scholar]
- Brierley I, Dos Ramos FJ 2006. Programmed ribosomal frameshifting in HIV-1 and the SARS-CoV. Virus Res 119: 29–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai R, Carpick B, Chun RF, Jeang KT, Williams BR 2000. HIV-I TAT inhibits PKR activity by both RNA-dependent and RNA-independent mechanisms. Arch Biochem Biophys 373: 361–367 [DOI] [PubMed] [Google Scholar]
- Chamond N, Locker N, Sargueil B 2010. The different pathways of HIV genomic RNA translation. Biochem Soc Trans 38: 1548–1552 [DOI] [PubMed] [Google Scholar]
- Chang YN, Kenan DJ, Keene JD, Gatignol A, Jeang KT 1994. Direct interactions between autoantigen La and human immunodeficiency virus leader RNA. J Virol 68: 7008–7020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charnay N, Ivanyi-Nagy R, Soto-Rifo R, Ohlmann T, López-Lastra M, Darlix J-L 2009. Mechanism of HIV-1 Tat RNA translation and its activation by the Tat protein. Retrovirology 6: 74 doi: 10.1186/1742-4690-6-74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin DJ, Selby MJ, Peterlin BM 1991. Human immunodeficiency virus type 1 Tat does not transactivate mature trans-acting responsive region RNA species in the nucleus or cytoplasm of primate cells. J Virol 65: 1758–1764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clerzius G, Gelinas JF, Gatignol A 2011. Multiple levels of PKR inhibition during HIV-1 replication. Rev Med Virol 21: 42–53 [DOI] [PubMed] [Google Scholar]
- Cullen BR 1986. Trans-activation of human immunodeficiency virus occurs via a bimodal mechanism. Cell 46: 973–982 [DOI] [PubMed] [Google Scholar]
- Dingwall C, Ernberg I, Gait MJ, Green SM, Heaphy S, Karn J, Lowe AD, Singh M, Skinner MA 1990. HIV-1 tat protein stimulates transcription by binding to a U-rich bulge in the stem of the TAR RNA structure. EMBO J 9: 4145–4153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinman JD, Wickner RB 1992. Ribosomal frameshifting efficiency and gag/gag-pol ratio are critical for yeast M1 double-stranded RNA virus propagation. J Virol 66: 3669–3676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorin D, Bonnet MC, Bannwarth S, Gatignol A, Meurs EF, Vaquero C 2003. The TAR RNA-binding protein, TRBP, stimulates the expression of TAR-containing RNAs in vitro and in vivo independently of its ability to inhibit the dsRNA-dependent kinase PKR. J Biol Chem 278: 4440–4448 [DOI] [PubMed] [Google Scholar]
- Doudna JA, Sarnow P 2007. Translation initiation by viral internal ribosome entry sites. In Translational control in biology and medicine (ed. MB Mathews et al.), pp. 129–153. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Dugre-Brisson S, Elvira G, Boulay K, Chatel-Chaix L, Mouland AJ, DesGroseillers L 2005. Interaction of Staufen1 with the 5′ end of mRNA facilitates translation of these RNAs. Nucleic Acids Res 33: 4797–4812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dulude D, Baril M, Brakier-Gingras L 2002. Characterization of the frameshift stimulatory signal controlling a programmed −1 ribosomal frameshift in the human immunodeficiency virus type 1. Nucleic Acids Res 30: 5094–5102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dulude D, Berchiche YA, Gendron K, Brakier-Gingras L, Heveker N 2006. Decreasing the frameshift efficiency translates into an equivalent reduction of the replication of the human immunodeficiency virus type 1. Virology 345: 127–136 [DOI] [PubMed] [Google Scholar]
- Fitzgerald KD, Semler BL 2009. Bridging IRES elements in mRNAs to the eukaryotic translation apparatus. Biochim Biophys Acta 1789: 518–528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii R, Okamoto M, Aratani S, Oishi T, Ohshima T, Taira K, Baba M, Fukamizu A, Nakajima T 2001. A role of RNA helicase A in cis-acting transactivation response element-mediated transcriptional regulation of human immunodeficiency virus type 1. J Biol Chem 276: 5445–5451 [DOI] [PubMed] [Google Scholar]
- Gatignol A 2007. Transcription of HIV: Tat and cellular chromatin. Adv Pharmacol 55: 137–159 [DOI] [PubMed] [Google Scholar]
- Gatignol A, Buckler-White A, Berkhout B, Jeang KT 1991. Characterization of a human TAR RNA-binding protein that activates the HIV-1 LTR. Science 251: 1597–1600 [DOI] [PubMed] [Google Scholar]
- Gaudin C, Mazauric MH, Traikia M, Guittet E, Yoshizawa S, Fourmy D 2005. Structure of the RNA signal essential for translational frameshifting in HIV-1. J Mol Biol 349: 1024–1035 [DOI] [PubMed] [Google Scholar]
- Gautier VW, Gu L, O'Donoghue N, Pennington S, Sheehy N, Hall WW 2009. In vitro nuclear interactome of the HIV-1 Tat protein. Retrovirology 6: 47 doi: 10.1186/1742-4690-6-47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geballe AP, Gray MK 1992. Variable inhibition of cell-free translation by HIV-1 transcript leader sequences. Nucleic Acids Res 20: 4291–4297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gebauer F, Hentze MW 2004. Molecular mechanisms of translational control. Nat Rev Mol Cell Biol 5: 827–835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendron K, Charbonneau J, Dulude D, Heveker N, Ferbeyre G, Brakier-Gingras L 2008. The presence of the TAR RNA structure alters the programmed −1 ribosomal frameshift efficiency of the human immunodeficiency virus type 1 (HIV-1) by modifying the rate of translation initiation. Nucleic Acids Res 36: 30–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendron K, Ferbeyre G, Heveker N, Brakier-Gingras L 2011. The activity of the HIV-1 IRES is stimulated by oxidative stress and controlled by a negative regulatory element. Nucleic Acids Res 39: 902–912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grentzmann G, Ingram JA, Kelly PJ, Gesteland RF, Atkins JF 1998. A dual-luciferase reporter system for studying recoding signals. RNA 4: 479–486 [PMC free article] [PubMed] [Google Scholar]
- Hellen CUT 2009. IRES-induced conformational changes in the ribosome and the mechanism of translation initiation by internal ribosomal entry. Biochim Biophys Acta 1789: 558–570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacks T, Power MD, Masiarz FR, Luciw PA, Barr PJ, Varmus HE 1988. Characterization of ribosomal frameshifting in HIV-1 gag-pol expression. Nature 331: 280–283 [DOI] [PubMed] [Google Scholar]
- Jackson RJ, Hellen CU, Pestova TV 2010. The mechanism of eukaryotic translation initiation and principles of its regulation. Nat Rev Mol Cell Biol 11: 113–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeang KT, Xiao H, Rich EA 1999. Multifaceted activities of the HIV-1 transactivator of transcription, Tat. J Biol Chem 274: 28837–28840 [DOI] [PubMed] [Google Scholar]
- Johri MK, Mishra R, Chhatbar C, Unni SK, Singh SK 2011. Tits and bits of HIV Tat protein. Expert Opin Biol Ther 11: 269–283 [DOI] [PubMed] [Google Scholar]
- Kim YS, Risser R 1993. TAR-independent transactivation of the murine cytomegalovirus major immediate-early promoter by the Tat protein. J Virol 67: 239–248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuppuswamy M, Subramanian T, Srinivasan A, Chinnadurai G 1989. Multiple functional domains of Tat, the trans-activator of HIV-1, defined by mutational analysis. Nucleic Acids Res 17: 3551–3561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurian L, Palanimurugan R, Godderz D, Dohmen RJ 2011. Polyamine sensing by nascent ornithine decarboxylase antizyme stimulates decoding of its mRNA. Nature 477: 490–494 [DOI] [PubMed] [Google Scholar]
- Leger M, Dulude D, Steinberg SV, Brakier-Gingras L 2007. The three transfer RNAs occupying the A, P and E sites on the ribosome are involved in viral programmed −1 ribosomal frameshift. Nucleic Acids Res 35: 5581–5592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao PY, Choi YS, Dinman JD, Lee KH 2011. The many paths to frameshifting: Kinetic modelling and analysis of the effects of different elongation steps on programmed −1 ribosomal frameshifting. Nucleic Acids Res 39: 300–312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Henao-Mejia J, Liu H, Zhao Y, He JJ 2011. Translational regulation of HIV-1 replication by HIV-1 Rev cellular cofactors Sam68, eIF5A, hRIP, and DDX3. J Neuroimmune Pharmacol 6: 308–321 [DOI] [PubMed] [Google Scholar]
- Lopez-Lastra M, Rivas A, Barria MI 2005. Protein synthesis in eukaryotes: The growing biological relevance of cap-independent translation initiation. Biol Res 38: 121–146 [DOI] [PubMed] [Google Scholar]
- Luo Y, Peterlin BM 1993. Juxtaposition between activation and basic domains of human immunodeficiency virus type 1 Tat is required for optimal interactions between Tat and TAR. J Virol 67: 3441–3445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miele G, Mouland A, Harrison GP, Cohen E, Lever AM 1996. The human immunodeficiency virus type 1 5′ packaging signal structure affects translation but does not function as an internal ribosome entry site structure. J Virol 70: 944–951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyauchi K, Komano J, Myint L, Futahashi Y, Urano E, Matsuda Z, Chiba T, Miura H, Sugiura W, Yamamoto N 2006. Rapid propagation of low-fitness drug-resistant mutants of human immunodeficiency virus type 1 by a streptococcal metabolite sparsomycin. Antivir Chem Chemother 17: 167–174 [DOI] [PubMed] [Google Scholar]
- Namy O, Moran SJ, Stuart DI, Gilbert RJ, Brierley I 2006. A mechanical explanation of RNA pseudoknot function in programmed ribosomal frameshifting. Nature 441: 244–247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacheco A, Martinez-Salas E 2010. Insights into the biology of IRES elements through riboproteomic approaches. J Biomed Biotechnol 2010: 458927 doi: 10.1155/2010/458927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkin NT, Cohen EA, Darveau A, Rosen C, Haseltine W, Sonenberg N 1988. Mutational analysis of the 5′ non-coding region of human immunodeficiency virus type 1: Effects of secondary structure on translation. EMBO J 7: 2831–2837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peden K, Emerman M, Montagnier L 1991. Changes in growth properties on passage in tissue culture of viruses derived from infectious molecular clones of HIV-1LAI, HIV-1MAL, and HIV-1ELI. Virology 185: 661–672 [DOI] [PubMed] [Google Scholar]
- Pestova TV, Lorsh JR, Hellen CUT 2007. The mechanism of translation initiation in eukaryotes. In Translational control in biology and medicine (ed. MB Mathews et al.), pp. 87–128. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Pugliese A, Vidotto V, Beltramo T, Petrini S, Torre D 2005. A review of HIV-1 Tat protein biological effects. Cell Biochem Funct 23: 223–227 [DOI] [PubMed] [Google Scholar]
- Ricci EP, Soto Rifo R, Herbreteau CH, Decimo D, Ohlmann T 2008. Lentiviral RNAs can use different mechanisms for translation initiation. Biochem Soc Trans 36: 690–693 [DOI] [PubMed] [Google Scholar]
- Romani B, Engelbrecht S, Glashoff RH 2009. Functions of Tat: The versatile protein of human immunodeficiency virus type 1. J Gen Virol 91: 1–12 [DOI] [PubMed] [Google Scholar]
- SenGupta DN, Berkhout B, Gatignol A, Zhou AM, Silverman RH 1990. Direct evidence for translational regulation by leader RNA and Tat protein of human immunodeficiency virus type 1. Proc Natl Acad Sci 87: 7492–7496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shehu-Xhilaga M, Crowe SM, Mak J 2001. Maintenance of the Gag/Gag-Pol ratio is important for human immunodeficiency virus type 1 RNA dimerization and viral infectivity. J Virol 75: 1834–1841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonenberg N, Hinnebusch AG 2009. Regulation of translation initiation in eukaryotes: Mechanisms and biological targets. Cell 136: 731–745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staple DW, Butcher SE 2005. Solution structure and thermodynamic investigation of the HIV-1 frameshift inducing element. J Mol Biol 349: 1011–1023 [DOI] [PubMed] [Google Scholar]
- Stauber RH, Pavlakis GN 1998. Intracellular trafficking and interactions of the HIV-1 Tat protein. Virology 252: 126–136 [DOI] [PubMed] [Google Scholar]
- Svitkin YV, Pause A, Sonenberg N 1994. La autoantigen alleviates translational repression by the 5′ leader sequence of the human immunodeficiency virus type 1 mRNA. J Virol 68: 7001–7007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takyar S, Hickerson RP, Noller HF 2005. mRNA helicase activity of the ribosome. Cell 120: 49–58 [DOI] [PubMed] [Google Scholar]
- Telenti A, Martinez R, Munoz M, Bleiber G, Greub G, Sanglard D, Peters S 2002. Analysis of natural variants of the human immunodeficiency virus type 1 gag-pol frameshift stem–loop structure. J Virol 76: 7868–7873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallejos M, Deforges J, Plank TD, Letelier A, Ramdohr P, Abraham CG, Valiente-Echeverria F, Kieft JS, Sargueil B, Lopez-Lastra M 2011. Activity of the human immunodeficiency virus type 1 cell cycle–dependent internal ribosomal entry site is modulated by IRES trans-acting factors. Nucleic Acids Res 39: 6186–6200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Duyne R, Kehn-Hall K, Carpio L, Kashanchi F 2009. Cell-type-specific proteome and interactome: Using HIV-1 Tat as a test case. Expert Rev Proteomics 6: 515–526 [DOI] [PubMed] [Google Scholar]
- Wei P, Garber ME, Fang SM, Fischer WH, Jones KA 1998. A novel CDK9-associated C-type cyclin interacts directly with HIV-1 Tat and mediates its high-affinity, loop-specific binding to TAR RNA. Cell 92: 451–462 [DOI] [PubMed] [Google Scholar]
- Wilkinson KA, Gorelick RJ, Vasa SM, Guex N, Rein A, Mathews DH, Giddings MC, Weeks KM 2008. High-throughput SHAPE analysis reveals structures in HIV-1 genomic RNA strongly conserved across distinct biological states. PLoS Biol 6: e96 doi: 10.1371/journal.pbio.0060096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yedavalli VS, Neuveut C, Chi YH, Kleiman L, Jeang KT 2004. Requirement of DDX3 DEAD box RNA helicase for HIV-1 Rev-RRE export function. Cell 119: 381–392 [DOI] [PubMed] [Google Scholar]
- Yilmaz A, Bolinger C, Boris-Lawrie K 2006. Retrovirus translation initiation: Issues and hypotheses derived from study of HIV-1. Curr HIV Res 4: 131–139 [DOI] [PMC free article] [PubMed] [Google Scholar]