Ferritin stores and detoxifies an excess of intracellular iron and thereby plays an important role in the metabolism of this metal. As unshielded iron promotes oxidative stress, ferritin is crucial in maintaining cellular redox balance and may also modulate cell growth, survival, and apoptosis. The expression of ferritin is controlled by transcriptional and post-transcriptional mechanisms. Here, the authors examined how ferritin mRNA translation responds to stress conditions. Under global inhibition of protein synthesis or specific inhibition of ferritin mRNA translation in cells overexpressing PKR or IRP1, respectively, they demonstrate association of ferritin mRNA with heavy polysomes. Further experiments revealed that the 5′ untranslated region (5′ UTR) of ferritin mRNA contains a bona fide internal ribosomal entry site (IRES).
Keywords: iron metabolism, PKR, eIF2α, eIF4G, poliovirus, IRP1, internal initiation, IRES
Abstract
Ferritin stores and detoxifies an excess of intracellular iron, and thereby plays an important role in the metabolism of this metal. As unshielded iron promotes oxidative stress, ferritin is crucial in maintaining cellular redox balance and may also modulate cell growth, survival, and apoptosis. The expression of ferritin is controlled by transcriptional and post-transcriptional mechanisms. In light of the well-established transcriptional induction of ferritin by inflammatory signals, we examined how ferritin mRNA translation responds to stress conditions. We first used HT1080 fibrosarcoma cells engineered for coumermycin-inducible expression of PKR, a stress kinase that inhibits protein synthesis in virus-infected cells by phosphorylating eIF2α. In this setting, iron triggered partial ferritin mRNA translation despite a PKR-induced global shutdown in protein synthesis. Moreover, iron-mediated ferritin synthesis was evident in cells infected with an attenuated strain of poliovirus; when eIF4GI was cleaved, eIF2α was phosphorylated, and host protein synthesis was inhibited. Under global inhibition of protein synthesis or specific inhibition of ferritin mRNA translation in cells overexpressing PKR or IRP1, respectively, we demonstrate association of ferritin mRNA with heavy polysomes. Further experiments revealed that the 5′ untranslated region (5′ UTR) of ferritin mRNA contains a bona fide internal ribosomal entry site (IRES). Our data are consistent with the existence of an alternative, noncanonical mechanism for ferritin mRNA translation, which may primarily operate under stress conditions to protect cells from oxidative stress.
INTRODUCTION
While iron is essential for cells and organisms, it may also promote oxidative stress by catalyzing formation of free radicals (Halliwell and Gutteridge 1990; Papanikolaou and Pantopoulos 2005). Therefore, iron metabolism has to be tightly regulated (Hentze et al. 2010; Wang and Pantopoulos 2011). Excess of intracellular iron is stored in the cytoplasm within ferritin, a multisubunit protein of H- and L-chains. Holo-ferritin forms a nanocage structure, capable of accommodating up to 4500 ferric ions; these are maintained in a nontoxic form and can be mobilized for metabolic purposes (Arosio et al. 2009).
The expression of ferritin is regulated in response to iron levels by a post-transcriptional mechanism (Hentze et al. 2010; Wang and Pantopoulos 2011). The mRNAs encoding H- and L-ferritins possess an iron responsive element (IRE) within their 5′ UTRs, in close proximity to the cap structure. This constitutes a target of two iron regulatory proteins, IRP1 and IRP2, which are activated for IRE binding in iron-deficient cells. The specific IRE/IRP interactions sterically inhibit translation of ferritin mRNAs (Gebauer and Hentze 2004).
Ferritin expression is also regulated transcriptionally by inflammatory cytokines, xenobiotics, oxidative stress, hormones, and growth factors (Torti and Torti 2002). Tumor necrosis factor α (TNFα) stimulates transcription of murine H-ferritin mRNA via NF-κB binding to the enhancer element FER2, located 4.8 kb upstream of the transcription start site (Kwak et al. 1995). Other cytokines, such as interleukin 1β (IL-1β) enhance H-ferritin mRNA translation via binding of poly(C)-binding proteins to a GC-rich acute box in the 5′ UTR (Thomson et al. 2005). Xenobiotics and oxidative stress induce ferritin mRNA transcription via binding of Nrf2/Maf or JunD to antioxidant response elements (AREs) within both H- and L-ferritin genes (Tsuji et al. 2000; Hintze and Theil 2005; Tsuji 2005).
The liver-specific disruption of mouse H-ferritin leads to oxidative tissue injury (Darshan et al. 2009), while mice with one functional H-ferritin allele (H-ferritin+/−) exhibit signs of oxidative stress in the brain (Thompson et al. 2003). On the other hand, the overexpression of an H-ferritin transgene in cell lines has been associated with decreased levels of redox-active “labile” iron and protection against oxidants (Epsztejn et al. 1999; Cozzi et al. 2000; Xie et al. 2005), while the up-regulation of endogenous H-ferritin protected cells against TNFα-induced apoptosis (Pham et al. 2004). Moreover, endogenous H-ferritin levels increased following UV irradiation in various cell lines (Fox et al. 2009) and in human skin (Applegate et al. 1998).
By responding to cytokines, oxidants, or UV irradiation, and by regulating cell survival, ferritin defines an important player in the context of inflammation and stress responses. The transcriptional induction of ferritin by cytokines and oxidants prompted us to investigate ferritin mRNA translation under stress conditions, considering that inflammation, oxidative stress, and UV irradiation are often associated with a global inhibition in protein synthesis. This is mediated via phosphorylation of eIF2α by the kinases PKR, PERK, GCN2, or HRI, which prevents GTP/GDP exchange and inhibits ternary complex formation, a critical step in translation initiation (Proud 2005; Wek et al. 2006).
By using a PKR-inducible cell system and an attenuated strain of poliovirus, we demonstrate that ferritin mRNA is partially translated following stimulation with iron, even under conditions where cap-dependent translation is strongly inhibited. We further show association of ferritin mRNA with polysomes despite the binding of its repressor IRP1 or a PKR-mediated global inhibition of protein synthesis. Finally, we provide evidence that the 5′ UTR of ferritin possesses properties of a bona fide IRES.
RESULTS
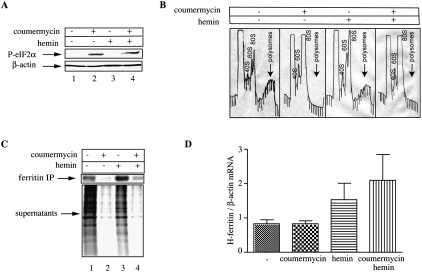
The eIF2α kinase PKR consists of a regulatory double-stranded RNA-binding domain and a catalytic domain. Binding of double-stranded RNA promotes dimerization and autophosphorylation, rendering the protein enzymatically active (Proud 2005). The replacement of the regulatory domain of PKR by the first 220 amino acids of the E. coli GyrB protein, allows dimerization and catalytic activation of the fusion protein upon addition of coumermycin (Ung et al. 2001). We utilized human HT1080 fibrosarcoma cells expressing coumermycin-inducible GyrB.PKR (Kazemi et al. 2004; Raven et al. 2008) as an established model to study ferritin mRNA translation under stress conditions. The addition of coumermycin activated GyrB.PKR and resulted in phosphorylation of eIF2α at Ser-51 (Fig. 1A), in agreement with previous observations (Kazemi et al. 2004; Raven et al. 2008). As expected, this response promoted a shut-off in protein synthesis, as judged by the disappearance of actively translating heavy polysomes in a sucrose gradient fractionation assay (Fig. 1B). The activation of GyrB.PKR sufficed to trigger eIF2α phosphorylation and translational arrest, even after treatment with hemin, an iron source, which may antagonize these effects via the heme-regulated inhibitor HRI (Chen 2000; Fillebeen et al. 2005).
FIGURE 1.
De novo ferritin synthesis despite global translational arrest upon eIF2α phosphorylation. HT1080-GyrB-PKR cells were treated with coumermycin (100 ng/mL) and hemin (100 μM) for 6 h. (A) Cell lysates were analyzed by Western blotting with an eIF2α phosphospecific (top) and a β-actin antibody (bottom). (B) Cytoplasmic extracts were fractionated on sucrose gradients with continuous monitoring of A260; arrows indicate the heavy polysomes. (C) The cells were metabolically labeled with [35S]methionine/cysteine for 2 h and 1000 μg of cell extracts were subjected to immunoprecipitation with 30 μg of ferritin antibody. Immunoprecipitated material and the IP supernatants were analyzed by SDS-PAGE on a 12% gel. The ferritin bands and the lanes showing global protein synthesis were visualized by autoradiography. (D) Analysis of H-ferritin mRNA from whole-cell lysates by qPCR.
Under these conditions, a challenge of the cells with excess iron would be expected to be detrimental if their capacity to store the metal within ferritin were compromised. The addition of 100 μM hemin for up to 6 h was not associated with any apparent toxicity, independently of the translation status of the cells (data not shown). This suggests that in translationally silenced cells, excess iron is detoxified either in pre-existing or in de novo synthesized ferritin; both scenarios are not mutually exclusive. To address whether de novo ferritin synthesis is possible under global translational arrest, the HT1080 cells were treated with coumermycin in the absence or presence of hemin, and metabolically labeled with [35S]methionine/cysteine; subsequently, ferritin mRNA translation was assessed by quantitative immunoprecipitation with a ferritin antibody. The addition of coumermycin profoundly inhibited the accumulation of [35S]-labeled polypeptides (Fig. 1C, bottom); nevertheless, newly synthesized [35S]-ferritin was partially recovered in the immunoprecipitate (Fig. 1C, top). This effect was more evident in hemin-treated cells (lane 4). Coumermycin did not trigger any statistically significant alterations in ferritin mRNA levels (Fig. 1D), while hemin stimulated ferritin mRNA expression, very likely at the transcriptional level (Coccia et al. 1992). These data suggest that ferritin mRNA possesses the capacity to, at least partially, bypass the translational blockade imposed by eIF2α phosphorylation.
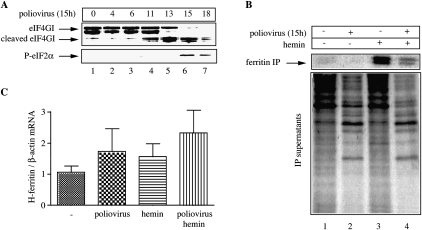
We then assessed whether ferritin synthesis is possible during inactivation of cap-dependent translation under more physiologically relevant conditions. To this end, we utilized an attenuated strain of Sabin-like, type I poliovirus (Svitkin et al. 1990). Polioviruses encode protease 2A that cleaves eIF4G, a scaffolding protein involved in the formation of a cap-binding complex on the mRNA and the recruitment of the 43S ribosome (Thompson and Sarnow 2000). The virus-mediated proteolytic cleavage of eIF4G accounts for inhibition of host protein synthesis, favoring cap-independent translation of viral proteins by internal initiation. HT1080 cells were infected with the attenuated poliovirus and the expression of eIF4GI, the major eIF4G isoform was analyzed by immunoblotting at different time intervals. The cleavage of eIF4GI was evident after 11 h (Fig. 2A, top, lanes 4–7), accompanied by phosphorylation of eIF2α after 15 h (Fig. 2A, bottom, lanes 6,7). We then analyzed ferritin synthesis 15 h post-infection, following metabolic labeling with [35S]methionine/cysteine. The poliovirus infection markedly inhibited host cap-dependent translation, concomitantly with the emergence of [35S]-labeled viral proteins (Fig. 2B). The synthesis of ferritin was also inhibited; however, in hemin-treated poliovirus-infected cells, ferritin synthesis was partially recovered. Even though the virus appeared to augment ferritin mRNA, by analogy to hemin, there was no statistically significant difference in ferritin mRNA levels among hemin-treated uninfected and infected cells (Fig. 2C). These findings provide additional evidence that de novo ferritin synthesis is possible under conditions where global protein synthesis is shut down.
FIGURE 2.
De novo ferritin synthesis in poliovirus-infected cells. (A) HT1080 cells were infected with the Sabin-like type I strain of poliovirus at a multiplicity of infection (MOI) of 10. At the indicated time intervals post-infection (h), cell lysates were prepared and analyzed by Western blotting with eIF4GI (top) and phospho-eIF2α (bottom) antibodies. (B) HT1080 cells were infected with the poliovirus (MOI of 10) for 15 h, treated with hemin for 6 h, and metabolically labeled with [35S]methionine/cysteine for 2 h. Cell extracts (1000 μg) were subjected to immunoprecipitation with 30 μg of ferritin antibody. Immunoprecipitated material and the IP supernatants were analyzed by SDS-PAGE on a 12% gel. The ferritin bands and the lanes corresponding to host and viral protein synthesis were visualized by autoradiography. (C) Analysis of H-ferritin mRNA from whole-cell lysates by qPCR.
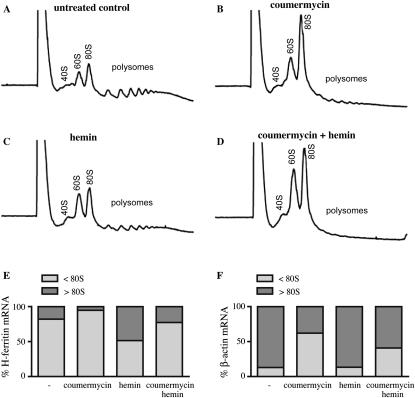
We analyzed polysome profiles in HT1080 cells containing phosphorylated eIF2α, and quantified ferritin mRNA in each fraction by real-time PCR. As expected, administration of coumermycin to activate eIF2α phosphorylation largely depleted the heavy polysomes and all of the ferritin mRNA shifted to the lighter fractions (Fig. 3A–D). Ferritin mRNA was translated considerably less efficiently than control β-actin mRNA, as judged by their distribution within heavy (>80 S) polysomes and light (<80 S) monosomes (Fig. 3E,F; Supplemental Fig. S1). Importantly, a combined hemin plus coumermycin treatment resulted in a ∼5% increase in the fraction of ferritin mRNA associated with heavy (>80 S) polysomes (as compared to no treatment), while the respective fraction of β-actin mRNA decreased by ∼28%. These results indicate an enhanced capacity of ferritin mRNA to bypass a translational blockade under stress conditions.
FIGURE 3.
Ferritin mRNA associates with the heavy polysomes following treatment with hemin, under conditions of global translational arrest. Cytoplasmic lysates from HT1080-GyrB-PKR cells were subjected to polysome profiling with continuous monitoring of A260. The cells were previously either left untreated (A) or treated with 100 ng/mL coumermycin (B), 100 μM hemin (C), or both coumermycin and hemin (D). H-ferritin and β-actin mRNA were analyzed by quantitative real-time PCR. The percentage of H-ferritin (E) and β-actin (F) mRNA in the fractions lighter or heavier than 80S is shown on the graphs corresponding to each treatment.
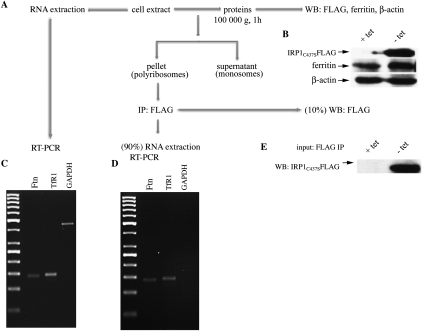
Next, we used human H1299 lung cancer cells, engineered for tetracycline-inducible overexpression of IRP1C437S. This constitutive IRP1 mutant is known to specifically inhibit translation of ferritin mRNA as well as of other mRNAs harboring an IRE in their 5′ UTR. However, in previous experiments, we reported that IRP1C437S failed to inhibit ferritin synthesis in dense cultures of H1299 cells (Wang and Pantopoulos 2002). Moreover, we found that after long-term maintenance of these cells, ferritin synthesis evaded the repressor activity of tetracycline-inducible IRP1C437S (or IRP2) even in low-density cultures (Maffettone et al. 2010). We recapitulated these findings and moved on to investigate whether IRP1C437S remains bound to ferritin mRNA or gets displaced during translation. For this purpose, cytoplasmic extracts from IRP1C437S-overexpressing cells were prepared and separated into two ribosomal populations, one containing polysomes and another one containing post-polysomes (monosomes) (Supplemental Fig. S2). IRP1C437S was immunoprecipitated from the polysomal fraction by using an antibody against the FLAG epitope tag (Fig. 4). One tenth of the FLAG immunoprecipitate was used for Western blotting and the rest for RNA analysis. The data in Figure 4D show that IRP1C437S remains bound to ferritin mRNA in the polysomal fraction when this mRNA is being translated. As expected, polysome-associated IRP1C437S also binds to TfR1 mRNA, but not GAPDH mRNA, which serve as positive and negative controls, respectively. All ferritin, TfR1, and GAPDH mRNAs are detectable in the whole-cell lysate (Fig. 4C). Thus, in this experimental setting, the translational machinery appears to bypass the IRP1C437S repressor to accomplish ferritin synthesis. Taken together, the data in Figures 1–4 raise the possibility for an alternative, noncanonical mechanism for ferritin mRNA translation, which may predominantly operate under conditions of cellular stress.
FIGURE 4.
Association of IRP1C437S with the polysome-bound ferritin mRNA. (A) The experimental procedure; cell lysates obtained from H1299 cells grown at high density in the presence or absence of tetracycline were subjected to either Western blotting with ferritin and β-actin antibodies (B), or qPCR with H-ferritin, TfR1, and GAPDH specific primers (C), or ultracentrifuged for 1 h to isolate the heavy polysomes from the monosomes. The polysomal pellet was subjected to immunoprecipitation with a FLAG antibody. Ninety pecent of the immunoprecipitate was used for RNA extraction and reverse transcription PCR with H-ferritin, TfR1, and GAPDH-specific primers (D), and 10% of the immunoprecipitate was subjected to Western blotting with a FLAG antibody to document the presence of FLAG-tagged IRP1C437S in the immunoprecipitate (E).
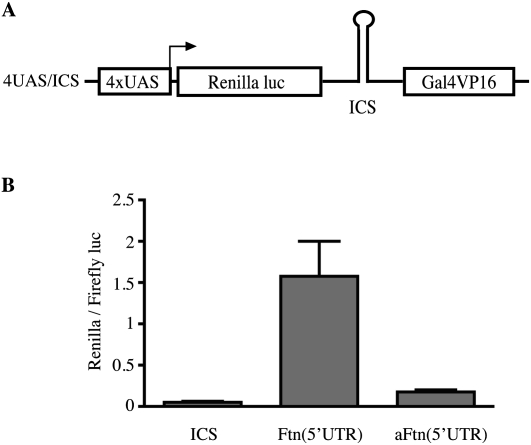
Cap-independent translation proceeds via internal ribosomal entry sites (IRESs) (Holcik and Sonenberg 2005). Such sequences were originally found in uncapped viral RNAs and subsequently in several capped eukaryotic mRNAs. They do not share considerable similarities in primary sequence or structure, and are rather defined by functional criteria. While IRESs were initially associated with complex RNA foldings, more recent data suggested that even very short sequences, for example, a 7-nt stretch within the 5′ UTR of the Gtx homeodomain mRNA (Dresios et al. 2006) may drive cap-independent translation initiation. We used a positive feedback vector to evaluate the capacity of the 5′ UTR of H-ferritin mRNA to function as an IRES. The vector encodes a bicistronic mRNA with Renilla luciferase in the first cistron and the yeast Gal4/viral protein 16 (Gal4/VP16) in the second cistron (Zhou et al. 2005). The 5′ UTR of H-ferritin mRNA was inserted within the intercistronic sequence (ICS) either in the sense (Ftn), or the antisense (aFtn) orientation (Fig. 5A), and the constructs were transfected into HT1080 cells. Because the transcription of the bicistronic mRNA is under the control of a minimal promoter containing four copies of the Gal4 upstream activator sequence (4xUAS), expression of Renilla luciferase is only possible via IRES-mediated, cap-independent translation of Gal4/VP16 to activate the promoter in a positive feedback loop. The data in Figure 5B demonstrate that the 5′ UTR of H-ferritin mRNA functions as an IRES, while it fails to do so when placed in reverse orientation.
FIGURE 5.
Evidence that the 5′ UTR of H-ferritin harbors a functional IRES. (A) Schematic diagram of the bicistronic, positive feedback vector used to test for putative IRESs. The transcription of the bicistronic mRNA depends on a minimal promoter containing four copies of the Gal4 upstream activator sequence (4×UAS). The first cistron encodes for Renilla luciferase and the second for the transactivator Gal4/VP16. The 5′ UTR of H-ferritin (Ftn) and the inverted sequence of H-ferritin 5′ UTR (aFtn) were cloned in the intercistronic sequence (ICS). (B) Renilla luciferase activity following transfection of HT1080 cells with control (ICS), Ftn (5′UTR), or aFtn (5′UTR) constructs. The values from three independent experiments (mean ± SD) were normalized with Firefly luciferase activity from a cotransfected construct.
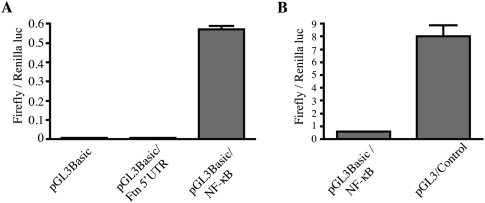
Subsequently, we examined whether H-ferritin 5′ UTR may possess a putative promoter activity that could stimulate Gal4/VP16 expression and promote Renilla luciferase expression (accounting for the data in Fig. 5B) in an IRES-independent manner. The 5′ UTR of H-ferritin was inserted within the promoterless pGL3Basic vector encoding Firefly luciferase. However, it failed to drive Firefly luciferase activity in transfected HT1080 cells (Fig. 6A). As expected, the insertion of an NF-κB promoter element within the pGL3Basic vector markedly induced Firefly luciferase activity (Fig. 6A). An even more robust induction was observed with a control vector (pGL3/Control) containing SV40 promoter and enhancer elements (Fig. 6B). Taken together, the data in Figures 5 and 6 are consistent with the presence of a functional IRES element within the 5′ UTR of H-ferritin mRNA.
FIGURE 6.
The 5′ UTR of H-ferritin does not possess transcriptional activity. Firefly luciferase activity following transfection of HT1080 cells with (A) the pGL3Basic, pGL3Basic/Ftn(5′UTR), or pGL3Basic/NF-κB constructs, and (B) the pGL3Basic/NF-κB or pGL3Control constructs. The values from three independent experiments (mean ± SD) were normalized with Renilla luciferase activity from cotransfected pRL-TK.
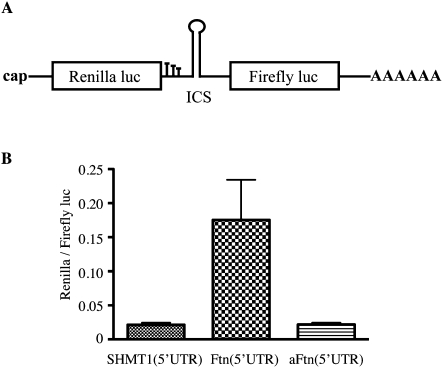
In light of the controversial nature of various proposed cellular IRESs (Gilbert 2010), we further examined the functionality of the putative ferritin IRES in a different context. We used a classic bicistronic construct giving rise to Renilla luciferase from the capped first cistron and Firefly luciferase from the second cistron, driven by the 5′ UTR of serine hydroxymethyltransferase 1 (SHMT1), which possesses IRES activity only in response to UV irradiation (Woeller et al. 2007; Fox et al. 2009). The H-ferritin 5′ UTR was cloned within the ICS in either sense (Ftn) or antisense (aFtn) orientation (Fig. 7A). In vitro transcribed bicistronic mRNAs were transfected into HT1080 cells, and luciferase activity was measured. Only the Ftn(5′UTR) construct exhibited Firefly luciferase activity (∼7.5-fold higher than background), illustrating the capacity of ferritin's 5′ UTR to drive cap-independent translation in this bicistronic system. These data provide further evidence for the existence of an alternative, noncanonical, and cap-independent mechanism for ferritin mRNA translation.
FIGURE 7.
The 5′ UTR of H-ferritin drives cap-independent translation of Firefly luciferase in a bicistronic construct. (A) Schematic diagram of the classic bicictronic construct. The first cistron encodes for Renilla luciferase, preceded by a cap structure and followed by three in-frame stop codons. The second cistron encodes for Firefly luciferase, followed by a poly(A) tail. An intracistronic sequence (ICS) is placed between the two cistrons (B). In vitro transcribed bicistronic RNAs with an ICS consisting of the 5′ UTR of SHMT1, or H-ferritin in sense (Ftn) or antisense orientation (aFtn) were transfected into HT1080 cells. Luciferase activity was monitored in cell lysates; values from three independent experiments (mean ± SD) are shown.
DISCUSSION
Ferritin plays a major role in iron detoxification and the regulation of its expression is critical for cell physiology. Alterations in ferritin levels are not only observed in disorders of iron metabolism, but also in a variety of inflammatory states or stress conditions (Torti and Torti 2002) often associated with a suppression of protein synthesis. We studied here how ferritin mRNA translation responds to such challenges. By utilizing the coumermycin-inducible GyrB.PKR system, we show that ferritin mRNA translation may, at least partially, evade global inhibition in protein synthesis, imposed by eIF2α phosphorylation (Fig. 1). Likewise, by utilizing an attenuated strain of poliovirus, we demonstrate ferritin synthesis in infected cells, under conditions of global translational arrest, as a result of eIF4GI cleavage (Fig. 2). These effects were more pronounced in iron-loaded cells, most likely due to iron-mediated inactivation of IRPs that liberates ferritin mRNA from their repressor activity (Gebauer and Hentze 2004; Wang and Pantopoulos 2011). The iron-stimulated synthesis of ferritin in translationally silenced cells was not complete, as compared to control cells. We speculate that this may be related to a limited availability of ferritin mRNA due to partial sequestration in stress granules (Anderson and Kedersha 2008).
Experiments with H1299 cells overexpressing IRP1C437S (Wang and Pantopoulos 2002) or IRP2 (Maffettone et al. 2010) demonstrated de novo ferritin synthesis despite excessive IRE-binding activity. High ferritin levels were also recorded in tumor xenografts derived from these cells (Chen et al. 2007; Maffettone et al. 2010). Mechanistic studies ruled out a scenario based upon alternative ferritin mRNA transcription from putative upstream or downstream cryptic sites (Wang and Pantopoulos 2002). This could either facilitate ferritin mRNA translation by increasing the distance of the IRE/IRP complex from the cap or by directly eliminating it, respectively. The data in Figure 4 suggest that the IRP repressor remains bound to ferritin mRNA in actively translating polysomes. These results exclude the possibility for displacement of IRP from its cognate IRE by the translation initiation machinery, and rather suggest a bypass of the IRE/IRP blockade.
Taken together, the data in Figures 1–4 provide strong evidence that ferritin mRNA can be translated by an alternative mechanism. Considering that cap-independent recruitment of ribosomes involves an internal entry site (Holcik and Sonenberg 2005; Komar and Hatzoglou 2005), we hypothesized that the 5′ UTR of ferritin mRNA may contain a functional IRES. By means of a positive feedback vector developed to identify IRESs (Zhou et al. 2005), and by transfection experiments with in vitro transcribed bicistronic RNAs (derived from a classic bicistronic vector), we show that the 5′ UTR of H-ferritin mRNA contains a bona fide IRES (Figs. 5–7). Transcriptional events can be excluded by the lack of promoter activity of H-ferritin 5′ UTR (Fig. 6) and by the direct usage of bicistronic RNAs (Fig. 7). As global protein synthesis was not inhibited in the experiments shown in Figures 5–7, these data suggest that a fraction of ferritin mRNA can be translated by the alternative mechanism, even under normal conditions. However, considering the sensitivity of the positive feedback assay and the low Firefly luciferase activity obtained with the classic bicistronic construct, we speculate that such a response is very likely negligible.
Apart from a high GC content (∼72%), the 5′ UTR of ferritin mRNA does not possess other features such as an extensive length and secondary structure, commonly found in most characterized viral and cellular IRESs (Holcik and Sonenberg 2005; Komar and Hatzoglou 2005); thus, the H-ferritin IRES appears to be rather atypical. Nonetheless, functional IRESs comprising minimal sequence elements have been reported (Dresios et al. 2006); such elements are thought to engage in direct interactions with ribosomes, thereby modulating mRNA translation in a specific manner (filter hypothesis) (Mauro and Edelman 2007). Defining the boundaries of H-ferritin IRES awaits further studies and is expected to provide important mechanistic insights. We did not formally evaluate the 5′ UTR of L-ferritin mRNA for the presence of an IRES. However, we consider this as a likely possibility, considering their sequence similarities and the fact that both H-ferritin and L-ferritin are readily synthesized de novo in poliovirus-infected cells (see high resolution image in Fig. 2B). The presence of IRESs may offer an explanation as to why uncapped ferritin mRNAs are translated almost as efficiently as capped counterparts in vitro (Dix et al. 1992).
Our results are consistent with the previously reported induction of ferritin synthesis in murine L929 cells infected with the picornavirus Mengo (Mulvey et al. 1996). This occurred despite a general arrest of host protein synthesis and in response to virus-mediated increase in “free” iron levels (Mulvey et al. 1996). An alternative IRES-dependent mechanism would also explain the enhanced ferritin synthesis in regenerated rat liver expressing high levels of active IRP2 (Cairo et al. 1998), but also the increased ferritin expression in mouse embryonic fibroblasts and livers with accumulated IRP2, due to ablation of its repressor FBXL5 (Moroishi et al. 2011). The evolution of an alternative mechanism for ferritin synthesis that bypasses a global, or an mRNA-specific blockade in translation, underscores a profound role of this protein in cell survival and proliferation.
MATERIALS AND METHODS
Plasmids
The 5′ UTR of H-ferritin was PCR-amplified from a human genomic DNA (BD Bioscience). EcoR1/BamH1 and BamH1/EcoR1 restriction sites were added for cloning in the sense or antisense orientation within the ICS of the positive feedback vector (4UAS/RLuc/ICS/Gal4VP16) for identification of nucleotide sequences that enhance translation (Zhou et al. 2005) and to create the constructs 4UAS/Ftn(5′UTR) and 4UAS/aFtn(5′UTR), respectively (Fig. 5A). The sequences of the PCR primer pairs were as follows: Ftn(5′UTR), 5′-AAAAGAATTCAGGGCCAGACGTTCTTCGC-3′ (forward) and 5′-AAAAGGATCCGGCGGCGACTAAGGAGAGGCG-3′ (reverse); aFtn(5′UTR), 5′-AAAAGGATCCAGGGCCAGACGTTCTTCGC-3′ (forward), and 5′-AAAAGAATTCGGCGGCGACTAAGGAGAGGCG-3′ (reverse). XhoI and HindIII restriction sites were added for cloning into the promoterless PGL3Basic vector (Promega) that encodes a modified Firefly luciferase, yielding the construct pGL3Basic/Ftn(5′UTR); the PCR primers used were: 5′-CTCGAGAGGGCCAGACGTTCTTCG-3′ (forward) and 5′-AAGCTTGGCGGCGACTAAGGAGAG-3′ (reverse). The pGL3Basic/NF-κB (Lin et al. 2006) and pGL3Control (Promega) constructs express Firefly luciferase under the control of the NF-κB and SV40 promoters, respectively. For cloning H-ferritin 5′ UTR in the sense and reverse orientation into the bicistronic construct depicted in Figure 7A, HindIII and NcoI restriction sites were added by PCR, using the following primers: Ftn(5′UTR), 5′-AAAAATTCGAAAGGGCCAGACGTTCTTCGC-3′ (forward) and 5′-AAAAACCATGGGGCGGCGACTAAGGAGAGGCG-3′ (reverse); aFtn(5′UTR), 5′-AAAAAGGTACCAGGGCCAGACGTTCTTCGC-3′ (forward) and 5′-AAAAAAGCTTGGCGGCGACTAAGGAGAGGCG-3′ (reverse).
Cell culture and transfections
HT1080 human fibrosarcoma cells engineered for coumermycin-inducible expression of GyrB.PKR (Kazemi et al. 2004) were maintained in DMEM supplemented with 10% fetal bovine serum, 2 mM glutamine, 100 U/mL penicillin, 0.1 mg/mL streptomycin, in the presence of 200 μg/mL Zeocin (Invitrogen). Infections with the Sabin-like type I poliovirus strain were performed as described in Baltzis et al. (2007). Transfections were performed using the Lipofectamine Plus reagent (Invitrogen) following the manufacturer's recommendations. A Firefly luciferase reporter plasmid was cotransfected with the bicistronic constructs; the pRL-TK Renilla luciferase vector (Promega) was cotransfected with the pGL3 constructs. H1299 cells expressing IRPC437S under the control of a tetracycline-inducible promoter were maintained in supplemented DMEM in the presence of 2 μg/mL tetracycline, 2 μg/mL puromycine, and 250 μg/mL G418, and cultured as previously described (Wang and Pantopoulos 2002).
In vitro transcription and transfection of bicistronic RNAs
A total of 10 μg of template DNA was linearized with EcoRI, treated with proteinase K, and purified with phenol/chloroform. The purified, linearized templates were in vitro transcribed with the SP6 mRNA mMessage mMachine kit (Ambion) following the manufacturer's recommendations. The crude mRNAs were treated with DNAse I (Ambion) for 15 min at 37°C and then purified with phenol/chloroform. The integrity of the transcripts was monitored by denaturing agarose gel electrophoresis. The purified bicistronic mRNAs were used to transfect HT1080 cells at ∼90% confluence on 12-well plates. For this purpose, 1:100 dilution of DMRIE-C transfection reagent (Invitrogen) in opti-Mem media and 5 μg/mL mRNA were incubated with the cells (washed twice with opti-Mem) for 4 h at 37°C. Subsequently, opti-Mem was replaced with DMEM and the cells were incubated for another 16 h, before the activities of the Renilla and Firefly luciferases were measured in the cell lysates.
Western blotting
Cells were washed twice in phosphate-buffered saline (PBS) and lysed in a buffer containing 20 mM Tris-Cl (pH 7.4), 40 mM Kcl, and 1% Triton X-100. The lysates were resolved by SDS-PAGE on 12% gels and transferred onto nitrocellulose filters. The blots were saturated with 10% nonfat milk in PBS and probed with P-eIF2α (BioSource), ferritin (Novus), eIF4GI (a kind gift from Dr. Nahum Sonenberg, McGill University), FLAG (Sigma), and β-actin (Sigma) antibodies. The antibodies were diluted 1:500 (ferritin) or 1:1000 (all others) in PBS containing 5% nonfat milk and 0.5% Tween-20. After overnight incubation at 4°C, the blots were washed three times with PBS containing 0.5% Tween-20 and then incubated for 2 h at room temperature with 1:5000 diluted peroxidase-coupled goat anti rabbit IgG. Detection was performed with the enhanced chemiluminescence method (Amersham).
Metabolic labeling and immunoprecipitation
Cells were metabolically labeled for 2 h with 50 μCi/mL Trans-[35S]-label (Amersham), a mixture of 70:30 [35S]methionine/cysteine. The cells were washed with PBS and lysed in a buffer containing 50 mM Tris-Cl (pH 7.4), 300 mM NaCl, and 1% Triton X-100. After removing the cell debris by centrifugation, the cell lysates were subjected to immunoprecipitation with ferritin antibody (30 μg of antibody was used for 1000 μg of lysates). Immunoprecipitated proteins as well as the supernatants were analyzed by SDS-PAGE on a 12% gel. Radioactive bands and lanes were visualized by autoradiography.
Quantitative real time PCR (qPCR) from whole-cell lysates
Total RNA was extracted from cells using Trizol reagent (Invitrogen). A total of 1 μg of RNA was reverse transcribed using the QuantiTect Reverse Transcription Kit (Qiagen) following the manufacturer's recommendations. qPCR was performed on a MX 3005 Real-time PCR system (Stratagene) with the SYBR Green QuantiTect PCR reagent (Qiagen). H-ferritin-specific primers: 5′-GAGACCACAAGCGACCCGCA-3′ (forward) and 5′-GAGGTGACGGAGGGCTGGCT-3′ (reverse) and β-actin specific primers: 5′-AGGATGCAGAAGGAGATCAC-3′ (forward) and 5′-GGGTGTAACGCAACTAAGTC-3′ (reverse) were used for each sample. The Pfaffl method for relative quantitation was used for this experiment and the results are expressed as amount of H-ferritin mRNA normalized to the amount of β-actin mRNA.
Polysome fractionation
Cycloheximide (100 μg/mL) was added to the culture medium before harvesting the cells. The cells were washed twice with ice-cold PBS containing 100 μg/mL cycloheximide, and lysed on the plate in a buffer containing 150 mM Tris-HCl (pH 8.0), 300 mM NaCl, 1% Triton-X100, 5 mM MgCl2, 0.5 mM dithiothreitol, 1 mg/mL heparin, and 100 μg/mL cycloheximide. The lysates were scraped, transferred to pre-chilled 1.5-mL eppendorf tubes, incubated on ice for 10 min, and centrifuged in the cold room at 12,000 rpm for 10 min. Cytoplasmic RNA was obtained by lysis of the cell pellet in 1 mL of polysome buffer containing 10 mM Tris-HCl (pH 8.0), 140 mM NaCl, 1.5 mM MgCl2, 0.5% Nonidet P-40, and the ribonuclease inhibitor RNasin. After the removal of nuclei, the cytosolic supernatant was supplemented with 100 μg/mL cycloheximide, 665 μg/mL heparin, 20 mM dithiothreitol, and 1 mM phenylmethanesulfonyl fluoride. Mitochondria and membrane debris were removed by centrifugation, and the post-mitochondrial supernatant was overlaid onto a 15%–50% sucrose gradient and spun at 40,000 rpm for 2.5 h at 4°C in a Beckman L7-65 Vacuum Ultracentrifuge, using a SW40Ti rotor. Fractions (750 μL) were collected from the top of each gradient and the A260 was continuously monitored.
Quantitative real time PCR from polysomal fractions
Total RNA was extracted from each polysomal fraction by using Trizol reagent (Invitrogen). A total of 200 ng of RNA was reverse transcribed by using the QuantiTect Reverse Transcription Kit (Qiagen), following the manufacturer's recommendations. Real-time PCR was performed on an ABI Prism 7500 Fast System (Applied Biosystems) using the SYBR Green QuantiTect PCR reagent (Qiagen) and H-ferritin and β-actin-specific primers (described above). Serially diluted standards of cDNA of known concentration were prepared in order to make a standard curve. Aliquots of the same stock cDNAs were used to generate a standard curve on each plate. The threshold cycle was recorded for each sample and the amount of target gene in each sample was interpolated from the standard curve (Wong and Medrano 2005). Results are expressed as percentage of mRNA in the fractions lighter than 80S and heavier than 80S (amount of RNA in the corresponding fractions divided by the total amount of mRNA, which was calculated as the sum of the 24 fractions).
Analysis of polysome-associated H-ferritin mRNA for IRP1C437S binding
Fractions containing polyribosomes or monosomes were obtained as described in Yang et al. (2006). Briefly, cells were lysed in a polysomal buffer containing 10 mm MOPS (pH 7.2), 250 mm of NaCl, 2.5 mm of MgOAc, 0.5% NP-40, 1 mm of phenylmethanesulfonyl fluoride, 200 g/mL heparin and 50 g/mL cycloheximide for 10 min. Cellular debris and nuclei were removed by centrifugation at 12,000g for 10 min. The polysomes were precipitated by ultracentrifugation at 100,000g for 1 h in a 75 Ti rotor (Beckman) at 4°C. The monosomes remained in the supernatant while the polysomes were present in the pellet. The protein concentration in the polyribosomal fraction was determined with the Bradford method, and 3 mg of protein were immunoprecipitated with 10 μL of FLAG antibody (Sigma). Ten percent of the immunoprecipitate was used for Western blotting with the FLAG antibody, while 90% was used for RNA extraction using Trizol reagent (Invitrogen). A total of 500 ng of RNA extracted from the polyribosome-bound FLAG immunoprecipitate was reverse transcribed using the QuantiTect Reverse Transcription Kit (Qiagen), following the manufacturer's recommendations. Reverse-transcription PCR was performed with specific primers for H-ferritin: 5′-AGGGCCAGACGTTCTTCGC-3′ (forward) and 5′-GGCGGCGACTAAGGAGAGGCG-3′ (reverse); TfR1: 5′-GCAAGTAGATGGCGATAACAG-3′ (forward) and 5′-GACGATCACAGCAATAGTCCC-3′ (reverse); and GAPDH: 5′-ACCACAGTCCATGCCATCAC-3′ (forward) and 5′-TCCACCACCCTGTTGCTGTA-3′ (reverse). Total RNA was extracted from the whole-cell lysate and subjected to reverse-transcription PCR with the same pairs of primers.
Luciferase assays
All luciferase assays were performed using the Dual Luciferase Reporter System (Promega), following the manufacturer's recommendations.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
ACKNOWLEDGMENTS
We thank Dr. Yuri Svitkin (McGill University) for providing us with the poliovirus strain, Dr. Vincent P. Mauro (The Scripps Research Institute) for the bicistronic positive feedback vector, and Dr. Patrik J. Stover (Cornell University) for the classic bicistronic vector. We also thank Dr. Lisa Lindqvist for advice with polysome fractionation experiments. This work was supported by a grant (MOP-86514) from the Canadian Institutes for Health Research (CIHR). A.D. is a recipient of a fellowship from the Fonds de la recherche en santé du Quebéc (FRSQ). A.E.K. is funded by CIHR. K.P. holds a Chercheur National career award from FRSQ.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.029322.111.
REFERENCES
- Anderson P, Kedersha N 2008. Stress granules: the Tao of RNA triage. Trends Biochem Sci 33: 141–150 [DOI] [PubMed] [Google Scholar]
- Applegate LA, Scaletta C, Panizzon R, Frenk E 1998. Evidence that ferritin is UV inducible in human skin: part of a putative defense mechanism. J Invest Dermatol 111: 159–163 [DOI] [PubMed] [Google Scholar]
- Arosio P, Ingrassia R, Cavadini P 2009. Ferritins: A family of molecules for iron storage, antioxidation and more. Biochim Biophys Acta 1790: 589–599 [DOI] [PubMed] [Google Scholar]
- Baltzis D, Pluquet O, Papadakis AI, Kazemi S, Qu LK, Koromilas AE 2007. The eIF2α kinases PERK and PKR activate glycogen synthase kinase 3 to promote the proteasomal degradation of p53. J Biol Chem 282: 31675–31687 [DOI] [PubMed] [Google Scholar]
- Cairo G, Tacchini L, Pietrangelo A 1998. Lack of coordinate control of ferritin and transferrin receptor expression during rat liver regeneration. Hepatology 28: 173–178 [DOI] [PubMed] [Google Scholar]
- Chen J-J. 2000. Heme-regulated eIF2α kinase. In Translational control of gene expression (ed. N. Sonenberg et al.), pp. 529–546. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York. [Google Scholar]
- Chen G, Fillebeen C, Wang J, Pantopoulos K 2007. Overexpression of iron regulatory protein 1 suppresses growth of tumor xenografts. Carcinogenesis 28: 785–791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coccia EM, Profita V, Fiorucci G, Romeo G, Affabris E, Testa U, Hentze MW, Battistini A 1992. Modulation of ferritin H-chain expression in friend erythroleukemia cells: transcriptional and translational regulation by hemin. Mol Cell Biol 12: 3015–3022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cozzi A, Corsi B, Levi S, Santambrogio P, Albertini A, Arosio P 2000. Overexpression of wild type and mutated human ferritin H-chain in HeLa cells. In vivo role of ferritin ferroxidase activity. J Biol Chem 275: 25122–25129 [DOI] [PubMed] [Google Scholar]
- Darshan D, Vanoaica L, Richman L, Beermann F, Kuhn LC 2009. Conditional deletion of ferritin H in mice induces loss of iron storage and liver damage. Hepatology 50: 852–860 [DOI] [PubMed] [Google Scholar]
- Dix DJ, Lin PN, Kimata Y, Theil EC 1992. The iron regulatory region (IRE) of ferritin mRNA is also a positive control element for iron-dependent translation. Biochem 31: 2818–2822 [DOI] [PubMed] [Google Scholar]
- Dresios J, Chappell SA, Zhou W, Mauro VP 2006. An mRNA-rRNA base-pairing mechanism for translation initiation in eukaryotes. Nat Struct Mol Biol 13: 30–34 [DOI] [PubMed] [Google Scholar]
- Epsztejn S, Glickstein H, Picard V, Slotki IN, Breuer W, Beaumont C, Cabantchik ZI 1999. H-Ferritin subunit overexpression in erythroid cells reduces the oxidative stress response and induces multidrug resistance properties. Blood 94: 3593–3603 [PubMed] [Google Scholar]
- Fillebeen C, Rivas-Estilla AM, Bisaillon M, Ponka P, Muckenthaler M, Hentze MW, Koromilas AE, Pantopoulos K 2005. Iron inactivates the RNA polymerase NS5B and suppresses subgenomic replication of hepatitis C virus. J Biol Chem 280: 9049–9057 [DOI] [PubMed] [Google Scholar]
- Fox JT, Shin WK, Caudill MA, Stover PJ 2009. A UV-responsive internal ribosome entry site enhances serine hydroxymethyltransferase 1 expression for DNA damage repair. J Biol Chem 284: 31097–31108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gebauer F, Hentze MW 2004. Molecular mechanisms of translational control. Nat Rev Mol Cell Biol 5: 827–835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert WV 2010. Alternative ways to think about cellular internal ribosome entry. J Biol Chem 285: 29033–29038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halliwell B, Gutteridge JMC 1990. The role of free radicals and catalytic metal ions in human disease: an overview. Methods Enzymol 186: 1–85 [DOI] [PubMed] [Google Scholar]
- Hentze MW, Muckenthaler MU, Galy B, Camaschella C 2010. Two to tango: regulation of Mammalian iron metabolism. Cell 142: 24–38 [DOI] [PubMed] [Google Scholar]
- Hintze KJ, Theil EC 2005. DNA and mRNA elements with complementary responses to hemin, antioxidant inducers, and iron control ferritin-L expression. Proc Natl Acad Sci 102: 15048–15052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holcik M, Sonenberg N 2005. Translational control in stress and apoptosis. Nat Rev Mol Cell Biol 6: 318–327 [DOI] [PubMed] [Google Scholar]
- Kazemi S, Papadopoulou S, Li S, Su Q, Wang S, Yoshimura A, Matlashewski G, Dever TE, Koromilas AE 2004. Control of α subunit of eukaryotic translation initiation factor 2 (eIF2 α) phosphorylation by the human papillomavirus type 18 E6 oncoprotein: implications for eIF2 α-dependent gene expression and cell death. Mol Cell Biol 24: 3415–3429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komar AA, Hatzoglou M 2005. Internal ribosome entry sites in cellular mRNAs: mystery of their existence. J Biol Chem 280: 23425–23428 [DOI] [PubMed] [Google Scholar]
- Kwak EL, Larochelle DA, Beaumont C, Torti SV, Torti FM 1995. Role for NF-κ B in the regulation of ferritin H by tumor necrosis factor-α. J Biol Chem 270: 15285–15293 [DOI] [PubMed] [Google Scholar]
- Lin R, Lacoste J, Nakhaei P, Sun Q, Yang L, Paz S, Wilkinson P, Julkunen I, Vitour D, Meurs E, et al. 2006. Dissociation of a MAVS/IPS-1/VISA/Cardif-IKKε molecular complex from the mitochondrial outer membrane by hepatitis C virus NS3-4A proteolytic cleavage. J Virol 80: 6072–6083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maffettone C, Chen G, Drozdov I, Ouzounis C, Pantopoulos K 2010. Tumorigenic properties of iron regulatory protein 2 (IRP2) mediated by its specific 73-amino acids insert. PLoS ONE 5: e10163 doi: 10.1371/journal.pone.0010163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauro VP, Edelman GM 2007. The ribosome filter redux. Cell Cycle 6: 2246–2251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moroishi T, Nishiyama M, Takeda Y, Iwai K, Nakayama KI 2011. The FBXL5-IRP2 axis is integral to control of iron metabolism in vivo. Cell Metab 14: 339–351 [DOI] [PubMed] [Google Scholar]
- Mulvey MR, Kuhn LC, Scraba DG 1996. Induction of ferritin synthesis in cells infected with Mengo virus. J Biol Chem 271: 9851–9857 [DOI] [PubMed] [Google Scholar]
- Papanikolaou G, Pantopoulos K 2005. Iron metabolism and toxicity. Toxicol Appl Pharmacol 202: 199–211 [DOI] [PubMed] [Google Scholar]
- Pham CG, Bubici C, Zazzeroni F, Papa S, Jones J, Alvarez K, Jayawardena S, De Smaele E, Cong R, Beaumont C, et al. 2004. Ferritin heavy chain upregulation by NF-κB inhibits TNFα-induced apoptosis by suppressing reactive oxygen species. Cell 119: 529–542 [DOI] [PubMed] [Google Scholar]
- Proud CG 2005. eIF2 and the control of cell physiology. Semin Cell Dev Biol 16: 3–12 [DOI] [PubMed] [Google Scholar]
- Raven JF, Baltzis D, Wang S, Mounir Z, Papadakis AI, Gao HQ, Koromilas AE 2008. PKR and PKR-like endoplasmic reticulum kinase induce the proteasome-dependent degradation of cyclin D1 via a mechanism requiring eukaryotic initiation factor 2α phosphorylation. J Biol Chem 283: 3097–3108 [DOI] [PubMed] [Google Scholar]
- Svitkin YV, Cammack N, Minor PD, Almond JW 1990. Translation deficiency of the Sabin type 3 poliovirus genome: association with an attenuating mutation C472 → U. Virology 175: 103–109 [DOI] [PubMed] [Google Scholar]
- Thompson SR, Sarnow P 2000. Regulation of host cell translation by viruses and effects on cell function. Curr Opin Microbiol 3: 366–370 [DOI] [PubMed] [Google Scholar]
- Thompson K, Menzies S, Muckenthaler M, Torti FM, Wood T, Torti SV, Hentze MW, Beard J, Connor J 2003. Mouse brains deficient in H-ferritin have normal iron concentration but a protein profile of iron deficiency and increased evidence of oxidative stress. J Neurosci Res 71: 46–63 [DOI] [PubMed] [Google Scholar]
- Thomson AM, Cahill CM, Cho HH, Kassachau KD, Epis MR, Bridges KR, Leedman PJ, Rogers JT 2005. The acute box cis-element in human heavy ferritin mRNA 5′-untranslated region is a unique translation enhancer that binds poly(C)-binding proteins. J Biol Chem 280: 30032–30045 [DOI] [PubMed] [Google Scholar]
- Torti FM, Torti SV 2002. Regulation of ferritin genes and protein. Blood 99: 3505–3516 [DOI] [PubMed] [Google Scholar]
- Tsuji Y 2005. JunD activates transcription of the human ferritin H gene through an antioxidant response element during oxidative stress. Oncogene 24: 7567–7578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuji Y, Ayaki H, Whitman SP, Morrow CS, Torti SV, Torti FM 2000. Coordinate transcriptional and translational regulation of ferritin in response to oxidative stress. Mol Cell Biol 20: 5818–5827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ung TL, Cao C, Lu J, Ozato K, Dever TE 2001. Heterologous dimerization domains functionally substitute for the double-stranded RNA binding domains of the kinase PKR. EMBO J 20: 3728–3737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Pantopoulos K 2002. Conditional de-repression of ferritin synthesis in cells expressing a constitutive IRP1 mutant. Mol Cell Biol 22: 4638–4651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Pantopoulos K 2011. Regulation of cellular iron metabolism. Biochem J 434: 365–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wek RC, Jiang HY, Anthony TG 2006. Coping with stress: eIF2 kinases and translational control. Biochem Soc Trans 34: 7–11 [DOI] [PubMed] [Google Scholar]
- Woeller CF, Fox JT, Perry C, Stover PJ 2007. A ferritin-responsive internal ribosome entry site regulates folate metabolism. J Biol Chem 282: 29927–29935 [DOI] [PubMed] [Google Scholar]
- Wong ML, Medrano JF 2005. Real-time PCR for mRNA quantitation. Biotechniques 39: 75–85 [DOI] [PubMed] [Google Scholar]
- Xie C, Zhang N, Zhou H, Li J, Li Q, Zarubin T, Lin SC, Han J 2005. Distinct roles of basal steady-state and induced H-ferritin in tumor necrosis factor-induced death in L929 cells. Mol Cell Biol 25: 6673–6681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang DQ, Halaby MJ, Zhang Y 2006. The identification of an internal ribosomal entry site in the 5′-untranslated region of p53 mRNA provides a novel mechanism for the regulation of its translation following DNA damage. Oncogene 25: 4613–4619 [DOI] [PubMed] [Google Scholar]
- Zhou W, Edelman GM, Mauro VP 2005. A positive feedback vector for identification of nucleotide sequences that enhance translation. Proc Natl Acad Sci 102: 6273–6278 [DOI] [PMC free article] [PubMed] [Google Scholar]