Genome sequencing of several pneumococcal strains has revealed valuable information about the potential proteins and genetic diversity of this prevalent human pathogen. However, little is known about its transcriptional regulation and its small regulatory noncoding RNAs. In this study, the authors performed deep sequencing of the Streptococcus pneumoniae TIGR4 strain RNome to identify small regulatory RNA candidates expressed in this pathogen. They discovered 1047 potential small RNAs including intragenic, 5′- and/or 3′-overlapping RNAs and 88 small RNAs encoded in intergenic regions. In addition to previously identified intergenic small RNAs, 68 novel candidates were identified, most of which are conserved in both sequence and genomic context in other S. pneumoniae strains.
Keywords: pneumococcus, small noncoding RNAs, transcriptional regulation, pathogenesis, competence
Abstract
Streptococcus pneumoniae is the main etiological agent of community-acquired pneumonia and a major cause of mortality and morbidity among children and the elderly. Genome sequencing of several pneumococcal strains revealed valuable information about the potential proteins and genetic diversity of this prevalent human pathogen. However, little is known about its transcriptional regulation and its small regulatory noncoding RNAs. In this study, we performed deep sequencing of the S. pneumoniae TIGR4 strain RNome to identify small regulatory RNA candidates expressed in this pathogen. We discovered 1047 potential small RNAs including intragenic, 5′- and/or 3′-overlapping RNAs and 88 small RNAs encoded in intergenic regions. With this approach, we recovered many of the previously identified intergenic small RNAs and identified 68 novel candidates, most of which are conserved in both sequence and genomic context in other S. pneumoniae strains. We confirmed the independent expression of 17 intergenic small RNAs and predicted putative mRNA targets for six of them using bioinformatics tools. Preliminary results suggest that one of these six is a key player in the regulation of competence development. This study is the biggest catalog of small noncoding RNAs reported to date in S. pneumoniae and provides a highly complete view of the small RNA network in this pathogen.
INTRODUCTION
Streptococcus pneumoniae, or pneumococcus, is a Gram-positive bacterium able to cause a diverse number of diseases (pneumonia, sinusitis, peritonitis, otitis media) and invasive processes (meningitis, septicemia), mainly in children, the elderly, and the immuno-compromised population. According to a recent report of the World Health Organization, pneumonia is the leading cause of death in children younger than 5 yr and is responsible for 1.6 million casualties each year, and S. pneumoniae and Haemophilus influenzae are the predominant causal agents (Howie and Adegbola 2006; Wardlaw et al. 2006; Scott 2008) (http://www.who.int). Moreover, pneumococci are responsible for 40% of the cases of chronic otitis media that, although less serious, affect approximately a third of all children during their first 3 yr of life. However, despite its importance as a pathogen, some relevant aspects concerning the pathophysiology of pneumococcal diseases remain unresolved.
S. pneumoniae often colonizes the nasopharynx of healthy individuals, behaving as harmless commensal bacteria (Margolis et al. 2010). The transition from commensal to opportunistic pathogen often occurs after a respiratory tract infection and is triggered by host and bacterial factors that are still unknown. Progression into the lungs or middle ear and the infection process exposes S. pneumoniae to numerous environmental changes and stress conditions, and the response to these different environments plays an important role in pneumococcal virulence.
In the last decade, small noncoding RNAs have emerged as crucial bacterial regulators that enable the cell to adjust its physiology to environmental changes (Wassarman et al. 1999; Waters and Storz 2009). These RNA regulators make up a very heterogeneous group of molecules that can be generated either via processing or as primary transcripts, with the latter being the most abundant class of known noncoding RNAs. They can be encoded as part of the mRNA that they regulate (cis-acting RNAs), such as riboswitches or 5′ untranslated regions (UTRs), or as independent transcripts (trans-acting RNAs). Additionally, trans-acting RNAs may be encoded in cis or in trans relative to their target transcript. Trans-acting small RNAs encoded in intergenic regions (sRNAs) have been the focus of most previous studies on small noncoding RNAs in bacteria and are considered the paradigm of these regulators. Most sRNAs exert their action by one of two mechanisms: (1) base-pairing with their mRNA target, modulating its translation and/or stability; or (2) binding to proteins, normally transcriptional regulators, antagonizing their function. In addition, there are sRNAs with other specific functions, such as tmRNA, 6S RNA, 4.5S RNA, or RNase P, which perform important housekeeping functions and are consequently highly conserved in bacteria (Gottesman 2002; Brantl 2009; Repoila and Darfeuille 2009; Waters and Storz 2009).
In recent years, many sRNAs have been discovered in various bacterial species using different approaches (Altuvia 2007; Livny and Waldor 2007; Landt et al. 2008; Livny et al. 2008; Sittka et al. 2008; Sharma and Vogel 2009; Sharma et al. 2010). Although most of these studies have been made in Escherichia coli and other Gram-negative bacteria, sRNAs have also recently been identified in some Gram-positive bacteria, including pathogens such as Bacillus anthracis (Passalacqua et al. 2009), Lysteria monocytogenes (Christiansen et al. 2006; Mandin et al. 2007), and Staphylococcus aureus (Boisset et al. 2007; Geissmann et al. 2009); and model systems such as Streptomyces coelicolor (Swiercz et al. 2008) and Bacillus subtilis (Rasmussen et al. 2009; Irnov et al. 2010). However, despite all of the available data and the dedicated efforts of researchers, the function of the majority of these sRNAs is still unknown. Nevertheless, it is clear that sRNAs coordinate complex networks of stress adaptation and play key roles in pathogenesis, colonization, and persistence, allowing the pathogen to adapt its metabolic needs during infection and to express its virulence genes when required. A variety of sRNAs have been identified in pathogenic bacteria, and some of them have been proven to directly modulate virulence (Pichon and Felden 2005; Chaturongakul et al. 2008; and reviews Romby et al. 2006; Toledo-Arana et al. 2007).
In S. pneumoniae, however, the study of small regulatory RNAs is still in its early stages. The first sRNAs to be described were found by serendipity in a search for genes directly regulated by the CiaRH two-component system in the R6 strain (Halfmann et al. 2007). CiaRH is involved in β-lactam resistance, autolysis, virulence, and competence development and is thought to regulate the expression of 15 genes, five of which encode intergenic sRNAs (named csRNAs1 to 5). CiaRH positively regulates the expression of these sRNAs, but their function, other than csRNA4 and 5, which affect stationary-phase autolysis (Halfmann et al. 2007), is still unknown. Additionally, a computational prediction of sRNAs in the TIGR4 strain (serotype 4) identified 63 putative sRNAs encoded in intergenic regions (IGRs) (Livny et al. 2006), nine of which were further confirmed by Northern blotting in the D39 strain (serotype 2) (Tsui et al. 2010). In the course of this study, 50 sRNAs were identified in S. pneumoniae TIGR4 using genome-tilling arrays, 13 of which were validated by RT-PCR (Kumar et al. 2010). However, only eight out of the 50 overlapped with the 63 computationally predicted sRNAs (Livny et al. 2006), indicating important discrepancies between the different sRNA data sets.
In this study, we describe the identification of small noncoding RNAs in S. pneumoniae serotype 4 clinical isolate TIGR4 using a deep-sequencing approach. We recovered many of the sRNAs previously detected in the S. pneumoniae TIGR4 strain and identified 68 novel candidates. This work significantly increases the number of small RNA regulators identified in S. pneumoniae. In addition, we provide evidence suggesting a role of one intergenic sRNA in modulating competence.
RESULTS AND DISCUSSION
S. pneumoniae small noncoding RNA sequencing
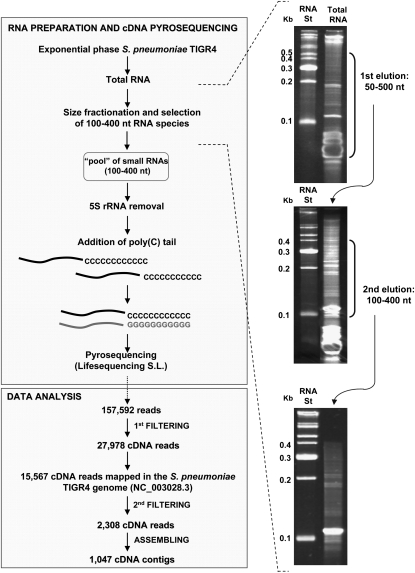
It was previously demonstrated that the gene expression pattern in S. pneumoniae cells grown in vitro in a liquid culture (planktonic cells) is similar to that found in pneumococci isolated from the blood (Oggioni et al. 2006). Therefore, to mimic the in vivo conditions as much as possible, we used in vitro planktonic growing cells to investigate the expression of small noncoding RNAs. For this purpose, capsulated S. pneumoniae TIGR4 cells were grown to mid-logarithmic phase, and total RNA was extracted. To enrich our sample for small-sized RNAs, the extract was size-fractionated, and the RNA fraction between 100 and 400 nt in length was selected (see workflow in Fig. 1). This size fractionation excluded most of the mature tRNA species, which are ∼75 nt in length (and are always <90 nt), but not the 5S rRNA (∼130 nt), which is the other highly abundant transcript in the size-selected fraction. To address this problem, we developed a 5S rRNA depletion step using hybridization with a complementary biotinylated oligonucleotide (see Materials and Methods). Such treatment effectively removed 87% of the 5S rRNA, as measured by quantitative RT-PCR (RT-qPCR) (data not shown) from downstream analysis. Further addition of a poly(C) tail, followed by ligation of specialized adapter molecules to the 5′ and 3′ ends, was used to synthesize a cDNA library by reverse transcription that was then analyzed by 454 pyrosequencing.
FIGURE 1.
Small RNA library generation and data analysis pipeline. Total RNA was processed to enrich the sample for small-sized RNAs by two consecutive PAA gel fractionation and elution steps. The 5S rRNA was depleted by hybridization with a complementary biotinylated oligonucleotide. The small RNA fraction was C-tailed with E. coli poly(A) polymerase and reverse-transcribed using a poly(G) oligonucleotide; the cDNA obtained was subjected to 454 sequencing on the Genome Sequencer FLX System from the Lifesequencing S.L. company. The total number of reads obtained, the reads that successfully mapped to the S. pneumoniae TIGR4 genome, and the total number of cDNA contigs finally obtained (≥50 nt) are indicated. Six percent PAA gels stained with ethidium bromide show RNA from the fractionation procedure. (St) The RNA marker used.
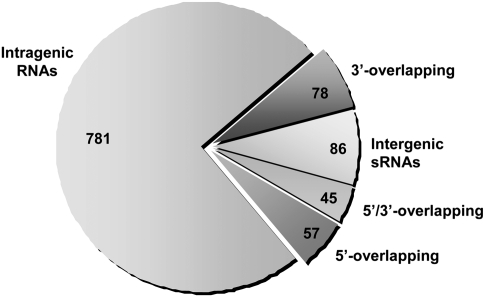
A total of 157,592 cDNA reads (≥30 nt) were obtained, of which 40,224 were shorter than 50 nt or did not meet the minimum quality criteria. The remaining 117,368 cDNA reads were filtered to eliminate those sequences derived from 16S and 23S rRNAs, as well as the 5S rRNA and tRNA that escaped the depletion steps. In the end, 27,978 cDNA sequences were obtained, of which 15,567 were successfully mapped to the S. pneumoniae TIGR4 genome (Tettelin et al. 2001) (∼90% of the total cDNA reads analyzed matched the S. pneumoniae genome). A second filtering step to remove highly repetitive or chimeric sequences, as well as RNAs that partially overlapped with one or more rRNAs or tRNAs, resulted in 2308 cDNA reads that were finally assembled into 1047 different contig sequences for potential RNAs (Supplemental Tables S1 and S2). Despite the RNA size selection performed to enrich our sample in RNAs from 100 to 400 nt in length, 840 reads corresponded to 539 contig sequences between 50 and 99 nt (Supplemental Table S2), while the remaining 1468 reads corresponded to 508 contig sequences ranging between 100 and 440 nt in length (Supplemental Table S1). The coding strand was determined for many, but not all, of the cDNA contig sequences by poly(C)-tail sequence detection. The 1047 cDNA contigs were classified into five types depending on their genomic location (Fig. 2; Supplemental Tables S1, S2). The most abundant class (781 contigs) corresponded to intragenic RNAs that mapped entirely (or with IGRs < 10 nt) within regions annotated as open reading frames (ORFs) in the TIGR4 genome. Many of the contig sequences encompassed both intergenic (≥10 nt) and intragenic (>10 nt) regions, partially overlapping the 5′ or 3′ ends of an ORF. Because the coding strand could not be determined for all of them, they were generally classified as overlapping RNAs—57 contigs corresponded to 5′-overlapping RNAs and 78 contigs corresponded to 3′-overlapping RNAs. Additionally, 45 cDNA contig sequences simultaneously overlapped with two contiguous genes, spanning the 3′ and the 5′ ends of the genes and the IGR between them. These contigs were classified as 5′/3′-overlapping RNAs, in which both overlapping regions are >10 nt. Finally, there was another class of cDNAs (86 contigs) that corresponded to RNAs that entirely mapped within IGRs (or with intragenic regions ≤10 nt). These are candidates for the so-called intergenic sRNAs, 44 of which were shorter than 100 nt (Tables 1, 2).
FIGURE 2.
Distribution of the small RNAs isolated from the S. pneumoniae TIGR4 strain. The sector diagram shows the distribution of the 1047 cDNA contigs obtained (after the removal of rRNAs and tRNAs) with regard to their origin and genetic context. The chart shows the number of RNAs assigned to each of the following categories: intragenic RNAs that overlap entirely with an ORF; 5′- or 3′-overlapping RNAs that overlap with the 5′ or 3′ end of an ORF; 5′/3′-overlapping RNAs that overlap with two adjacent genes simultaneously; and intergenic sRNAs, RNAs entirely encoded in intergenic regions.
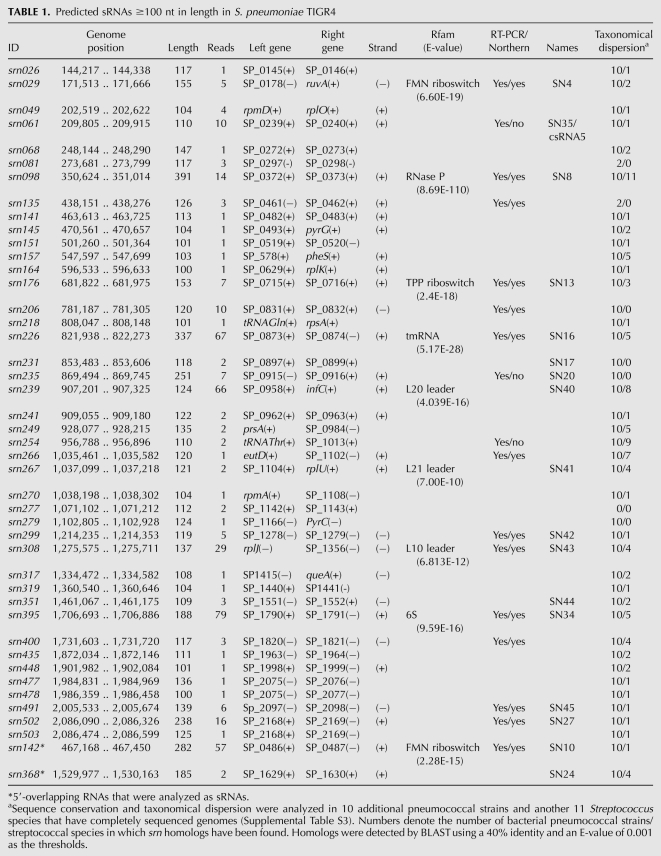
TABLE 1.
Predicted sRNAs ≥100 nt in length in S. pneumoniae TIGR4
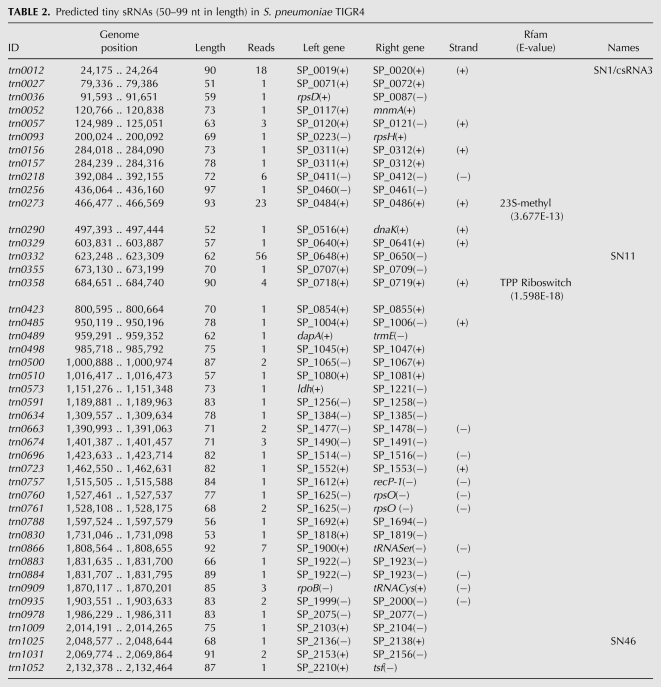
TABLE 2.
Predicted tiny sRNAs (50–99 nt in length) in S. pneumoniae TIGR4
In total, 13% of the cDNA sequences obtained (135 contigs) corresponded to RNAs that partially overlap with one ORF. Transcript leaders, or UTRs, may have specialized functions that affect the stability, the fate or, more often, the transcription/translation of cis-encoded genes (Irnov et al. 2006). In these cases, the formation of a secondary structure is normally required within the UTR. Sixteen out of the 135 overlapping RNAs harbored intergenic regions longer than 100 nt (seven were 5′ overlapping and nine were 3′ overlapping), which might support stable secondary structure elements (Supplemental Fig. S1). Whether any of these overlapping RNAs may act as a cis-regulator is a very interesting question that remains to be answered.
Small RNAs encoded in intergenic regions
Small RNAs encoded in the intergenic regions are the paradigm of regulatory RNAs in bacteria. In this study, we identified a total of 86 candidates for small regulatory RNAs encoded in IGRs (sRNAs). Of those, 18 had been previously identified by genome tilling arrays (Kumar et al. 2010), and 68 are novel candidates (Tables 1, 2). Only six of the 18 sRNAs previously identified were detected by RT-PCR, and none of them was studied further. In addition, we recovered eight of the 63 sRNAs predicted by computational analysis (srn029, srn068, srn239, srn254, srn270, srn308, trn0696, and trn0761) (Livny et al. 2006), of which three (srn029, srn239, and srn308) overlapped with the 18 candidates detected by tilling arrays. We also identified the previously known csRNA3 (trn0012) and csRNA5 (srn061), which are two of the five most studied sRNAs in S. pneumoniae (Halfmann et al. 2007). However, csRNA1, 2, and 4 were not detected. The expression of these five sRNAs is controlled by the CiaRH two-component system, and they are likely to be expressed in similar conditions. The five csRNAs are highly similar in sequence, mainly the csRNAs1 to 4, which differ only at a few bases (Halfmann et al. 2007), and it is likely that the Reference Mapper software used for the analysis was not able to discriminate between them, thereby assigning all reads to the same csRNA3 contig. srn142 and srn368 were previously identified as intergenic sRNAs (SN10 and SN24) (Kumar et al. 2010). These RNAs appeared to be 3′ overlapping in our analysis (the former is an antisense RNA, and the latter is a 3′-UTR RNA) because they are longer than the previous study reported and overlap with the downstream gene. However, because the sequencing procedure we used does not allow us to determine the exact transcription initiation and termination sites, these RNAs were studied as sRNAs (Table 1). Therefore, in this study, we analyzed a total of 88 small RNAs.
To gain some clues about the possible function of these sRNAs, we performed a search with all RNA sequences against the Rfam database (http://rfam.sanger.ac.uk/), a collection of sequence alignments and profiles of noncoding RNA families (Gardner et al. 2009). We found 11 sRNAs with homology with well-characterized RNA families (see Tables 1, 2). Three of them—srn098, srn226, and srn395—were identified as the housekeeping sRNAs RNase P, tmRNA, and 6S RNA, respectively. These RNAs are known to be widespread in bacteria and play essential, specific roles related to stress and stationary-phase adaptation and tRNA maturation. Four other sRNAs (srn029, srn142, srn176, and trn0358) were predicted to be FMN or TPP riboswitches, common 5′ UTRs that are able to bind a target molecule (e.g., a metabolite), change its secondary structure, and affect the transcription or translation of downstream genes (Nudler and Mironov 2004). The srn239, srn267, and srn308 appeared to be ribosomal protein leader autoregulatory structures (L20, L21, and L10 leader regions), commonly found in the 5′ UTRs of ribosomal protein genes, and trn0273 was shown to be a conserved RNA motif located upstream of rRNA methyltransferase-encoding genes (Weinberg et al. 2007). With this analysis, we could assign putative functions to 11 of the pneumococcal sRNAs identified, eight of which were predicted to be cis-regulators. The remaining 77 sRNAs, however, corresponded to unknown RNAs with no homology with any family in the Rfam database and appear to represent a novel set of sRNAs in pneumococci.
Comparative genomics and computational analysis of sRNAs
sRNAs appear to be quite conserved among Gram-negative bacteria, especially in the γ-proteobacterial class, probably because they have common mRNA targets or specially optimized secondary structures (Irnov et al. 2010). In stark contrast, such conservation is much more vague in Gram-positive bacteria, where it is mostly confined to the genus or even to the species level (Geissmann et al. 2009; Rasmussen et al. 2009; Marx et al. 2010). To explore the conservation of the sRNA candidates identified in this study, we used BLAST to analyze their taxonomical distribution in various pneumococcal strains and other related and unrelated bacterial species (Supplemental Table S3). To ensure confidence in the analysis, we selected only those candidates ≥100 nt in length. The results obtained revealed that seven sRNAs were unique to S. pneumoniae, because they could not be identified in other streptococcal species (Table 1). The remaining 37 appeared to also be present in other streptococci, although most of them (17 sRNAs) were only found in Streptococcus mitis, the closest species to pneumococcus. Interestingly, only srn029 was identified in a nonstreptococcal bacterium, the Gram-positive B. subtilis, but not in any Gram-negative species. Therefore, it appears that most of the sRNAs identified are widespread in pneumococcal strains but that they are quite species- and genus-specific.
In total, 41 sRNAs showed 90%–100% sequence identity in all S. pneumoniae strains and could be considered universal pneumococcal sRNAs (Supplemental Fig. S2). However, srn081 and srn135 were present only in two strains, and srn277 appeared to be exclusive to TIGR4. Additionally, 34 out of the 41 pneumococcal sRNAs shared one or, in most cases, both flanking genes in all strains (Supplemental Table S4). The high degree of sequence and genomic context conservation observed is suggestive of a high evolutionary pressure to maintain these sRNAs in the pneumococcal lineage.
Expression and target prediction of the intergenic sRNAs
The first step to demonstrate that these sRNAs are, indeed, functionally required for gene regulation is to confirm their independent expression. We concentrated on those RNAs that were longer than 99 nt and chose several of the most highly expressed as well as some apparently interesting sRNA candidates for experimental validation by both RT-PCR and Northern blot analysis. All 17 sRNAs selected for validation (Table 1) were detected by RT-PCR, and 14 of them were also successfully detected by Northern blotting (Figs. 3–6, see below). In every case, the Northern blot showed a discrete band with a size similar to the predicted sRNA, and in some cases, the blot also showed additional bands.
FIGURE 3.
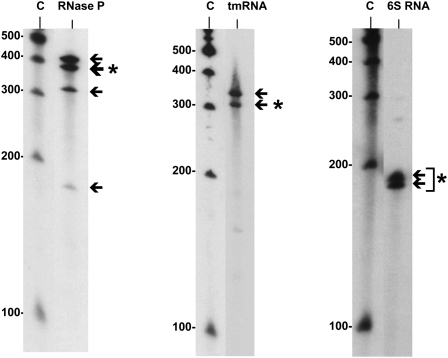
Northern blot analysis of three highly conserved housekeeping sRNAs that were isolated from the S. pneumoniae TIGR4 strain: RNase P, tmRNA, and 6S RNA. (Arrows) The specific bands detected by Northern blotting; (asterisks) the band of sRNA detected, which agrees with the size predicted by sequencing.
FIGURE 6.
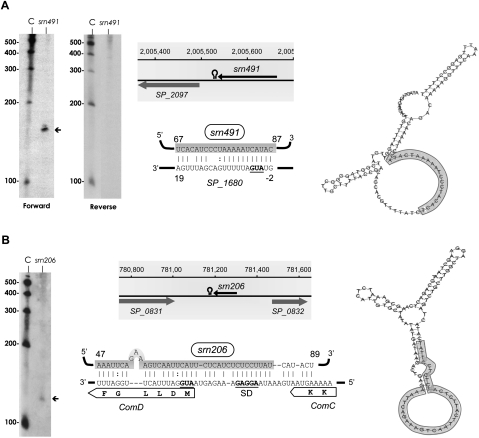
Detection, genomic context, predicted secondary structure, and target prediction of sRNAs isolated from the S. pneumoniae TIGR4 strain. (A) srn491 and (B) srn206. srn206 was detected with a probe labeled internally with biotin, whereas srn491 was hybridized to either forward or reverse biotinylated oligonucleotides, as indicated. The arrows indicate the size of the sRNA detected by Northern blotting, which agrees with the size predicted by sequencing. (C) RNA molecular weight marker. The shadowed rectangles show the coordinates in the genome and the flanking genes for each sRNA. The figure also shows candidate mRNA targets for srn491 and srn206 (P-values of 3.63 × 10−3 and 4.88 × 10−5, respectively) predicted by TargetRNA software. The secondary structures were predicted using RNAfold server (ΔG values of −29.8 and −26.9 kcal/mol for srn491 and srn206, respectively). The sRNA region predicted to associate with the target mRNA is highlighted in gray. The start codons and SD are in bold, underlined letters.
sRNAs belonging to known families
The three housekeeping sRNAs identified—the RNase P (srn098), the tmRNA (srn226), and the 6S RNA (srn395)—are known to undergo processing to produce the mature form in other bacteria (Fig. 3; Sakamoto et al. 1983; Lin-Chao et al. 1999; Kim et al. 2005; Trotochaud and Wassarman 2005; Barrick et al. 2008). RNase P is the catalytic RNA component of the ribonuclease P, a ubiquitous, essential ribonucleoprotein complex found in all three domains of life. It is one of the only two examples of RNA-based enzymes whose role is to catalyze the 5′-end maturation of the pre-tRNA as well as the cleavage of other naturally occurring substrates, such as the 4.5S RNA, tmRNA, operon mRNAs, or some transient structures in riboswitches (for review, see Kazantsev and Pace 2006; Hartmann et al. 2009; Esakova and Krasilnikov 2010). The tmRNA is the putative transfer messenger RNA responsible for ribosome rescue and aberrant peptide tagging in the trans-translation process (Gillet and Felden 2001). The 6S RNA binds to σ70 RNA polymerase holoenzyme to globally regulate gene expression in adaptation to stationary phase. The Northern blot for these three sRNAs showed bands that correspond to the predicted size of the mature transcripts as well as bands corresponding to preprocessed forms and/or stable degradation products (Fig. 3). Whether all of these forms or only one of them is functional in pneumococcus remains to be elucidated.
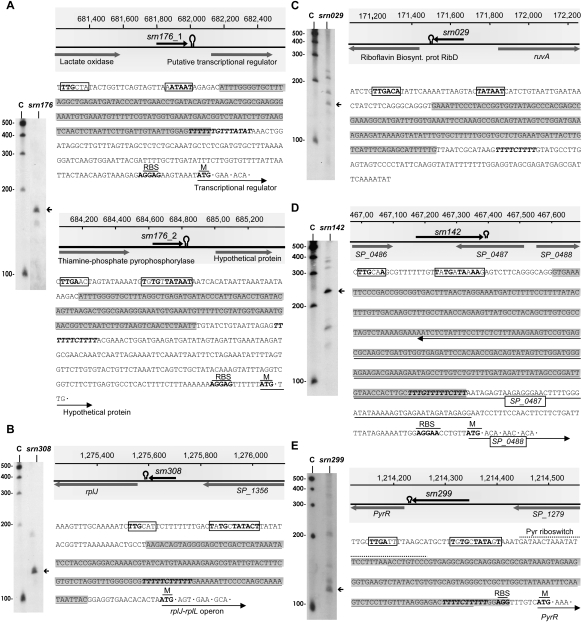
srn176, srn308, and srn029 appear to be cis-acting regulatory elements that are able to regulate the expression of downstream genes upon binding to metabolites or proteins. The mechanism of regulation typically involves a conformational change in the secondary structure of the RNA that serves to regulate the expression of a downstream gene through a variety of mechanisms. Often, cis-regulatory elements include an intrinsic terminator or an attenuator structure in their sequence, which is usually formed upon binding of the target molecule, thereby terminating transcription. Expression regulation by premature transcription termination results in the synthesis of the corresponding small transcript. In agreement with this, Northern blot of srn176 showed a specific band that was slightly longer than the predicted size (Fig. 4A). Interestingly, srn176 is encoded by two relatively close regions of the genome, named srn176_1 (also identified by Kumar et al. 2010) and srn176_2, which shares 90% sequence identity (Fig. 4A). Both srn176_1 and srn176_2 appear by Rfam prediction to be TPP cis-acting regulatory elements that are able to bind thiamine pyrophosphate (vitamin B1), an essential cofactor for several important enzymes in carbohydrate metabolism, and to regulate the expression of the downstream gene (Winkler et al. 2002). Sequence inspection of the srn176 coding regions revealed a putative promoter at −10 (differing only in one base from the consensus sequence) and −35 for srn176_1 and a consensus putative extended promoter for srn176_2 (Fig. 4A). Additionally, in both srn176_1 and srn176_2, the presence of a stretch of T's at the 3′ end suggests the formation of a Rho-independent transcriptional terminator structure, which would terminate transcription, causing the accumulation of the srn176 transcript.
FIGURE 4.
Detection, genomic context, and sequence of sRNAs isolated from the S. pneumoniae TIGR4 strain. (A) srn176, (B) srn308, (C) srn029, (D) srn142, and (E) srn299. The arrows indicate the size of the sRNA detected by Northern blotting, which agrees with the size predicted by sequencing. (C) RNA molecular weight marker. (Shadowed rectangles) The coordinates in the genome and the flanking genes for each sRNA. (Open circles) A potential intrinsic terminator. The nucleotide sequence of each sRNA (shadowed) is also shown, where the putative promoters at −10, or −10 extended, and −35 boxes are labeled with an empty rectangle, and the consensus nucleotides are depicted in bold letters. Potential terminators are shown in italic, bold letters, and putative ribosomal binding sites (RBS) and start codons of downstream genes (M) are in bold, underlined letters.
Similarly, srn308 was identified as an L10 leader region, a family of autoregulatory structures found in the 5′ UTRs of the operon mRNAs encoding ribosomal proteins L10 and L12 (rplJ-rplL) (Zengel and Lindahl 1994). These proteins form a complex that is able to bind specifically to the leader sequence of the rplJ-rplL mRNA, disturbing a specific secondary structure and inhibiting translation of the encoded ribosomal proteins (Johnsen et al. 1982). Multiple sequence alignments of several bacterial L10 leader regions that are available in Rfam revealed a stretch of ∼135 nt with four highly conserved sequence motifs that are all present in srn308, including a Rho-independent transcriptional terminator. Accordingly, Northern blot analysis of srn308 revealed the accumulation of a small ∼130-nt transcript in the conditions tested (Fig. 4B), which may be the result of transcription termination at the putative Rho-independent terminator.
srn029 appears to be an FMN riboswitch (also known as a RFN), a highly conserved RNA element frequently found in the 5′ UTRs of prokaryotic mRNAs that encode flavin mononucleotide (FMN) biosynthesis and transport proteins (Vitreschak et al. 2002). In fact, srn029 is located 75 nt upstream of a riboflavin biosynthesis protein gene. This element is a metabolite-dependent riboswitch that binds FMN in the absence of proteins and controls gene expression by causing premature transcription termination or precluding access to the ribosome-binding site (Mironov et al. 2002). Northern blotting revealed several specific bands with similar intensity (Fig. 4C). The shortest band detected likely corresponds to the 155-nt sRNA identified by pyrosequencing, whereas the other bands may correspond to longer species, which may be derived from alternative terminator structures or may be srn029 precursors. Whether all of these forms or only one of them are functional remains unknown.
As mentioned above, srn142 appears, from our analysis, to be an antisense RNA whose 3′ end overlaps with the 3′ end of the adjacent gene (SP_0487) in a tail-to-tail manner (Fig. 4D). However, srn142 corresponds to SN10 RNA, which was previously identified as a shorter intergenic sRNA that lacked the overlapping region (Kumar et al. 2010). Often, the criteria used for ORF predictions are not sufficiently restricted, and several misannotations can be found in certain genomes. Indeed, it is suspected that 30% of genomes are overannotated (Frishman 2007), suggesting that a substantial portion of the ORFs shorter than 300 nt may not actually encode polypeptides. This might be the case with SP_0487, which is only 219 nt in length and theoretically encodes a 73-amino-acid peptide. SP_0487 has an uncommon start (UUG) and stop (UAG) codon and no putative Shine-Dalgarno sequence. Moreover, SP_0487 exhibits a fivefold overabundance of rare pneumococcal codons (≤0.5% occurrence, ≤12% of all isocodons) and has not been identified as an ORF in pneumococcal genomes other than S. pneumoniae TIGR4. Additionally, a sequence search against the Rfam database confidently indicated that srn142 is a member of the FMN family of cis-regulators. Northern blots for srn142 showed a prominent band slightly longer than the predicted size, and sequence inspection revealed a putative promoter and a Rho-independent terminator localized inside the putative SP_0487 gene (Fig. 4D). Therefore, all data strongly suggest that SP_0487 is not a polypeptide-encoding ORF and that srn142 is, indeed, an intergenic sRNA, which acts as a cis-regulator modulating the expression of the SP_0488 downstream gene through premature transcription termination.
srn299 is encoded in the IGR between the bifunctional pyrimidine regulatory protein PyrR uracil phosphoribosyltransferase (SP_1278) and the endonuclease III (SP_1279)–encoding genes. According to our analysis, srn299 is 119 nt long and has no significant homology within the Rfam database. However, this sRNA overlaps with SN42, which was previously identified as a member of the PyrR family (Kumar et al. 2010), an RNA element found upstream of genes involved in pyrimidine biosynthesis and transport. srn299 is shorter than SN42 and lacks the 20-nt sequence at the 5′ end that contains the PyrR motif (Fig. 4E), which explains the absence of Rfam homology. In B. subtilis, binding of the PyrR protein to three attenuation sites at the 5′ end of different pyr genes allows a terminator hairpin to form, which represses the downstream pyr genes of the pyr operon (Bonner et al. 2001). Our Northern blot showed a prominent band corresponding to the predicted size of srn299, although other larger and less intense bands were also detected (Fig. 4E). Therefore, several srn299 RNA species may coexist inside the cell at certain conditions, and all of them may be functional.
The evidence suggests that srn176, srn308, srn142, srn029, and srn299, although encoded in IGRs, are indeed cis-regulatory RNAs that probably modulate the expression of downstream genes by premature transcription termination. Such a mechanism may lead to the accumulation of small transcripts inside the cell under certain conditions, which would then have the potential to function in trans on other mRNAs. Very recently, a riboswitch element was discovered in L. monocytogenes that, in addition to cis-regulating the downstream gene, has the ability to act in trans to modulate the expression of a distantly encoded mRNA (Loh et al. 2009; Xayarath and Freitag 2009). Taking into account the intracellular accumulation of srn176, srn308, and srn142, as detected by Northern blot analysis, it is tempting to speculate about the possibility of these RNAs acting either in cis or in trans to regulate the expression of different mRNAs under certain conditions, as occurs in L. monocytogenes. However, further studies will be necessary to investigate this possibility.
sRNAs of unknown families
Six other intergenic sRNAs were analyzed by Northern blot: srn135, srn206, srn266, srn400, srn491, and srn502. These sRNAs did not show homology with the Rfam database, and four of them were novel candidates (srn135, srn206, srn266, and srn400). srn135, srn206, and srn502 showed a single and specific band by Northern blotting that corresponded to their predicted sizes (Figs. 5A,C, 6B), while srn266 and srn400 exhibited several bands (Fig. 5B,D). With regard to srn491, its coding strand could not be identified because a poly(C) tail was not detected in any of the cDNA reads assembled. Therefore, to assess its orientation, probes complementary to both strands were used in the Northern blot analysis. A band corresponding to the predicted size was detected only with the forward oligonucleotide, indicating that srn491 is encoded on the negative strand, as are the flanking genes (Fig. 6A).
FIGURE 5.
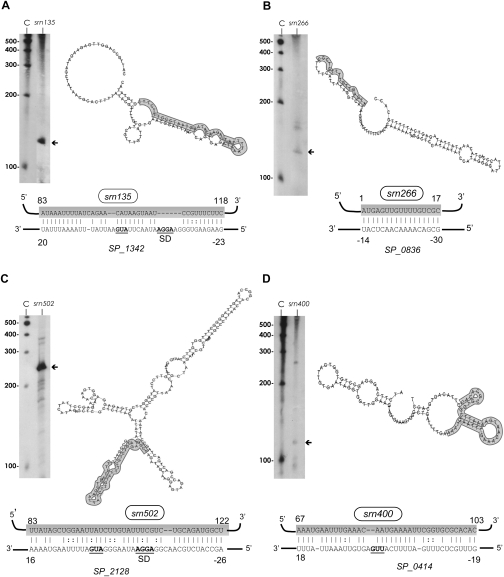
Detection, predicted secondary structure, and target prediction of sRNAs isolated from the S. pneumoniae TIGR4 strain. (A) srn135, (B) srn266, (C) srn502, and (D) srn400. The arrows indicate the size of the sRNA detected by Northern blotting, which agrees with the size predicted by sequencing. (C) RNA molecular weight marker. The putative mRNA targets for srn135, srn266, srn502, and srn400 (P-values of 9.08 × 10−4, 1.77 × 10−4, 1.39 × 10−4, and 1.15 × 10−3, respectively) predicted by TargetRNA software are shown. The secondary structures were predicted using RNAfold server (ΔG values of −14.6, −28.2, −79.5, and −23.1 kcal/mol for srn135, srn266, srn502, and srn400, respectively). The sRNA region predicted to associate with the target mRNAs is highlighted in gray. The start codons and SD are in bold, underlined letters.
To understand the possible roles of these six sRNAs, they were subjected to target prediction by TargetRNA analysis. This program is designed to identify putative mRNA targets by searching for interrupted base-pair interactions within intergenic regions (Tjaden et al. 2006). In addition, their secondary structure was also analyzed using RNAfold prediction (http://rna.tbi.univie.ac.at/cgi-bin/RNAfold.cgi). TargetRNA analysis of srn135, srn266, srn400, and srn502 resulted in at least one confident putative mRNA target for each of them (P-values between 1.15E-3 and 1.39E-4), and some of these targets appear to be interesting. For instance, srn135 appears to base-pair with the initiating translation region of the permease protein of the drug efflux ABC transporter (SP_1342), which has been theorized to have a role in antibiotic resistance, while the best targets for srn266 are a set of transposase-encoding genes (IS1167). However, the four sRNAs were predicted to be highly structured (ΔG between −23.10 and −79.47 kcal/mol), and in all cases, the region involved in base-pairing with their mRNA targets was included in a highly stable stem structure (Fig. 5A–D), thus raising doubt on the target predictions. Therefore, additional experiments will be required to assess the function of these sRNAs.
In the case of srn491, the secondary structure prediction and TargetRNA analysis predicted a base-pairing between the sequence in the bubble of srn491, which was therefore available for interaction, and the translation start site of the SP_1680 mRNA in the TIGR4 genome (Fig. 6A). SP_1680 encodes a hypothetical protein that, upon BLAST analysis, appeared to belong to the DUF386 superfamily, a group of conserved, hypothetical proteins of unknown function. Therefore, association between srn491 and its putative mRNA target through an antisense mechanism would negatively affect SP_1680 translation.
Interestingly, TargetRNA analysis of srn206 rendered several putative mRNA targets with very low P-values, although the strongest candidate (P-value=4.88E-5) corresponded to comD mRNA. ComD protein is the histidine kinase that senses the competence-stimulating peptide (CSP), which is responsible for the induction of competence for genetic transformation in S. pneumoniae. Pneumococcus is a naturally competent bacterium that is able to incorporate exogenous DNA into the genome through an uptake and recombination process controlled by the competence machinery. The extracellular concentration of CSP, an exported peptide pheromone derived from the precursor protein ComC, is sensed by the ComD receptor. Binding of CSP to ComD results in phosphorylation of ComD, which transfers the phosphate group to the cognate response regulator ComE, which then activates transcription of early competence genes (Claverys et al. 2009; Johnsborg and Havarstein 2009). ComC, ComD, and ComE are cotranscribed in a long mRNA transcript, and its transcription is activated by P-ComE. srn206 is predicted to associate with this mRNA transcript in the translation start region of ComD, sequestering its start codon and ribosome-binding site (Fig. 6B). Moreover, secondary structure prediction of srn206 by RNAfold revealed that, although highly structured, the region predicted to base-pair with ComD mRNA is primarily located in a bubble and is therefore free to interact with its target (Fig. 6B). Inhibition of translation of ComD by srn206 would affect the activation of the competence cascade, preventing transformation of pneumococcal cells.
srn206 and competence development
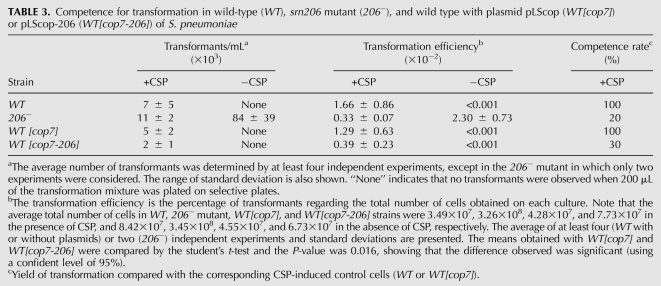
To test whether srn206 was, in fact, involved in modulating competence, a mutant strain lacking the srn206 gene was constructed through insertion/deletion of a Cm resistance gene. The transformation efficiency of wild-type (wt) and mutant strains was assayed using streptomycin-resistant (Smr) chromosomal DNA. Six independent experiments were performed with both strains. The results showed that, as expected, the wt strain became competent only in response to exogenous CSP, and no transformants were obtained without the inducer (Table 3). In contrast, in two of the six experiments, the TIGR4srn206- mutant was able to accept exogenous DNA in the absence of CSP to levels that were sevenfold higher than those reached in the presence of the inducer (Table 3). However, in the following four experiments, the srn206- mutant behaved like the wt, and no transformants were obtained without CSP. The spontaneous competence phenotype displayed by the mutant strain in some experiments would be consistent with the postulated role of srn206 in ComD translational repression. The uptake of DNA in the absence of inducer never occurs in the wild type, and, therefore, the capacity of the mutant to become spontaneously competent in some experiments is suggestive of a role of srn206 in competence modulation. Competence induction is highly stressful for the cell and needs to be tightly controlled. Therefore, it is likely that the mutant strain has suffered a secondary mutation that compensates the detrimental effect of having the competence system constantly on.
TABLE 3.
Competence for transformation in wild-type (WT), srn206 mutant (206−), and wild type with plasmid pLScop (WT[cop7]) or pLScop-206 (WT[cop7-206]) of S. pneumoniae
To overcome this problem, we cloned a chromosomal region containing the srn206 gene in the pLS1cop7 plasmid (named as pLScop herein), a high-copy-number mutant derivative of the promiscuous streptococcal plasmid pLS1 (del Solar et al. 1987). The constructed plasmid, pLScop-206, which allows the expression of srn206 from its natural promoter, was introduced in wild-type TIGR4 cells through transformation. Quantification by RT-qPCR revealed that the expression of srn206 in TIGR4[pLScop-206] cells was 38-fold higher than that obtained in the control cells (TIGR4[pLScop]). Such overexpression caused approximately a threefold reduction in the transformation efficiency of pLScop-206-containing cells in a competence assay (Table 3), and this reduction was significant, as shown by the Student's t-test (P-value=0.016). These results are in agreement with our predictions and suggest that srn206 is involved in competence modulation. Whether the action of snr206 is mediated through ComD translational repression or not needs to be investigated, and further experiments are required to address the precise function of srn206. However, although preliminary, our data point to this sRNA as one of the factors involved in the maintenance of the competence on–off switch in pneumococcus.
CONCLUSIONS
In this study, we describe the identification of a repertoire of small regulatory RNAs in the human pathogen S. pneumoniae TIGR4 that can be generally classified as 5′- or 3′-overlapping RNAs and sRNAs from IGRs. The 88 sRNAs identified include several previously described sRNAs and 68 novel candidates. Combining all previous and present data, a total of 118 sRNA candidates have been identified to date in the S. pneumoniae TIGR4 strain, and 50 others have been computationally predicted to be encoded in IGRs. Therefore, it appears that, although smaller, the pneumococcal genome encodes a similar number of sRNAs as B. subtilis (Irnov et al. 2010), the classical model microorganism for Gram-positive bacteria. These numbers are also close to the 200–300 sRNAs that have been estimated to be present in the average bacterial genome (Hershberg et al. 2003).
In this study, we have combined high-throughput pyrosequencing and low-throughput analyses for complete detection of the sRNA pool of pneumococcus. A set of mRNA targets was predicted for some of the sRNAs, and interesting findings were obtained. For instance, the sRNA srn206 is suggested to modulate the expression of the histidine kinase sensor that allows entrance into a competent state in pneumococci. Competence is a pivotal mechanism in S. pneumoniae and is involved in virulence and antibiotic resistance (Lau et al. 2001; Oggioni et al. 2004; Prudhomme et al. 2006; Kowalko and Sebert 2008), and its modulation would certainly affect many important aspects in pneumococcal pathogenesis. The core portion of the pneumococcal competence regulatory cascade is well understood. However, the signals that trigger competence are still unknown, and deciphering this question may help us better understand the role of competence. Our results suggest that srn206 may be one of the players involved in the maintenance of the competence on–off switch in pneumococcus, and its relationship with the ComD protein needs to be explored.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions
All studies were performed in the TIGR4 strain of S. pneumoniae (serotype 4). Cloning experiments using the pLS1cop7 plasmid kindly provided by Dr. G. del Solar (del Solar et al. 1987), named as pLScop herein, were carried out in the R6 strain of S. pneumoniae. For cloning in the pGEM-T Easy vector (from Promega), the E. coli DH5α [F−, ø80dlacZΔM15, Δ(lacZYA-argF)U169, deoR, recA1, endA1, hsdR17(rK−, mK+), phoA, supE44, λ−, thi-1, gyrA96, relA1] was used. For 454 pyrosequencing, RT-PCR, and Northern blot analyses, pneumococcal cells were grown as a static culture in Todd-Hewitt medium supplemented with 0.5% yeast extract (THY) to an optical density at 600 nm (OD600) of 0.4–0.5. Then the culture was diluted 12-fold in the same medium (preheated), and growth continued to OD600=0.2. Cells were cooled on ice, centrifuged, and used for RNA extraction.
RNA isolation
For total RNA extraction, pneumococcal cultures were centrifuged at 6000g for 10 min, and the cellular pellet was resuspended in 0.15 culture volumes of Lysis Solution (50 mM Tris at pH 8.0, 10 mM EDTA, and 0.1% sodium deoxycholate) and lysed by incubation for 5 min at 37°C. Total RNA was isolated using TRIzol Reagent (Invitrogen), following the instructions from the manufacturer. The integrity of the rRNAs was checked by gel electrophoresis through a 0.8% agarose gel, and the RNA concentration was determined using a NanoDrop ND-1000 Spectrophotometer. Total RNA was treated with DNase I (Roche) to remove residual genomic DNA, precipitated, and stored at −80°C in nuclease-free water.
For pyrosequencing, the total RNA was further processed to enrich the sample for small-sized RNAs and to deplete the 5S rRNA as follows: Total RNA isolated from 24 mL of culture was subjected to two consecutives steps of size fractionation and elution on denaturing 8% polyacrylamide (PAA)/7 M urea gels in 1× TBE buffer. In the first step, RNAs between 50 and 500 nt in length were selected, eluted from the gel, and ethanol-precipitated as described (Conrad et al. 1998). The small RNA pool recovered (50–500 nt) was again gel-fractionated on a PAA/7 M urea gel, and the RNA fraction between 100 and 400 nt in length was extracted following the same procedure. To deplete the 5S rRNA from the small RNA pool (the predominant RNA species within the size-range selected), a complementary, biotinylated oligonucleotide [5′-(T15)-GGCTTAACTTCTGTGTTCGGCATGGGTA-3′] and streptavidin magnetic beads (Roche Applied Science) were used following a procedure previously described (Pang et al. 2004). Briefly, 125 μL of streptavidin magnetic beads was washed three times with 0.5× SSC (75 mM NaCl and 7.5 mM Na citrate at pH 7), mixed with 720 pmol of 5S rRNA biotin-labeled oligonucleotide, and incubated at room temperature for 30 min. RNA was resuspended in 0.5× SSC, incubated for 5 min at 70°C for denaturing, and chilled on ice. Then the RNA sample was transferred to previously prepared biotin-labeled oligonucleotide magnetic beads and incubated for 30 min at 68°C to allow hybridization to occur. A magnet was applied to the bottom of the tube, and the supernatant was carefully transferred into a clean, RNase-free microcentrifuge tube, precipitated and resuspended in 60 μL of water. This procedure was repeated twice, and the remaining 5S rRNA in the sample was measured by RT-qPCR using the specific primers 5S-F: 5′-GTGACGATAGCCTAGGAGATACACCTG-3′ and 5S-R: 5′-CCCTATCTCACAGGGGGCAAC-3′.
cDNA synthesis and pyrosequencing analysis
To synthesize the cDNA for pyrosequencing, the small RNA pool remaining after 5S rRNA depletion was first C-tailed using 3.5 U of E. coli poly(A) polymerase (Ambion) and 1 mM CTP for 60 min at 37°C in a 105-μL volume. Unincorporated nucleotides were removed from the mixture using NucAway Spin columns (Ambion). For first-strand cDNA synthesis, the SuperScript Plasmid System with Gateway Technology from Invitrogen was used. For this purpose, 9.6 μg of C-tailed RNA was split into three tubes, and each 3.2-μg aliquot of RNA was hybridized with 1 μg of a poly(G) oligonucleotide (5′-TCGCGAGCGGCCGCG15-3′) for 10 min at 70°C in 9.5 μL. Then 200 U/μg SuperScript II reverse transcriptase (Invitrogen), 0.2 mM dNTPs, and 0.25 mM DTT were added to each tube and incubated for 1 h at 45°C for reverse transcription. For second-strand synthesis, 25 mM Tris-HCl (pH 7.5); 100 mM KCl; 5 mM MgCl2; 10 mM (NH4)2SO4; 0.15 mM β-NAD+; 250 μM each of dATP, dCTP, dGTP, and dTTP; 1.2 mM DTT; 65 U/mL DNA ligase; 250 U/mL DNA polymerase I; and 13 U/mL RNase H were added to the mixtures to a final volume of 150 μL. After gentle mixing, the reaction was incubated for 2 h at 16°C. Then 2 μL (10 U) of T4 DNA polymerase was added, and incubation continued for 5 min at 16°C. After that, cDNA was purified using a QIAquick PCR Purification Kit from QIAGEN. Finally, 3.7 μg of cDNA was obtained and subjected to pyrosequencing using 454 sequencing on the Genome Sequencer FLX System from the Lifesequencing S.L. company. The resulting cDNA sequences were mapped onto the S. pneumoniae genome (Tettelin et al. 2001) using Reference Mapper software from Roche.
Real-time PCR
PCR primers were designed (listed in Supplemental Table S5) using Oligo Perfect Designer software (Invitrogen). cDNA synthesis was performed with the Transcriptor First Strand cDNA Synthesis Kit from Roche Applied Science following the instructions from manufacturers. Briefly, 1 or 2 μg of total RNA and 60 μM of random hexamers were mixed in a 13-μL reaction volume, incubated for 10 min at 65°C, and immediately annealed for 10 min at 4°C. Reverse transcription was carried out in a 20-μL reaction volume with 10 U of Transcriptor Reverse Transcriptase (+RT), 10 mM each dNTP, and 20 U of Protector RNase Inhibitor in the reaction buffer. The mixture was incubated for 10 min at 25°C, followed by 30 min of incubation at 55°C. Finally, the reverse transcriptase was inactivated by incubation for 5 min at 85°C. A mock cDNA synthesis reaction was performed, with the reverse transcriptase replaced by water (−RT). For each sRNA, at least two different RT reactions were set up.
For validating the expression of sRNAs, PCR reactions were performed with the LightCycler Carousel-Based System for Real-Time PCR (Roche Applied Science) using primers specific for each sRNA. Reactions were carried out using 1 μL of cDNA (+RT and −RT), 0.25 μM each primer, and 11 μL of SYBRPremix Ex Taq (Takara) in a 22-μL reaction volume. Ten-microliter duplicates were loaded on the LightCycler capillaries. A three-step PCR protocol (5 min at 95°C, then 45 cycles of 10 sec at 95°C, 20 sec at 60°C, and 15 sec at 72°C) was used. The temperature transition rates were programmed at 20°C/sec. After PCR completion, a melting curve was recorded by cooling to 65°C at 20°C/sec, holding for 15 sec at 65°C, and then heating slowly at 0.1°C/sec until 95°C was reached. Samples were then cooled for 30 sec to 40°C.
For quantification of the srn206 expression, reactions were performed as described above using the LightCycler 480 System for Real-Time PCR (Roche Applied Science). 16S rRNA was used as the internal reference gene. The gene-specific primer pairs used were srn206q and 16Sq for the srn206 and the 16S rRNA, respectively (Supplemental Table S5). A calibration curve using the PCR product obtained with srn206q primers was used for quantification. To estimate variation in the technique, two technical and two biological replicates were carried out.
Northern blot
Total RNA samples (10 or 20 μg) were heated for 10 min at 80°C in 1× formamide containing Gel Loading Buffer II (Ambion) and fractionated in a 6% PAA/7 M urea gel. RNAs were transferred to Hybond-N+ membrane (Amersham) by electroblotting, using the Trans-blot cell from Bio-Rad in 1× TAE buffer according to the manufacturer's instructions. The blots were UV-cross-linked in a CL-1000 UV Crosslinker (UVP, Inc.) by two cycles of irradiation at 254 nm and 1200 J and then prehybridized for at least 10 min with PerfectHyb Plus buffer (Sigma-Aldrich). Hybridization was performed overnight with either 20 ng/mL internally labeled biotinylated DNA (0.1 mL/cm of membrane) or 50 pmol of 5′-biotin-labeled oligonucleotides as probes (listed in Supplemental Table S5) at the appropriate hybridization temperature for each probe in the same hybridization buffer. Detection was performed by autoradiography using a Phototope-Star Detection kit from New England Biolabs. Internally labeled biotinylated DNA probes were synthesized using the NEBlot Phototope Kit from New England Biolabs through a random primer reaction with biotinylated dATP. The template for the labeling reaction was generated by standard PCR using 10 pmol of the specific primers for each sRNA (listed in Supplemental Table S5), 200 μM dNTPs, and FastStart Taq DNA polymerase from Roche. All PCRs were purified using the QIAquick PCR Purification Kit from QIAGEN. Labeling was then performed with 100 ng of each specific PCR for each sRNA following the instructions from the manufacturer and further purified with NucAway Spin columns from Ambion, Inc. to remove unincorporated nucleotides.
Transformation of pneumococcal cells
Cultures of S. pneumoniae TIGR4 grown in a casein hydrolase–based medium (AGCH) with 0.2% sucrose (Suc) and 0.001% CaCl2 containing 1.5 × 107 CFU/mL were treated with 75 ng/mL CSP-2 for 20 min at 30°C. After that, DNA was added, and the culture was incubated for 40 min at 30°C, then transferred to a 37°C water bath and incubated for 90 min to allow expression of the antibiotic resistance marker. When the antibiotic was Cm, after 70 min at 37°C, 0.5 μg/mL Cm was added to the culture to induce the expression of the antibiotic-resistance gene, and the cells were incubated for 20 min more. After that, transformants were plated on media plates (AGCH medium with 1% agar plus 0.3% Suc and 0.2% yeast extract) containing the corresponding antibiotic. Transformants were grown at 37°C in a 5% CO2 atmosphere.
Construction of the srn206 mutant strain of S. pneumoniae
The srn206-deficient mutant was created through allelic replacement mutagenesis (Song et al. 2005). A fragment upstream of the srn206 gene was amplified with primers S206-F (5′-TCCTCCGAAAGCTGTCGAA-3′) and S206-CAT-R (5′-TTCTTTGTTCCACGCTTACTACGAGATTTCGCTTGAGTCTGC-3′), and a fragment downstream from srn206 was amplified using primers S206-CAT-F (5′-TACAGCTCCAGATCCATATCAGTCTGCTTGGCTAGGAAG-3′) and S206-R (5′-CTTGCTGAAGGTGCCTTTC-3′) from the TIGR4 genome. The chloramphenicol-resistance cassette (Cmr) was amplified from the pJS3 plasmid (Ballester et al. 1986) using CAT-F (5′-GTAGTAAGCGTGGAACAAAGAA-3′) and CAT-R (5′-GATATGGATCTGGAGCTGTA-3′). The three PCR reactions were mixed together and amplified using the S206-F and S206-R oligonucleotides. Finally, a 3266-bp PCR fragment in which a 69-nt internal region of the srn206 encoding gene was replaced by a Cmr marker was obtained. This fragment was used to transform TIGR4 competent cells of S. pneumoniae. One Cmr transformant was selected, and the insertion/deletion mutation was confirmed by DNA sequencing.
Competence assay
Cultures of pneumococcal cells were started by inoculating AGCH plus 0.3% Suc and 0.2% yeast extract (YE) with 1/1000 volume of frozen stock cells. Growth was carried out at 37°C in a static water bath. At an OD650 of 0.175, the cells were diluted in AGCH plus 0.3% Suc and 0.001% CaCl2 to an OD650 of 0.017 and allowed to continue to grow. When the cultures reached an OD650 of 0.05, they were split in half. One-half was induced by addition of 75 ng/mL CSP2. After 14 min at 37°C, 100 ng/mL streptomycin-resistance (Smr) chromosomal DNA from the RF46SKNE strain (Lopez et al. 1984) was added to both the induced and uninduced cultures, and then the cultures were incubated for 1 h at 37°C. Then 0.1 mL of serial dilutions was plated on AGCH medium with 1% agar plus 0.3% Suc and 0.2% YE either with or without 100 μg/mL streptomycin. Transformants were counted after 20 h at 37°C in a 5% CO2 atmosphere.
Cloning of srn206-encoding gene
A 718-bp genomic region containing the srn206 gene was amplified from TIGR4 chromosomal DNA by standard PCR using Expand High Fidelity (Roche) and primers RNA206F2 (5′-GCGAAGCTTGAAGTGACTGAACGTGTTAAA-3′) and RNA206-R4 (5′-CGCGAATTCATCATCCGCTTGAGCTGG-3′). The resulting PCR product was cloned into a pGEM-T Easy vector using the pGEM-T Easy vector system I from Promega and transformed into E. coli DH5α-competent cells. The resulting plasmid, pG206, was digested with EcoRI, and the 726-bp fragment containing the srn206 gene was then cloned into the pLScop previously cut with EcoRI. The resulting plasmid was designated pLScop-206 and was introduced into wild-type pneumococcal cells by transformation. Transformants were selected using 1 μg/mL Tc, and their sequence was confirmed.
Bioinformatics
Genome sequences with ORF annotation and coordinates were downloaded from the NBI FTP repository site (http://www.ncbi.nlm.nih.gov/genomes/lproks.cgi). cDNA contigs were mapped on all genomes using BLAST (Altschul et al. 1990) called by in-house Perl scripts. Equivalent loci between pneumococcal strains were assigned as the best mutual BLAST hit in their genomes. Free energies of the secondary structures were calculated with RNAfold from the Vienna suite (Hofacker et al. 1994)
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
ACKNOWLEDGMENTS
We thank Dr. Adela G de la Campa for critical reading and scientific discussion and an anonymous reviewer for helpful comments on earlier versions of this manuscript. We also thank Dr. G. del Solar (Centro de Investigaciones Biológicas, CSIC, Spain) for providing the pLS1cop7 plasmid. This work was supported by the Fondo de Investigación Sanitaria (FIS) (PI08/0442 and PI11/00656) and the Ministerio de Ciencia e Innovación (MICINN) (Ramón y Cajal program, RYC-2007-00179). A.J.M. was the recipient of a postdoctoral Miguel Servet contract from the Sistema Nacional de Salud funded by the FIS. During the first part of this work, M.A. was the recipient of a junior researcher contract funded by the Ramon y Cajal program from MICINN. S.N. was the recipient of a grant from CIBER Enfermedades Respiratorias (an initiative of the Instituto de Salud Carlos III).
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.027359.111.
REFERENCES
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ 1990. Basic local alignment search tool. J Mol Biol 215: 403–410 [DOI] [PubMed] [Google Scholar]
- Altuvia S 2007. Identification of bacterial small non-coding RNAs: Experimental approaches. Curr Opin Microbiol 10: 257–261 [DOI] [PubMed] [Google Scholar]
- Ballester S, Lopez P, Alonso JC, Espinosa M, Lacks SA 1986. Selective advantage of deletions enhancing chloramphenicol acetyltransferase gene expression in Streptococcus pneumoniae plasmids. Gene 41: 153–163 [DOI] [PubMed] [Google Scholar]
- Barrick JE, Sudarsan N, Weinberg Z, Ruzzo WL, Breaker RR 2008. 6S RNA is a widespread regulator of eubacterial RNA polymerase that resembles an open promoter. RNA 11: 774–784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boisset S, Geissmann T, Huntzinger E, Fechter P, Bendridi N, Possedko M, Chevalier C, Helfer AC, Benito Y, Jacquier A, et al. 2007. Staphylococcus aureus RNAIII coordinately represses the synthesis of virulence factors and the transcription regulator Rot by an antisense mechanism. Genes Dev 21: 1353–1366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonner ER, D'Elia JN, Billips BK, Switzer RL 2001. Molecular recognition of pyr mRNA by the Bacillus subtilis attenuation regulatory protein PyrR. Nucleic Acids Res 29: 4851–4865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brantl S 2009. Bacterial chromosome-encoded small regulatory RNAs. Future Microbiol 4: 85–103 [DOI] [PubMed] [Google Scholar]
- Chaturongakul S, Raengpradub S, Wiedmann M, Boor KJ 2008. Modulation of stress and virulence in Listeria monocytogenes. Trends Microbiol 16: 388–396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christiansen JK, Nielsen JS, Ebersbach T, Valentin-Hansen P, Sogaard-Andersen L, Kallipolitis BH 2006. Identification of small Hfq-binding RNAs in Listeria monocytogenes. RNA 12: 1383–1396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claverys JP, Martin B, Polard P 2009. The genetic transformation machinery: Composition, localization, and mechanism. FEMS Microbiol Rev 33: 643–656 [DOI] [PubMed] [Google Scholar]
- Conrad C, Rauhut R, Klug G 1998. Different cleavage specificities of RNases III from Rhodobacter capsulatus and Escherichia coli. Nucleic Acids Res 26: 4446–4453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Solar G, Puyet A, Espinosa M 1987. Initiation signals for the conversion of single stranded to double stranded DNA forms in the streptococcal plasmid pLS1. Nucleic Acids Res 15: 5561–5580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esakova O, Krasilnikov AS 2010. Of proteins and RNA: The RNase P/MRP family. RNA 16: 1725–1747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frishman D 2007. Protein annotation at genomic scale: The current status. Chem Rev 107: 3448–3466 [DOI] [PubMed] [Google Scholar]
- Gardner PP, Daub J, Tate JG, Nawrocki EP, Kolbe DL, Lindgreen S, Wilkinson AC, Finn RD, Griffiths-Jones S, Eddy SR, et al. 2009. Rfam: Updates to the RNA families database. Nucleic Acids Res 37: D136–D140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geissmann T, Chevalier C, Cros MJ, Boisset S, Fechter P, Noirot C, Schrenzel J, Francois P, Vandenesch F, Gaspin C, et al. 2009. A search for small noncoding RNAs in Staphylococcus aureus reveals a conserved sequence motif for regulation. Nucleic Acids Res 37: 7239–7257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillet R, Felden B 2001. Emerging views on tmRNA-mediated protein tagging and ribosome rescue. Mol Microbiol 42: 879–885 [DOI] [PubMed] [Google Scholar]
- Gottesman S 2002. Stealth regulation: Biological circuits with small RNA switches. Genes Dev 16: 2829–2842 [DOI] [PubMed] [Google Scholar]
- Halfmann A, Kovacs M, Hakenbeck R, Bruckner R 2007. Identification of the genes directly controlled by the response regulator CiaR in Streptococcus pneumoniae: Five out of 15 promoters drive expression of small non-coding RNAs. Mol Microbiol 66: 110–126 [DOI] [PubMed] [Google Scholar]
- Hartmann RK, Gossringer M, Spath B, Fischer S, Marchfelder A 2009. The making of tRNAs and more—RNase P and tRNase Z. Prog Mol Biol Transl Sci 85: 319–368 [DOI] [PubMed] [Google Scholar]
- Hershberg R, Altuvia S, Margalit H 2003. A survey of small RNA-encoding genes in Escherichia coli. Nucleic Acids Res 31: 1813–1820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofacker IL, Fontana W, Stadler PF, Bonhoeffer S, Tacker M, Schuster P 1994. Fast folding and comparison of RNA secondary structures. Monatsh Chem 125: 167–188 [Google Scholar]
- Howie SR, Adegbola RA 2006. Pneumonia and child mortality. Lancet 368: 1646. [DOI] [PubMed] [Google Scholar]
- Irnov I, Kertsburg A, Winkler WC 2006. Genetic control by cis-acting regulatory RNAs in Bacillus subtilis: General principles and prospects for discovery. Cold Spring Harb Symp Quant Biol 71: 239–249 [DOI] [PubMed] [Google Scholar]
- Irnov I, Sharma CM, Vogel J, Winkler WC 2010. Identification of regulatory RNAs in Bacillus subtilis. Nucleic Acids Res 38: 6637–6651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnsborg O, Havarstein LS 2009. Regulation of natural genetic transformation and acquisition of transforming DNA in Streptococcus pneumoniae. FEMS Microbiol Rev 33: 627–642 [DOI] [PubMed] [Google Scholar]
- Johnsen M, Christensen T, Dennis PP, Fiil NP 1982. Autogenous control: Ribosomal protein L10–L12 complex binds to the leader sequence of its mRNA. EMBO J 1: 999–1004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazantsev AV, Pace NR 2006. Bacterial RNase P: A new view of an ancient enzyme. Nat Rev Microbiol 4: 729–740 [DOI] [PubMed] [Google Scholar]
- Kim KS, Sim S, Ko JH, Lee Y 2005. Processing of m1 RNA at the 3′ end protects its primary transcript from degradation. J Biol Chem 280: 34667–34674 [DOI] [PubMed] [Google Scholar]
- Kowalko JE, Sebert ME 2008. The Streptococcus pneumoniae competence regulatory system influences respiratory tract colonization. Infect Immun 76: 3131–3140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar R, Shah P, Swiatlo E, Burgess SC, Lawrence ML, Nanduri B 2010. Identification of novel non-coding small RNAs from Streptococcus pneumoniae TIGR4 using high-resolution genome tiling arrays. BMC Genomics 11: 350 doi: 10.1186/1471-2164-11-350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landt SG, Abeliuk E, McGrath PT, Lesley JA, McAdams HH, Shapiro L 2008. Small non-coding RNAs in Caulobacter crescentus. Mol Microbiol 68: 600–614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau GW, Haataja S, Lonetto M, Kensit SE, Marra A, Bryant AP, McDevitt D, Morrison DA, Holden DW 2001. A functional genomic analysis of type 3 Streptococcus pneumoniae virulence. Mol Microbiol 40: 555–571 [DOI] [PubMed] [Google Scholar]
- Lin-Chao S, Wei CL, Lin YT 1999. RNase E is required for the maturation of ssrA RNA and normal ssrA RNA peptide-tagging activity. Proc Natl Acad Sci 96: 12406–12411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livny J, Waldor MK 2007. Identification of small RNAs in diverse bacterial species. Curr Opin Microbiol 10: 96–101 [DOI] [PubMed] [Google Scholar]
- Livny J, Brencic A, Lory S, Waldor MK 2006. Identification of 17 Pseudomonas aeruginosa sRNAs and prediction of sRNA-encoding genes in 10 diverse pathogens using the bioinformatic tool sRNAPredict2. Nucleic Acids Res 34: 3484–3493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livny J, Teonadi H, Livny M, Waldor MK 2008. High-throughput, kingdom-wide prediction and annotation of bacterial non-coding RNAs. PLoS ONE 3: e3197 doi: 10.1371/journal.pone.0003197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loh E, Dussurget O, Gripenland J, Vaitkevicius K, Tiensuu T, Mandin P, Repoila F, Buchrieser C, Cossart P, Johansson J 2009. A trans-acting riboswitch controls expression of the virulence regulator PrfA in Listeria monocytogenes. Cell 139: 770–779 [DOI] [PubMed] [Google Scholar]
- Lopez P, Espinosa M, Lacks SA 1984. Physical structure and genetic expression of the sulfonamide-resistance plasmid pLS80 and its derivatives in Streptococcus pneumoniae and Bacillus subtilis. Mol Gen Genet 195: 403–410 [DOI] [PubMed] [Google Scholar]
- Mandin P, Repoila F, Vergassola M, Geissmann T, Cossart P 2007. Identification of new noncoding RNAs in Listeria monocytogenes and prediction of mRNA targets. Nucleic Acids Res 35: 962–974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margolis E, Yates A, Levin BR 2010. The ecology of nasal colonization of Streptococcus pneumoniae, Haemophilus influenzae and Staphylococcus aureus: The role of competition and interactions with host's immune response. BMC Microbiol 10: 59 doi: 10.1186/1471-2180-10-59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marx P, Nuhn M, Kovacs M, Hakenbeck R, Bruckner R 2010. Identification of genes for small non-coding RNAs that belong to the regulon of the two-component regulatory system CiaRH in Streptococcus. BMC Genomics 11: 661 doi: 10.1186/1471-2164-11-661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mironov AS, Gusarov I, Rafikov R, Lopez LE, Shatalin K, Kreneva RA, Perumov DA, Nudler E 2002. Sensing small molecules by nascent RNA: A mechanism to control transcription in bacteria. Cell 111: 747–756 [DOI] [PubMed] [Google Scholar]
- Nudler E, Mironov AS 2004. The riboswitch control of bacterial metabolism. Trends Biochem Sci 29: 11–17 [DOI] [PubMed] [Google Scholar]
- Oggioni MR, Iannelli F, Ricci S, Chiavolini D, Parigi R, Trappetti C, Claverys JP, Pozzi G 2004. Antibacterial activity of a competence-stimulating peptide in experimental sepsis caused by Streptococcus pneumoniae. Antimicrob Agents Chemother 48: 4725–4732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oggioni MR, Trappetti C, Kadioglu A, Cassone M, Iannelli F, Ricci S, Andrew PW, Pozzi G 2006. Switch from planktonic to sessile life: A major event in pneumococcal pathogenesis. Mol Microbiol 61: 1196–1210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang X, Zhou D, Song Y, Pei D, Wang J, Guo Z, Yang R 2004. Bacterial mRNA purification by magnetic capture-hybridization method. Microbiol Immunol 48: 91–96 [DOI] [PubMed] [Google Scholar]
- Passalacqua KD, Varadarajan A, Ondov BD, Okou DT, Zwick ME, Bergman NH 2009. Structure and complexity of a bacterial transcriptome. J Bacteriol 191: 3203–3211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichon C, Felden B 2005. Small RNA genes expressed from Staphylococcus aureus genomic and pathogenicity islands with specific expression among pathogenic strains. Proc Natl Acad Sci 102: 14249–14254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prudhomme M, Attaiech L, Sanchez G, Martin B, Claverys JP 2006. Antibiotic stress induces genetic transformability in the human pathogen Streptococcus pneumoniae. Science 313: 89–92 [DOI] [PubMed] [Google Scholar]
- Rasmussen S, Nielsen HB, Jarmer H 2009. The transcriptionally active regions in the genome of Bacillus subtilis. Mol Microbiol 73: 1043–1057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Repoila F, Darfeuille F 2009. Small regulatory non-coding RNAs in bacteria: Physiology and mechanistic aspects. Biol Cell 101: 117–131 [DOI] [PubMed] [Google Scholar]
- Romby P, Vandenesch F, Wagner EG 2006. The role of RNAs in the regulation of virulence-gene expression. Curr Opin Microbiol 9: 229–236 [DOI] [PubMed] [Google Scholar]
- Sakamoto H, Kimura N, Shimura Y 1983. Processing of transcription products of the gene encoding the RNA component of RNase P. Proc Natl Acad Sci 80: 6187–6191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott JA 2008. The global epidemiology of childhood pneumonia 20 years on. Bull World Health Organ 86: 494–496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma CM, Vogel J 2009. Experimental approaches for the discovery and characterization of regulatory small RNA. Curr Opin Microbiol 12: 536–546 [DOI] [PubMed] [Google Scholar]
- Sharma CM, Hoffmann S, Darfeuille F, Reignier J, Findeiss S, Sittka A, Chabas S, Reiche K, Hackermuller J, Reinhardt R, et al. 2010. The primary transcriptome of the major human pathogen Helicobacter pylori. Nature 464: 250–255 [DOI] [PubMed] [Google Scholar]
- Sittka A, Lucchini S, Papenfort K, Sharma CM, Rolle K, Binnewies TT, Hinton JC, Vogel J 2008. Deep sequencing analysis of small noncoding RNA and mRNA targets of the global post-transcriptional regulator, Hfq. PLoS Genet 4: e1000163 doi: 10.1371/journal.pgen.1000163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song JH, Ko KS, Lee JY, Baek JY, Oh WS, Yoon HS, Jeong JY, Chun J 2005. Identification of essential genes in Streptococcus pneumoniae by allelic replacement mutagenesis. Mol Cells 19: 365–374 [PubMed] [Google Scholar]
- Swiercz JP, Hindra, Bobek J, Bobek J, Haiser HJ, Di Berardo C, Tjaden B, Elliot MA 2008. Small non-coding RNAs in Streptomyces coelicolor. Nucleic Acids Res 36: 7240–7251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tettelin H, Nelson KE, Paulsen IT, Eisen JA, Read TD, Peterson S, Heidelberg J, DeBoy RT, Haft DH, Dodson RJ, et al. 2001. Complete genome sequence of a virulent isolate of Streptococcus pneumoniae. Science 293: 498–506 [DOI] [PubMed] [Google Scholar]
- Tjaden B, Goodwin SS, Opdyke JA, Guillier M, Fu DX, Gottesman S, Storz G 2006. Target prediction for small, noncoding RNAs in bacteria. Nucleic Acids Res 34: 2791–2802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toledo-Arana A, Repoila F, Cossart P 2007. Small noncoding RNAs controlling pathogenesis. Curr Opin Microbiol 10: 182–188 [DOI] [PubMed] [Google Scholar]
- Trotochaud AE, Wassarman KM 2005. A highly conserved 6S RNA structure is required for regulation of transcription. Nat Struct Mol Biol 12: 313–319 [DOI] [PubMed] [Google Scholar]
- Tsui HC, Mukherjee D, Ray VA, Sham LT, Feig AL, Winkler ME 2010. Identification and characterization of noncoding small RNAs in Streptococcus pneumoniae serotype 2 strain D39. J Bacteriol 192: 264–279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitreschak AG, Rodionov DA, Mironov AA, Gelfand MS 2002. Regulation of riboflavin biosynthesis and transport genes in bacteria by transcriptional and translational attenuation. Nucleic Acids Res 30: 3141–3151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wardlaw T, Salama P, Johansson EW, Mason E 2006. Pneumonia: The leading killer of children. Lancet 368: 1048–1050 [DOI] [PubMed] [Google Scholar]
- Wassarman KM, Zhang A, Storz G 1999. Small RNAs in Escherichia coli. Trends Microbiol 7: 37–45 [DOI] [PubMed] [Google Scholar]
- Waters LS, Storz G 2009. Regulatory RNAs in bacteria. Cell 136: 615–628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberg Z, Barrick JE, Yao Z, Roth A, Kim JN, Gore J, Wang JX, Lee ER, Block KF, Sudarsan N, et al. 2007. Identification of 22 candidate structured RNAs in bacteria using the CMfinder comparative genomics pipeline. Nucleic Acids Res 35: 4809–4819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler W, Nahvi A, Breaker RR 2002. Thiamine derivatives bind messenger RNAs directly to regulate bacterial gene expression. Nature 419: 952–956 [DOI] [PubMed] [Google Scholar]
- Xayarath B, Freitag NE 2009. A bacterial pathogen flips the riboswitch. Cell Host Microbe 6: 400–402 [DOI] [PubMed] [Google Scholar]
- Zengel JM, Lindahl L 1994. Diverse mechanisms for regulating ribosomal protein synthesis in Escherichia coli. Prog Nucleic Acid Res Mol Biol 47: 331–370 [DOI] [PubMed] [Google Scholar]