The efficiency of trans-splicing ribozymes, which is currently too low for therapeutic applications, can be improved by careful choice of the targeted splice sites on the mRNA substrate. Here, the authors show that binding free energies derived from predictions of RNA secondary structure correlate well with the trans-splicing efficiencies experimentally determined on chloramphenicol acetyl transferase mRNA. Thus, the proposed computation of binding free energies provides a rapid and inexpensive method to identify efficient splice sites for trans-splicing ribozymes.
Keywords: group I intron, ribozyme, trans-splicing, secondary structure, free energy
Abstract
Group I introns have been engineered into trans-splicing ribozymes capable of replacing the 3′-terminal portion of an external mRNA with their own 3′-exon. Although this design makes trans-splicing ribozymes potentially useful for therapeutic application, their trans-splicing efficiency is usually too low for medical use. One factor that strongly influences trans-splicing efficiency is the position of the target splice site on the mRNA substrate. Viable splice sites are currently determined using a biochemical trans-tagging assay. Here, we propose a rapid and inexpensive alternative approach to identify efficient splice sites. This approach involves the computation of the binding free energies between ribozyme and mRNA substrate. We found that the computed binding free energies correlate well with the trans-splicing efficiency experimentally determined at 18 different splice sites on the mRNA of chloramphenicol acetyl transferase. In contrast, our results from the trans-tagging assay correlate less well with measured trans-splicing efficiency. The computed free energy components suggest that splice site efficiency depends on the following secondary structure rearrangements: hybridization of the ribozyme's internal guide sequence (IGS) with mRNA substrate (most important), unfolding of substrate proximal to the splice site, and release of the IGS from the 3′-exon (least important). The proposed computational approach can also be extended to fulfill additional design requirements of efficient trans-splicing ribozymes, such as the optimization of 3′-exon and extended guide sequences.
INTRODUCTION
Group I introns are catalytic RNAs (ribozymes) that excise themselves from primary transcripts (Kruger et al. 1982). These ribozymes have been well-characterized both biochemically and structurally, with five high-resolution crystal structures solved to date (Adams et al. 2004; Guo et al. 2004; Golden et al. 2005; Lipchock and Strobel 2008). Furthermore, it has been possible to convert the Tetrahymena cis-splicing group I intron into an artificial trans-splicing ribozyme that can modify the sequence of an arbitrary mRNA substrate. This was achieved by removing the 5′ portion of the wild-type Tetrahymena group I intron, thus exposing the intron's internal guide sequence (IGS) at the 5′ terminus (Inoue et al. 1985; Sullenger and Cech 1994). The resulting ribozyme can hybridize its IGS to a complementary target site on a substrate mRNA, forming a helix equivalent to the P1 duplex in the wild-type intron (Been and Cech 1986; Waring et al. 1986). The ribozyme then catalyzes cleavage of the substrate at the target site and transfer of the ribozyme 3′-exon to the remaining 5′ portion of the substrate (Zarrinkar and Sullenger 1998).
The ability to replace a portion of a substrate mRNA with the 3′-exon carried by the ribozyme has created potential roles for trans-splicing ribozymes in therapeutic applications (Fiskaa and Birgisdottir 2010). These ribozymes could be used to treat genetic disorders by repairing the sequence of a mutated mRNA (Sullenger and Cech 1994; Lan et al. 1998; Phylactou et al. 1998), to selectively kill cancer cells by splicing a sequence that encodes a toxic peptide into cancer-specific mRNAs (Ayre et al. 1999; Kwon et al. 2005; Lee et al. 2010), and to kill virally infected cells with the same strategy (Ayre et al. 1999; Ryu et al. 2003; Carter et al. 2010).
However, technical difficulties have so far prevented the medical use of trans-splicing ribozymes. One problem is the localized and efficient delivery of therapeutic RNAs to the affected tissues (Guo et al. 2010; Gao et al. 2011), though solutions involving viral vectors and modified Salmonella strains appear to be promising (Thomas et al. 2003; Bai et al. 2011). Another problem of trans-splicing ribozymes is their low efficiency in cells. Here, trans-splicing efficiency denotes the fraction of target mRNA that is converted to product by the ribozymes. Trans-splicing efficiency is typically 10% or less in cells (Fiskaa and Birgisdottir 2010). Although efficiencies of up to 50% have been reported (Byun et al. 2003), the necessary intracellular ribozyme concentrations appear too high to be acceptable in a clinical setting. Therefore, an outstanding task for the development of trans-splicing ribozymes is to increase their efficiency, allowing a sufficient fraction of mRNA substrate to be trans-spliced at low ribozyme concentrations.
The trans-splicing efficiency varies strongly with the location of the splice site within the mRNA substrate (Campbell and Cech 1995; Lan et al. 2000). In particular, secondary structures within the substrate can render potential target sites inaccessible to the ribozyme. The trans-splicing efficiency may also vary between splice sites owing to different stabilities of the P1 duplex formed between ribozyme and substrate and to different base-pairing interactions between the ribozyme IGS and the ribozyme 3′-exon. Thus, the identification of efficient splice sites, i.e., those sites that mediate a high trans-splicing efficiency, is a nontrivial problem in the design of trans-splicing ribozymes.
An elegant experimental method, known as the trans-tagging assay, has been developed to identify efficient splice sites (Jones et al. 1996). In this assay, the mRNA substrate is incubated in vitro with a pool of trans-splicing ribozymes whose IGS is randomized. This pool of ribozymes is thus able to recognize any potential splice site on the substrate. Ribozymes targeting efficient splice sites are able to trans-splice at those sites. The resulting products are detected by reverse transcription, subcloning, and sequencing. The sequences from several clones are then used to deduce the positions of efficient splice sites (Jones et al. 1996; Lan et al. 1998; Einvik et al. 2004). This trans-tagging assay has been successfully applied—without explicitly reported limitations—to uncover efficient splice sites on various mRNA substrates (Lan et al. 1998; Watanabe and Sullenger 2000; Rogers et al. 2002; Park et al. 2003; Ryu and Lee 2003; Jung and Lee 2005; Fiskaa et al. 2006; Jung and Lee 2006; Kim et al. 2007).
The problem of identifying efficient splice sites for trans-splicing ribozymes is, in concept, similar to that of finding accessible or optimal binding sites on mRNA substrates targeted by antisense DNA oligonucleotides (Chan et al. 2006), siRNAs (Shao et al. 2007a; Walton et al. 2010), and other non-protein-coding RNAs (Pichon and Felden 2008). In these cases, several computational methods have been proposed and found to be effective at predicting the relevant target sites (Ding and Lawrence 2001; Wang and Drlica 2004; Far et al. 2005; Shao et al. 2006; Tafer et al. 2008; Backofen and Hess 2010; Thomas et al. 2010). One recurring theme is the use of RNA secondary structure prediction algorithms to calculate the free energy changes, ΔGbind, associated with binding an oligonucleotide to each possible target site on the mRNA substrate. Target sites with strongly negative values of ΔGbind are then predicted to be the most accessible sites on the substrate (Mathews et al. 1999; Walton et al. 1999; Busch et al. 2008; Lu and Mathews 2008; Tafer et al. 2008).
Here, we have adapted this approach of computing ΔGbind to the problem of finding efficient splice sites for trans-splicing ribozymes. The mRNA of chloramphenicol acetyl transferase (CAT) was used as a model substrate for both computations and experiments. In particular, we computed ΔGbind for all 186 splice sites on this substrate, and we experimentally determined the trans-splicing efficiency in vitro for 18 of those splice sites. Furthermore, to map the accessible splice sites on CAT mRNA, we carried out the trans-tagging assay, which differs substantially from direct measurements of trans-splicing efficiency. Surprisingly, the experimental trans-splicing efficiencies at the 18 tested splice sites were found to correlate better with the computed ΔGbind values than with the results of the trans-tagging assay. These results suggest that the proposed calculation of ΔGbind could be used to predict efficient splice sites for trans-splicing ribozymes more quickly, cheaply, and accurately than with the presently used experimental methods.
RESULTS
Calculation of ΔGbind for trans-splicing ribozymes
We defined ΔGbind as the free energy change associated with the binding of the ribozyme's IGS to a given splice site on the mRNA substrate. Our hypothesis was that a very negative ΔGbind corresponds to a high trans-splicing efficiency. To calculate ΔGbind, the binding process was modeled as consisting of three idealized molecular events that involve only local changes in RNA secondary structure: the unfolding of the target site on the substrate, the release of the IGS on the ribozyme, and the hybridization between target site and IGS to form the P1 duplex (Fig. 1A). The corresponding component free energy changes were denoted by ΔGunfold-target, ΔGrelease-IGS, and ΔGhybrid, respectively. By “release” of the IGS, we mean the breaking up of any base pairs that may form between the IGS and the 3′-exon. This release is necessary to make the IGS available for binding to the target site. We thus computed ΔGbind by summing the above components, i.e., ΔGbind = ΔGunfold-target + ΔGrelease-IGS + ΔGhybrid. These components can, in turn, be computed using several available RNA folding algorithms, which predict the possible secondary structures for a given RNA sequence as well as the free energy changes of those structures relative to the unfolded state (Condon and Jabbari 2009).
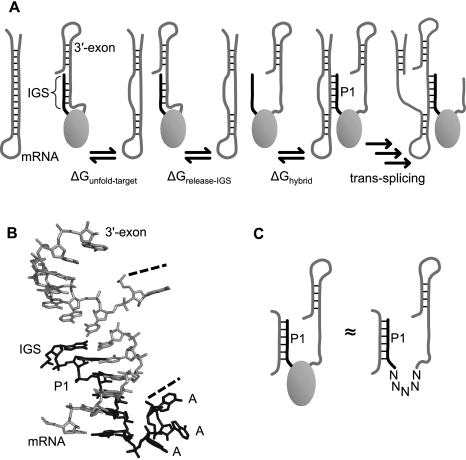
FIGURE 1.
Interactions within and between the mRNA substrate and the ribozyme, and their treatment during computation. (A) Schematic of the substrate binding step in trans-splicing. The mRNA substrate is illustrated as a hairpin structure, and the body of the ribozyme is illustrated as a gray oval. Unfolding of the target site on the mRNA (first step) and release of the internal guide sequence (IGS) (second step) allow the mRNA target site and the IGS to hybridize (third step). (B) The 3D structure of the Tetrahymena ribozyme (Lehnert et al. 1996) indicates that the AAA linker (black) is not stacked on the P1 duplex. Additionally, the 3′-exon (gray) is close enough to the IGS (black) for interactions if the mRNA (gray) is absent. (C) To facilitate computational treatment of the binding process, the body of the ribozyme was replaced by a linker sequence joining the IGS to the 3′-exon. This linker does not include the natural AAA linker but consists of five unspecified nucleotides (N), which are ignored in the calculation of base-pairing and π-stacking interactions.
To compute the components of ΔGbind for trans-splicing ribozymes that interact with a long mRNA substrate, we adapted the ribozyme and mRNA sequences before submission to RNA folding algorithms. The 3D model of the Tetrahymena ribozyme (Lehnert et al. 1996) suggests that, before pairing to an mRNA target site, the IGS is sterically allowed to base-pair with the 3′-exon but does not base-pair with the body of the folded intron (Fig. 1B). Thus, to calculate ΔGrelease-IGS, we used a ribozyme sequence in which the intron portion downstream from the IGS was replaced by a 5-nt linker region (Fig. 1C). This shortened sequence allowed the IGS to interact with the 3′-exon, while permitting rapid computation of ΔGrelease-IGS by RNA folding algorithms. To choose an appropriate linker sequence, we noted that, in the Tetrahymena group I intron, the IGS is followed by three unpaired adenosines, and the 3′-exon is preceded by one unpaired guanosine (Cech et al. 1994). Although these nucleotides seem natural candidates for inclusion in the linker, the 3D structure of the ribozyme shows that these nucleotides are conformationally prohibited from pairing with the substrate and from engaging in π-stacking interactions with the IGS or the 3′-exon (Fig. 1B). Therefore, we chose the linker sequence to consist of five unspecified nucleotides, which are effectively ignored by RNA folding algorithms in the calculation of ΔGrelease-IGS and ΔGhybrid. Varying the length of the linker from four to seven unspecified nucleotides did not significantly change the computed ΔG values (data not shown).
To predict ΔGunfold-target from the local secondary structure around each splice site on a long mRNA substrate, we directed the RNA folding algorithms to analyze subsets, or windows, of the entire substrate sequence. Thus, only the sequence within each window containing a given splice site was used to calculate ΔGunfold-target for that site. For the 678-nt sequence of our model mRNA (see below), the resulting values of ΔGunfold-target were found to vary by >1 kcal/mol over different window sizes (data not shown). This variation may be due to the somewhat arbitrary exclusion of base pairs involving nucleotides outside of a given window. To minimize the influence of window size on the predicted ΔGbind, we computed ΔGunfold-target using window sizes of 100, 200, 300, 400, 500, and 600 nt, and then we averaged the resulting values of ΔGbind.
Splice sites chosen on a model mRNA substrate, CAT mRNA
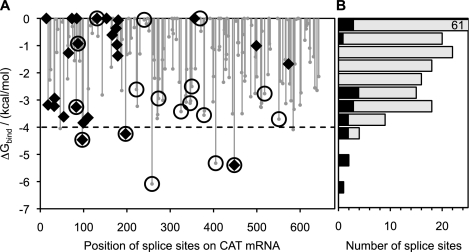
As a model mRNA substrate for our computational and experimental investigations of trans-splicing efficiency, we chose the mRNA of chloramphenicol acetyl transferase. This mRNA sequence is 678 nt long and contains 186 uridines downstream from the AUG translation start codon. Each of these uridines represents a potential splice site for trans-splicing ribozymes (Doudna et al. 1989). We thus computed ΔGbind for each of these splice sites. Figure 2A plots the resulting ΔGbind values as a function of splice site position relative to the adenine of the AUG start codon. These values are distributed over the range from 0 to −6.5 kcal/mol, with most values closer to 0 kcal/mol (Fig. 2B). In particular, only seven out of the 186 available splice sites had ΔGbind < −4.0 kcal/mol, suggesting that this value of ΔGbind could be used as a threshold to predict efficient splice sites.
FIGURE 2.
Computed values of ΔGbind for splice sites on CAT mRNA. (A) Plot of ΔGbind against splice site position relative to the adenosine (position 1) of the AUG transcription start codon. Only splice sites downstream from this codon were mapped by the trans-tagging assay in this study. Circles with or without a diamond inside indicate splice sites chosen for experimental determination of trans-splicing efficiency. Diamonds indicate splice sites found among 66 product sequences obtained by trans-tagging assay. (B) Distribution of the 186 splice sites (gray bars) of CAT mRNA over the computed values of ΔGbind. Black bars represent the 18 splice sites that were chosen for experimental testing. The topmost bar represents 61 splice sites and is shown truncated.
To determine experimentally whether the computed values of ΔGbind can be used to identify efficient splice sites, we chose a set of 18 different target splice sites on CAT mRNA. To reveal possible correlations between trans-splicing efficiency and ΔGbind, as well as between trans-splicing efficiency and position on mRNA substrate, these 18 sites were chosen to cover the full range of ΔGbind values (Fig. 2B) and were distributed over the length of the CAT mRNA (Fig. 2A). Two splice sites with ΔGbind slightly below −4.0 kcal/mol were omitted in order to cover the wide range of the predicted values for ΔGbind with the available resources.
Experimental trans-splicing efficiencies on CAT mRNA
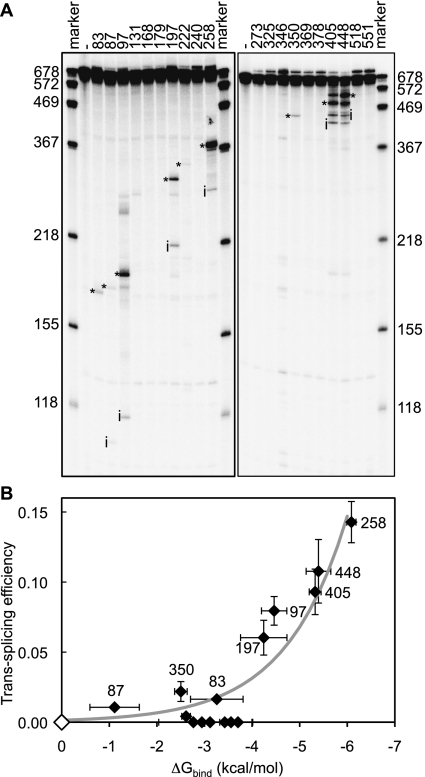
To measure experimentally the trans-splicing efficiency of the 18 chosen splice sites, we generated 18 trans-splicing ribozymes that contained different IGSs but were otherwise identical. Each IGS was designed to target a particular one of the chosen splice sites (Supplemental Table S1). Each of the 18 ribozymes was incubated with 5′-radiolabeled CAT mRNA, and the reaction products were analyzed by polyacrylamide gel electrophoresis and autoradiography (Fig. 3A). As a measure of trans-splicing efficiency, we quantified the fraction of radiolabeled substrates converted to trans-splicing products after 4 h of incubation. Bands consistent with the expected trans-splicing products were observed for nine of the 18 tested splice sites, namely for splice sites at uridines 83, 87, 97, 197, 222, 258, 350, 405, and 448. Product fractions for these splice sites ranged from 0.4% of the substrate at uridine 222 to 14.3% at uridine 258 (Supplemental Table S1). Nine of the 18 splice sites did not yield quantifiable products.
FIGURE 3.
Trans-splicing efficiency measured on 18 splice sites on CAT mRNA. (A) Autoradiogram of reaction products from trans-splicing reactions with radiolabeled substrate. Lanes are labeled with the targeted splice sites, or with (−) where ribozyme was absent. RNA marker sizes in nucleotides are indicated. The unreacted substrate had a length of 678 nucleotides, whereas the length of reaction intermediates (i) and trans-splicing products (*) varied with the splice sites. Note that the ribozymes targeting splice sites 405 and 448 also spliced at each other's splice site. (B) Correlation between computed binding free energies and experimental trans-splicing efficiencies. Each diamond represents a specific splice site. Splice sites 131, 240, and 369 have ΔGbind ≥ 0 kcal/mol and are represented by a white diamond at the origin. The data points near or over the ΔGbind-axis around the value of −3 kcal/mol correspond, from left to right, to splice sites 222, 518, 273, 346, 325, 378, and 551. The thick gray line represents a least-mean-squares exponential fit, y = A exp(B x), to the data points, with a coefficient of determination R2 = 0.87. Alternatively, a linear fit (data not shown) to the same data points yields a correlation coefficient R = −0.75, with a probability p = 0.00033 that the values are not correlated. Horizontal error bars are standard deviations over three to six different window sizes (100–600 nt). Vertical error bars are standard deviations from three independent experiments.
The experimental trans-splicing efficiencies were found to agree with the computed ΔGbind values (Fig. 3B). In particular, the efficiency of splice sites with ΔGbind > −4 kcal/mol was relatively low, reaching, at most, 2.2%. On the other hand, the efficiency of splice sites with ΔGbind < −4 kcal/mol increased steadily with increasingly more negative values of ΔGbind, reaching 14.3% at ΔGbind = −6.1 kcal/mol. Only the five most efficient splice sites had a computed ΔGbind < −4.0 kcal/mol, again suggesting that efficient splice sites could be predicted by comparing the computed ΔGbind to the empirical threshold of ΔGbind ≈ −4.0 kcal/mol.
To test the identity of the observed trans-splicing products, we reverse-transcribed, cloned, and sequenced the products of all nine reactions that had resulted in quantifiable bands on polyacrylamide gels (sequence data not shown). The expected product sequences were obtained for eight of the nine tested splice sites. Product sequences for splice site 350, which did not yield the expected product, indicated that the corresponding ribozyme had spliced primarily at uridine 307. Therefore, the actual trans-splicing efficiency at splice site 350 may be lower than its measured trans-splicing efficiency (2.2%), possibly improving further the correlation between computed ΔGbind and trans-splicing efficiency seen in Figure 3B. In summary, the sequencing results confirmed all but one of the nine products observed in the trans-splicing reactions.
Experimental trans-splicing efficiency with unstructured RNA substrates
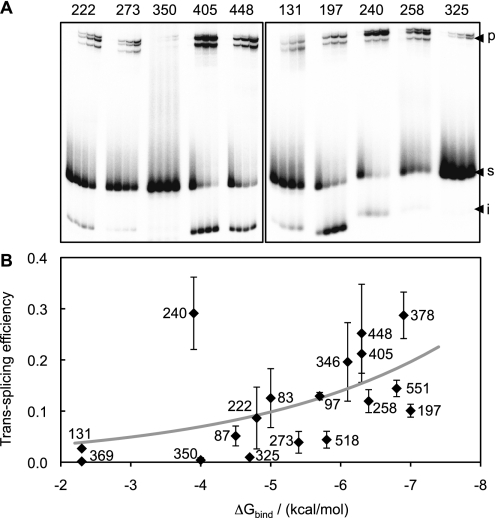
To confirm that all of the 18 ribozymes employed in experiments with CAT mRNA were catalytically active, we measured their trans-splicing efficiency on short substrates that were designed to prevent formation of secondary structure. Hence, this experiment also allowed us to examine the effects of substrate secondary structure on trans-splicing efficiency. Each short substrate consisted of a 13-nt sequence containing one of the 18 splice sites already targeted on CAT mRNA. The single-stranded conformation predicted by RNA folding algorithms for these substrates effectively abolishes the hypothesized ΔGunfold-target component of ΔGbind. The reaction conditions were the same as with the CAT mRNA, except that the incubation time was 30 times shorter and the ribozyme concentration was 15 times lower. The reaction products were then analyzed by polyacrylamide gel electrophoresis and autoradiography (Fig. 4A). The trans-splicing product fractions ranged from 0.16 ± 0.07%, for splice site 369, to 29 ± 7%, for splice site 240 (Supplemental Table S1), confirming that all ribozymes were, indeed, active in the absence of substrate secondary structure. Because the short substrates required considerably shorter reaction times and lower ribozyme concentrations to yield product fractions comparable to those obtained with CAT mRNA, our results reinforce the notion that substrate folding can significantly reduce trans-splicing efficiency.
FIGURE 4.
Trans-splicing efficiency with short, 13-nt substrates. (A) Representative autoradiograms of products from trans-splicing reactions. Each 13-mer substrate contains one of the 18 splice sites that were targeted on CAT mRNA. Each group of lanes shows samples taken after 1, 8, 16, and 31 min of reaction and is labeled with the position of the targeted splice site. The positions of the unreacted substrate (s; 13 nt), reaction intermediate (i; 8 nt), and trans-splicing product (p; 85 nt) are indicated. The size of the product was confirmed by comparison with an 85-nt RNA marker in separate experiments (data not shown). (B) Plot of experimental trans-splicing efficiency as a function of computed ΔGbind. The thick gray line represents a least-mean-squares exponential fit to the data points, with a coefficient of determination R2 = 0.27. Only splice site 240 (−3.9 kcal/mol; 29% reacted) results in a strong deviation from this trend. Alternatively, a linear fit (data not shown) to the same data points yields a correlation coefficient R = −0.51, with a probability p = 0.030 that the values are not correlated. Error bars are standard deviations from three independent experiments.
For almost all ribozymes, the expected trans-splicing product of 85 nt (8 nt of cleaved substrate plus 77 nt of 3′-exon) was accompanied by two other products, whose lengths were ∼80 and 90 nt (Fig. 4A). The additional products most likely resulted from one or more of the possible side reactions in which group I intron ribozymes are known to engage (Kay and Inoue 1987; Johnson et al. 2005; Dotson et al. 2008). These side reactions depend on the same process modeled in this study, i.e., the binding of ribozyme's IGS to substrate target site. Nevertheless, because the identities of the side products were not determined experimentally and because our focus was on the correct trans-splicing product, we took the fraction of the 85-nt product alone as a measure of trans-splicing efficiency for splice sites on the short substrates. The ensuing qualitative correlation of computed ΔGbind with experimentally measured efficiency (see below) was not strongly affected when the sum of all three product fractions was used as a measure of trans-splicing efficiency (data not shown).
To determine whether the two remaining energetic contributions, P1 duplex stability, modeled by ΔGhybrid, and base-pairing interactions between IGS and 3′-exon, modeled by ΔGrelease-IGS, were able to explain the observed variation in trans-splicing efficiency for splice sites on the short substrates, we plotted the trans-splicing efficiency measured for each splice site as a function of ΔGbind computed for that site (Fig. 4B). This plot reveals a correlation between calculations and experiments that is less tight than the correlation for the full-length CAT mRNA, suggesting that factors other than P1 duplex stability and IGS/3′-exon interactions might influence trans-splicing efficiency. One reason why these factors become important for unstructured substrates (Fig. 4B) but not for structured substrates (Fig. 3B) may be the faster trans-splicing kinetics with short substrates. Several processes may play a stronger role at faster reaction kinetics, such as the initial folding of the ribozyme (Pan et al. 1997; Shi et al. 2009) and later steps in the trans-splicing process (Karbstein et al. 2002; Bell et al. 2004), which are not captured by the thermodynamic model used in this study.
Computed energetic contributions to trans-splicing efficiency
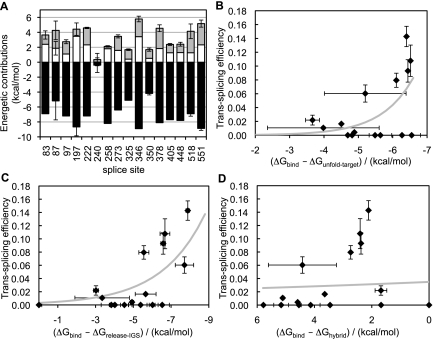
The good correlation observed between computed values of ΔGbind and experimental trans-splicing efficiencies for CAT mRNA supports the notion that the ribozyme-substrate binding process can be approximately modeled by assuming three idealized molecular events: unfolding of target site, release of the IGS, and IGS-target site hybridization. Here, we examine the relative energetic contributions of these hypothetical molecular events to the observed efficiency of a given splice site.
The thermodynamic model (Fig. 1A) suggests that a very negative hybridization energy, ΔGhybrid, would favor trans-splicing, whereas very positive energies of substrate unfolding, ΔGunfold-target, and of IGS release, ΔGrelease-IGS, would disfavor trans-splicing. Figure 5A and Supplemental Table S2 show the three energy components of ΔGbind for each of the experimentally tested splice sites on CAT mRNA. The average of ΔGunfold-target over all splice sites is 1.8 ± 0.8 kcal/mol, the average of ΔGrelease-IGS is 1.8 ± 0.9 kcal/mol, and the average of ΔGhybrid is −7.2 ± 1.4 kcal/mol. Thus, both the strongest energetic contribution (−7.2 kcal/mol) and the largest variation among splice sites (±1.4 kcal/mol) come from the hybridization of IGS with the target sites. These results indicate that the free energy of IGS-target site hybridization is more important, on average, than the free energies of target unfolding and IGS release in modulating ΔGbind.
FIGURE 5.
Energetic contributions to the computed binding free energy on CAT mRNA, from the three molecular events described in Figure 1A. (A) The computed energetic contributions from target site unfolding (ΔGunfold-target, gray), ribozyme IGS release (ΔGrelease-IGS, white), and IGS-target site hybridization (ΔGhybrid, black) are shown for each tested splice site. Three of the 18 tested splice sites were omitted (131, 240, 369) because their computed energetic values were not among the strongest 10,000 interactions reported by IntaRNA. Error bars are standard deviations of the energies calculated with three to six different window sizes as in Figure 3. (B–D) Plots of trans-splicing efficiency as a function of computed ΔGbind values when the contribution of (B) ΔGunfold-target, (C) ΔGrelease-IGS, and (D) ΔGhybrid is omitted in turn. The thick gray lines in B–D represent exponential fits, as in Figures 3B and 4B, with coefficients of determination R2 = 0.38, 0.57, and 0.076 for B, C, and D, respectively. For additional details, see Figure 3.
To assess the importance of the energetic components ΔGunfold-target, ΔGrelease-IGS, and ΔGhybrid toward the correlation observed between experimental trans-splicing efficiency and ΔGbind (Fig. 3B), we recomputed ΔGbind by omitting each component in turn, and we calculated the coefficient of determination associated with a least-mean-squares exponential fit to the experimental trans-splicing efficiency as a function of computed ΔGbind. For splice sites on CAT mRNA, omitting any of the energetic components resulted in a poorer correlation and a consequent loss of a reliable threshold bounding values of ΔGbind associated with efficient splice sites (Fig. 5B–D). In particular, omission of ΔGhybrid resulted in a nonnegative ΔGbind for all splice sites (Fig. 5D), whereas omission of either ΔGunfold-target or ΔGrelease-IGS resulted in the appearance of up to six false-positives, i.e., splice sites with ΔGbind < −4 kcal/mol but without any experimentally detected trans-splicing product (Fig. 5B,C). A poor correlation also ensued when omitting pairs of ΔG components from ΔGbind (Supplemental Fig. S2; Supplemental Table S2). Comparing the coefficients of determination (Figs. 3B, 5B–D) indicates that the importance of individual energetic contributions toward the observed correlation between trans-splicing efficiency and ΔGbind follows the order ΔGhybrid > ΔGunfold-target > ΔGrelease-IGS, but these energetic contributions are all necessary to achieve a good correlation.
When the same analysis was performed for splice sites on the short substrates, we found that omitting ΔGunfold-target made no significant difference because this component was always <0.4 kcal/mol, as expected (Supplemental Fig. S1A,B). Omitting either of the other components resulted in a worse correlation (Supplemental Fig. S1C,D). The values of ΔGrelease-IGS for the 13-mers were identical to the values for corresponding splice sites on CAT mRNA, which was expected because ΔGrelease-IGS does not depend on the substrate. Values of ΔGhybrid, however, differed by up to ∼1 kcal/mol between corresponding splices sites on 13-mers and CAT mRNA. These small energetic differences arose from the different nucleotides flanking the target sites in the two contexts. Such nucleotides vary with splice site on CAT mRNA (Supplemental Table S1) but do not vary with splice site on the short substrates (see Materials and Methods).
Trans-tagging assay on CAT mRNA
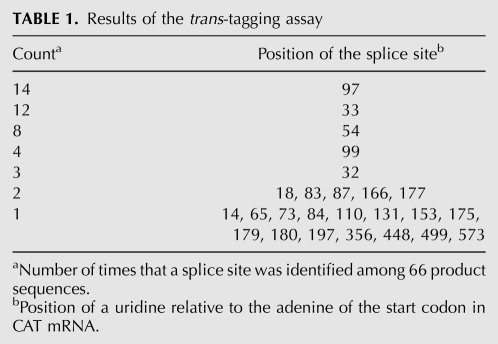
To determine whether the results of the trans-tagging assay also generate a good correlation with the experimentally determined trans-splicing efficiencies, we performed this assay using CAT mRNA as the substrate. The output of the trans-tagging assay is the number of times a given splice site is identified in a set of product sequences obtained from in vitro trans-splicing reactions with randomized IGSs (Jones et al. 1996). We obtained a total of 66 product sequences which were consistent with trans-splicing at 25 different uridines among the 186 uridines that follow the start codon in the CAT mRNA sequence (Table 1). The splice sites detected most frequently were at uridines 97 and 33, with 14 and 12 occurrences, respectively. On the other hand, 15 of the 25 detected splice sites were found only once. The splice sites detected by the trans-tagging assay are represented by diamonds in Figure 2A.
TABLE 1.
Results of the trans-tagging assay
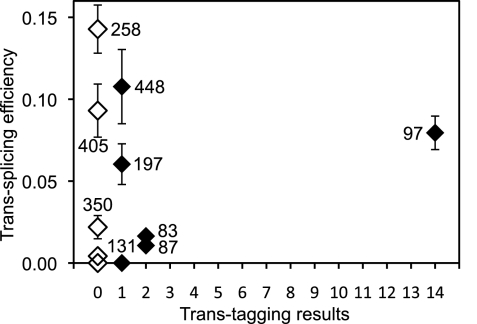
When the experimentally determined trans-splicing efficiencies were plotted as a function of the splice site counts in the trans-tagging assay, no correlation was evident (Fig. 6). More strikingly, the assay did not find splice sites 258 and 405, even though these splice sites were more efficient than splice site 97, which was detected 14 times by the trans-splicing assay. Similarly, the assay found each of the splice sites 448 and 197 only once in 66 product sequences, even though these splice sites were two of the five most efficient ones among the 18 tested splice sites. Therefore, the number of times that a given splice site is found by the trans-tagging assay does not necessarily reflect the trans-splicing efficiency measured for that splice site.
FIGURE 6.
Comparison between trans-splicing efficiency and trans-tagging results. The measured trans-splicing efficiency for each splice site is plotted against the number of times that the splice site was found in 66 product sequences obtained with the trans-tagging assay (filled diamonds). Open diamonds denote splice sites that were not found by the trans-tagging assay. Note that eight of the 18 sequences are clustered at the origin of the plot.
Possible biases of the trans-tagging assay
Why did the trans-tagging results only partially reflect the measured splice site efficiency? One possible explanation is that the PCR step in the assay disfavors long RT-PCR products, a well-known phenomenon in quantitative PCR (Bustin 2000). In our trans-tagging experiments, this phenomenon may have caused the pattern seen in Figure 2A. Here, the forward PCR primer was designed to identify splice sites between positions 14 and 643 on CAT mRNA, but only four out of the 25 identified splice sites were found at positions beyond uridine 197 (Table 1). In contrast, the five most efficient splice sites (also with ΔGbind below the threshold of −4 kcal/mol) were more evenly distributed over the CAT mRNA (positions 97, 197, 258, 405, and 448). We reasoned that if the PCR step, indeed, caused more splice sites to be identified near the forward PCR primer than elsewhere, then moving this primer farther downstream from the mRNA 5′ terminus should reveal a different pattern of detected splice sites, now crowded near the new position of the forward primer. Therefore, we repeated the trans-tagging experiment on CAT mRNA, this time using a forward primer designed to identify only splice sites at positions 207 to 643. The resulting nine product sequences yielded eight new splice sites, those at uridines 248, 271, 273, 321, 350, 378, 384, and 405 (data not shown in Fig. 2A). None of these splice sites had been found among the previous set of 66 trans-tagging product sequences. The new splice sites were again crowded near the forward primer. These results suggest that the PCR step may cause the trans-tagging assay to miss efficient splice sites located 200 nt or more downstream from the forward PCR primer.
A second possible explanation for a bias in the trans-tagging assay lies in the different rates at which ribozymes with different IGSs are transcribed by T7 RNA polymerase in vitro. The assay employs a pool of trans-splicing ribozymes whose IGS differs between ribozymes. Because the IGS represents the 5′ terminus of the transcript and because the transcription efficiency of T7 RNA polymerase is strongly dependent on the sequence of the first 6 nt in the transcript (Milligan et al. 1987; Milligan and Uhlenbeck 1989) we hypothesized that the difference in transcription efficiency achieved for different ribozymes may bias the results of the trans-tagging assay.
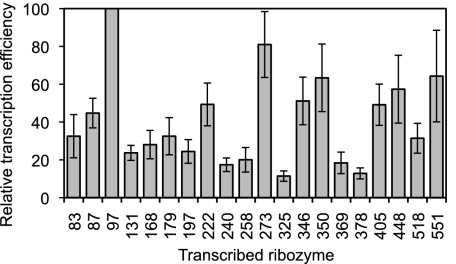
To test this second hypothesis, we measured the transcription efficiency for the 18 studied ribozymes. Specifically, the ribozymes were individually transcribed in the presence of α-[32P]-GTP, the transcription products were separated by denaturing polyacrylamide gel electrophoresis, and the transcription efficiency was measured by quantitating the relevant bands on autoradiograms. The measured transcription efficiency varied eightfold among the different ribozymes (Fig. 7; Supplemental Table S1). Notably, ribozyme 97 had the highest transcription efficiency, fivefold higher than that of ribozyme 258. These measured differences in ribozyme amounts represent lower bounds for the actual differences affecting the trans-tagging assay. Specifically, in the individual transcription reactions, partial saturation may have been reached for the more efficient transcriptions but not for the less efficient ones. This partial saturation would have reduced the observed difference between efficient and less efficient transcriptions. On the other hand, in the trans-tagging assay, all ribozymes were transcribed together from equimolar amounts of templates under the same reaction conditions, leading to equal extents of product saturation for all ribozymes. Hence, the difference in the amounts of ribozymes 97 and 258 was likely greater than fivefold in the trans-tagging assay. Therefore, the observed bias in transcription efficiency for ribozymes with different IGSs could explain why splice site 97 was found most frequently in the trans-tagging assay, while splice site 258, which was the most efficient in trans-splicing experiments with CAT mRNA, was absent in all 75 product sequences that we obtained with the trans-tagging assay.
FIGURE 7.
Transcription efficiencies of ribozymes analyzed in this study. All values are relative to the transcription efficiency of the ribozyme targeting splice site 97. Transcription efficiencies were determined from band intensities of internally [32P]-labeled ribozymes, which had been transcribed in vitro and separated by denaturing polyacrylamide gel electrophoresis. The targeted splice site is indicated at the bottom of each bar. Error bars are standard deviations from seven independent transcriptions.
A third potential source of bias for the outcome of the trans-tagging assay is the tendency of group I intron variants to lose their 3′-exon via side reactions known as 3′-specific hydrolysis and G-exchange (Inoue et al. 1986; Thompson and Herrin 1991; van der Horst and Inoue 1993; Haugen et al. 2004). Indeed, a significant loss of 3′-exons was revealed during the separation of in vitro transcribed ribozymes on denaturing polyacrylamide gels (above). Specifically, for each band of a full-length ribozyme, we detected a faster migrating band, which corresponded in size to ribozymes that had lost their 3′-exon. If the loss of 3′-exons during transcription and during trans-splicing depends on the IGS, then the pool of transcribed ribozymes used in the trans-tagging assay will be biased against specific splice sites.
To determine whether the loss of 3′-exons varied with IGS under in vitro transcription conditions, we quantitated the relevant bands on the same autoradiograms that were used to measure transcription efficiency. The resulting fractions of lost 3′-exons varied from 12 ± 4% to 39 ± 4%, with an average of 28 ± 7% over all 18 ribozymes (Supplemental Table S1). This means that, among the studied ribozymes, between 88% and 61% of the transcribed ribozymes retained their 3′-exon, generating only a 1.4-fold bias for the trans-tagging assay. Therefore, 3′-exon loss during transcription was not a major contributor to the large bias observed in the trans-tagging assay.
The loss of 3′-exons could also have taken place during the trans-splicing reaction. Because these conditions were different from the transcription conditions, we also measured the loss of 3′-exons under trans-splicing conditions. Internally radiolabeled ribozymes with a 3′-exon were size-purified by denaturing PAGE, then incubated under trans-splicing conditions, and the fraction of ribozymes that lost their 3′-exon was determined by a second denaturing PAGE and quantitation by phosphorimaging. The fraction of 3′-exons lost after four hours varied from 22 ± 4% to 65 ± 7% with an average of 40 ± 10% over all ribozymes (Supplemental Table S1). Therefore, we estimate that, during the 1-h incubation under trans-tagging conditions, between ∼80% and ∼95% of the ribozymes retained their 3′-exon, suggesting that the small proportion of ribozymes that lost their 3′-exon should not constitute a major bias in the trans-tagging assay.
In summary, we found that the results of the trans-tagging assay were skewed by experimental biases. The strongest influences appeared to originate from a product-size bias in the PCR step and from different efficiencies in the transcription of ribozymes, whereas a smaller influence came from the loss of 3′-exons by ribozymes during transcription and trans-splicing.
DISCUSSION
We have developed and experimentally tested a computational approach to identify efficient splice sites for trans-splicing ribozymes. This approach is based on the computation of the free energy change, ΔGbind, for the hybridization and secondary structure rearrangements accompanying the first step of trans-splicing, i.e., the binding of IGS to substrate. The computed values of ΔGbind were found to correlate well with experimentally determined trans-splicing efficiencies at 18 different splice sites on CAT mRNA. The correlation was significantly better than the correlation of experimental trans-splicing efficiencies with the results of the trans-tagging assay, suggesting that the proposed computational approach could provide an alternative solution to the identification of efficient splice sites for trans-splicing ribozymes.
In particular, our results suggest that a set of candidate efficient splice sites for trans-splicing ribozymes could be determined by selecting those sites with ΔGbind < −4 kcal/mol. Although other mRNAs may show different values for ΔGbind that better discriminate between efficient and inefficient splice sites, a threshold of ΔGbind < −4 kcal/mol may provide a rough guideline for choosing candidate splice sites on other mRNA substrates. Our results also suggest that the unfolding of substrate mRNA at the target site, the hybridization of the substrate to the ribozyme, and the release of IGS from its secondary interactions with the 3′-exon all contribute toward the efficiency of a given splice site on long mRNAs, albeit to varying degrees.
The above results may seem unsurprising at first because computations similar to ours have already been applied to identify target sites for different types of RNA-binding molecules. For example, a thermodynamic cycle equivalent to our calculation of ΔGbind predicted antisense oligonucleotides with high binding affinity for rabbit β-globin and mouse tumor necrosis factor-α mRNAs, achieving 60% accuracy and a significant correlation with experimental data (Walton et al. 1999). Analogous calculations included the concentration-dependent effects of oligonucleotide dimerization, predicting oligonucleotide-target affinities consistent with results from an RNase-H mapping assay on the AT1 receptor mRNA and from a trans-tagging assay on sickle β-globin mRNA (Mathews et al. 1999). More recently, a target site accessibility measure provided by the program RNAplfold and related to our ΔGunfold-target was used to improve the prediction of effective siRNAs (Tafer et al. 2008). A similar calculation also improved and simplified the prediction of effective microRNA targets in Drosophila melanogaster tissue culture cells (Kertesz et al. 2007). Another study used a statistical sampling technique to calculate ΔG values analogous to our ΔGbind (Shao et al. 2007b). These values were found to correlate significantly with experimentally measured trans-cleavage activities of 15 hammerhead ribozymes targeting transcripts of human breast cancer resistance protein. Contrary to our results, this study found that ΔGunfold-target contributes more than ΔGhybrid toward the observed correlation, underscoring the structural and mechanistic differences between trans-cleaving and trans-splicing ribozymes (Scott 2007). Lastly, the program IntaRNA, which we used to calculate ΔGbind, was more effective than other software in predicting 18 targets of small bacterial regulatory RNAs (Busch et al. 2008).
Despite the previous achievements by similar methods, it was not obvious at the outset whether our proposed computations of ΔGbind would correlate well with the efficiency of trans-splicing ribozymes, because these molecules are more complex than those investigated in most of the previous reports. Although Mathews et al. (1999) found some agreement between their ΔGbind calculations and the counts of accessible splice sites reported by Lan et al. (1998), this comparison considered only a short 70-nt region of the mRNA substrate and did not directly measure trans-splicing efficiency. In contrast, our study individually tested the trans-splicing efficiency on 18 different splice sites, distributed over the full length of the CAT mRNA, and compared the experimental results to the computed values of ΔGbind. Our study also included the ribozyme 3′-exon into the calculations. The good correlation we observed between experiments and computation confirms that a calculation of ΔGbind based on established RNA folding algorithms can be used to identify efficient splice sites for trans-splicing ribozymes.
The scope of the present study was intentionally limited to providing a first assessment of the proposed calculation of ΔGbind as a tool for predicting efficient splice sites. Therefore, we carried out experiments using a single model mRNA substrate, and we targeted a subset of the possible splice sites on this substrate. On the other hand, the trans-tagging assay has already been successfully applied to map efficient splice sites on at least eight different mRNA substrates (Lan et al. 1998; Watanabe and Sullenger 2000; Rogers et al. 2002; Park et al. 2003; Ryu and Lee 2003; Jung and Lee 2005; Fiskaa et al. 2006; Kim et al. 2007). Moreover, the proposed calculation of ΔGbind relies on secondary structure prediction algorithms that are very fast but not always accurate because they do not take into account all possible RNA interactions and they are sensitive to errors in experimental energy parameters (Layton and Bundschuh 2005; Condon and Jabbari 2009). Thus, future experimental studies on additional mRNA substrates will show whether calculations of ΔGbind provide a reliable means of predicting efficient splice sites.
The proposed method is attractive because the calculations of ΔGbind do not require bench-work and can be performed in one afternoon. In contrast, the experimental assay, from the preparation of ribozymes and substrate mRNA to the analysis of trans-splicing product sequences, requires at least one week of work. Thus a computational approach, once proven reliable, will likely be cheaper and quicker than the experimental route. Moreover, the computational approach can be developed further to satisfy additional design requirements. For example, if the aim of the ribozymes is to repair a mutated mRNA sequence, then the sequence of the 3′-exon must be adjusted according to the targeted splice site (Long and Sullenger 1999). This adjustment, which is likely important based on our analysis of energetic contributions toward trans-splicing efficiency, would be difficult to achieve with the trans-tagging assay, whose pool of ribozymes presently carries the same 3′-exon sequence for all targeted splice sites. On the other hand, the proposed computational approach can easily be modified to adjust the 3′-exon sequence in accordance with the splice site.
Another design requirement might be to avoid trans-splicing ribozymes that react at multiple splice sites. This behavior was observed in the present study for ribozymes targeting splice sites 405 and 448 (Fig. 3A). Such cross-reactivity was likely caused by the similarity of the corresponding IGSs, which differ in only one out of 6 nt (GGGGAA for splice site 405 versus GGGGAU for splice site 448). A computational approach can easily overcome this problem, as a simple comparison of the predicted IGSs suffices to detect and discard potentially cross-reactive ribozymes.
The computational approach could also accommodate the design of 5′-extended guide sequences (EGSs), which have been shown to strongly increase trans-splicing efficiency (Kohler et al. 1999; Fiskaa and Birgisdottir 2010). The design of these EGSs depends critically on the mRNA sequence immediately downstream from the splice site. If the trans-tagging assay were to be used for the simultaneous optimization of IGSs and EGSs, then the sequence of the EGSs would have to covary with the randomized IGS. This would be a very laborious task because each sequence would have to be synthesized individually. Therefore, EGS optimization cannot be easily achieved with the trans-tagging assay. In contrast, our computational approach can be modified to include a splice site-specific EGS optimization, which would allow one not only to identify efficient splice sites on a given mRNA but also to maximize their efficiency.
In conclusion, the proposed computational approach, together with future algorithmic extensions aimed at optimizing 3′-exon and EGS, could facilitate the development of efficient trans-splicing ribozymes while elucidating the interactions between trans-splicing ribozymes and their mRNA substrates.
MATERIALS AND METHODS
Prediction of ΔGbind
The values of the binding free energy change, ΔGbind, and its components were initially computed using the Vienna RNA package (Hofacker et al. 1994). Specifically, we used RNAeval to calculate ΔGhybrid, and RNAfold to calculate ΔGunfold-target and ΔGrelease-IGS, using the partition function approach, which yields an ensemble average of ΔG values associated with all possible pseudo knot-free secondary structures achievable with a given RNA sequence (McCaskill 1990). We then found that the same calculations can be carried out more conveniently using the IntaRNA software ((Busch et al. 2008); http://rna.informatik.uni-freiburg.de:8080/IntaRNA.jsp), which was developed to find putative target sites for bacterial small regulatory RNAs (sRNAs, typically 50–250 nt in length) on long mRNA sequences. This software takes as input two sequences: a short sRNA and a long target mRNA. The software outputs a list of possible base-pairing interactions between the two RNAs, together with the corresponding ΔGbind = ΔGhybrid + ΔGunfold-mRNA + ΔGunfold-sRNA, where ΔGhybrid is the hybridization free energy component of the interaction, and ΔGunfold-mRNA/sRNA is the cost in free energy for locally unfolding the region of mRNA/sRNA involved in the base-pairing interaction. To calculate ΔGbind for the possible splice sites on CAT mRNA, the 678-nt sequence of this substrate was specified as the input mRNA sequence, and the shortened ribozyme sequence GXXXXXNNNNNACGCACGTCAATTGGCCGCTGGATGGGGCCCCTGTGAAGTGTTGCTGAGCAACGCGCTGGCGCGGCTCAGAGGCTTC was specified as the input sRNA sequence, where GXXXXX denotes a specific IGS, NNNNN is a linker replacing the body of the ribozyme, and the rest of the sequence is the 77-nt 3′-exon used for the trans-tagging and trans-splicing experiments in this study. To calculate ΔGbind for the short 13-mer substrates, the input sRNA sequence was the shortened ribozyme sequence shown above, and the input mRNA sequence was the 13-mer substrate sequence GGYYYYYUAAAAA, where YYYYY is the reverse complement of the five IGS bases XXXXX. For all calculations, the maximum length of the hybridized region was seven, the upper energy threshold was 10 kcal/mol, the exact number of seed base pairs was three, the maximum number of reported suboptimal interactions was 10,000, while all other parameters of the software had default values. Requesting 10,000 suboptimal interactions was necessary to obtain ΔGbind values for as many as possible of the least efficient splice sites tested in this study. However, requesting 40 suboptimal interactions was sufficient to predict the five most efficient of the tested splice sites. Only interactions involving each IGS and the corresponding target site on the substrate were extracted from the program's output. The above procedure was automated using a short Perl script, which is available from G.A. upon request. Values of ΔGbind for CAT mRNA were computed using window sizes of 100, 200, 300, 400, 500, and 600 nt, which cover uniformly the length of the substrate. Because each window size produced different outliers in the plot of experimental trans-splicing efficiency versus ΔGbind (data not shown), we averaged ΔGbind over all six window sizes, thus obtaining a more consistent trend (Fig. 3B). Error bars of ΔGbind in Figure 3B are standard deviations over the six window sizes. Splice sites whose ΔGbind value was reported for less than three window sizes were not assessed for energetic contributions and are not shown in Figure 3B. When no ΔGbind values were returned for specific combinations of window size and splice site, the value zero was used for ΔGbind in Figure 2. All calculations for the 13-mer substrates were done with a window size of 13 nt.
Ribozymes
To prepare DNA templates for transcription of trans-splicing ribozymes with specific IGSs (Supplemental Table S1), a DNA fragment for each such ribozyme was amplified by PCR, using forward primer GCGTAATACGACTCACTATAGXXXXXAAAAGTTATCAGGCATGCACC and reverse primer GAAGCCTCTGAGCCGCGCCAG (PR1), from a plasmid containing the Tetrahymena ribozyme gene linked to a 77-nt segment of the alpha-mannosidase gene as the 3′-exon (Einvik et al. 2004). The above forward primer contains the promoter for T7 RNA polymerase, the desired IGS nucleotides GXXXXX, and the first 21 nucleotides downstream from the IGS in the Tetrahymena ribozyme sequence. All PCR products were cloned into the EcoRI / BamHI sites of pUC19 using appropriate PCR primers, and the ribozyme sequences were confirmed by sequencing. DNA templates for ribozyme transcription were obtained from these ribozyme-encoding plasmids by PCR amplification using forward primer GCGTAATACGACTCACTATAG and reverse primer PR1. To minimize the loss of 3′-exons, ribozyme transcriptions were carried out at 30°C for 20–30 min. The transcripts were purified by denaturing polyacrylamide gel electrophoresis (PAGE) and eluted from gel slices. Ribozyme concentrations were determined from absorbance measurements at 260 nm using an extinction coefficient of 4.0 μM−1cm−1.
Substrates
The template for run-off transcription of CAT mRNA was amplified by PCR from plasmid pLysS (Novagen), in two consecutive PCRs. The first PCR used forward primer CAGGAGCTAAGGAAGCTAAAATG and reverse primer CGCCCCGCCCTGCCACTCATC; the second PCR used forward primer GCGTAATACGACTCACTATAGCAGGAGCTAAGGAAGCTAAAATG and the same reverse primer. After transcription, the full-length CAT mRNA was purified on Micro Bio-Spin 6 columns (Bio-Rad), dephosphorylated with Antarctic phosphatase (NEB), and radiolabeled at the 5′ terminus using γ-[32P]-ATP (Perkin Elmer) and polynucleotide kinase (NEB). The radiolabeled substrate was purified by denaturing 5% PAGE and eluted from gel slices using RNA elution solution (20– 50 mM MOPS/NaOH pH 7.0, 0.2% SDS, 300 mM NaCl). The 13-mer substrates were prepared as described previously (Milligan and Uhlenbeck 1989) to obtain the sequences GGYYYYYUAAAAA, where YYYYY is the reverse complement of the five IGS bases XXXXX adjacent to the 5′-terminal G of the ribozyme. Templates for transcription were obtained by annealing the sense oligonucleotide GCTAATACGACTCACTATAG, which contains a T7 RNA polymerase promoter, with antisense DNA oligonucleotides TTTTTAXXXXXCCTATAGTGAGTCGTATTAGC. In vitro transcription was carried out at 37°C for 1 h in the presence of α-[32P]-UTP (Perkin Elmer), using 500 nM DNA template and ∼1 unit/μL of T7 RNA polymerase. The radiolabeled transcripts were purified by denaturing 20% PAGE.
Trans-splicing reactions
Ribozymes and substrates were mixed in a buffer containing 1 mM MgCl2, 135 mM KCl, 50 mM MOPS/NaOH pH 7.0, 20 μM GTP, and 2 mM spermidine and incubated at 37°C. Before mixing with the substrate, the ribozymes were preincubated for 10 min at 37°C in the absence of magnesium. In reactions with CAT mRNA substrate, ribozyme concentrations were 1.5 μM, substrate concentration was ∼50 nM, and the incubation time was 4 h. This reaction time was chosen as a measure for trans-splicing efficiency because, after this time, the signal for the most efficient splice site (258) began saturating, while the signals for most other splice sites were relatively weak or absent. In reactions with 13-mer substrates, ribozyme concentrations were 100 nM, substrate concentrations were ∼10 nM, and the incubation time was 8 min. At this time, none of the reaction products appeared saturated, and all ribozyme-substrate pairs yielded bands sufficiently strong for quantitation. The samples were separated by denaturing 20% PAGE and visualized by phosphorimaging (Bio-Rad PMI). Band intensities in the resulting digital images were quantified using custom computer software that employs previously described curve-fitting procedures (Shadle et al. 1997; Mitov et al. 2009). Trans-splicing efficiencies were calculated using the formula (trans-splicing efficiency) = (trans-splicing product) / (trans-splicing product + 5′-cleavage product + unreacted substrate), where the amount of each species was assumed proportional to the corresponding band intensity. Error bars were calculated as standard deviations over three independent experiments.
Confirmation of specific trans-splicing products
Samples from trans-splicing reactions that yielded visible bands on autoradiograms of polyacrylamide gels were reverse-transcribed using AMV reverse transcriptase (NEB) with reverse primer PR2. The RT products were amplified by PCR using the nested reverse primer PR3 and one of the forward primers PF1, CGGAATTCGATATGGGATAGTGTTCACCC, CGGAATTCCCGTTGATATATCCCAATGGC, CGGAATTCTGGCCTATTTCCCTAAAGGG, and GCCTTTATTCACATTCTTGCC, which anneal at different positions on the CAT mRNA sequence. The PCR products were cloned into the plasmid pUC19 and sequenced.
Trans-tagging assay
The trans-tagging assay was carried out essentially as described (Einvik et al. 2004). All ribozymes were synthesized by run-off in vitro transcription using T7 RNA polymerase, from DNA templates that were generated by PCR. Preparation of the ribozyme pool was as described above for ribozymes with specific IGSs, except that here the forward primer encoding the T7 promoter contained five randomized nucleotides at the positions denoted with X. The trans-tagging reactions contained 100 nM substrate, 10 nM ribozyme, 0.2 mM GTP, 5 mM MgCl2, 50 mM MOPS/KOH pH 7.0, 135 mM KCl, and 2 mM spermidine. After the substrate and the ribozyme were preincubated in separate tubes in the absence of MgCl2 for 10 min at 37°C, all reagents were mixed and incubated for 1 h at 37°C. Reaction products were reverse transcribed with AMV reverse transcriptase (NEB) using primer GAAGCCTCTGAGCCGCG (PR2). The template RNA was hydrolyzed by incubation in 200 mM NaOH at 90°C for 10 min. RT products were amplified by PCR using nested reverse primer GCGGATCCTGCTCAGCAACACTTCACAGG (PR3) and forward primers CGAATTCCAGGAGCTAAGGAAGCTAAAATG (PF1) or CGGAATTCCATTCTTGCCCGCCTGATGAATGC, cloned into pUC19, and sequenced. The resulting sequences were compared with the ribozyme 3′-exon sequence and the CAT mRNA sequence to determine the positions of the splice sites on CAT mRNA.
Ribozyme transcription efficiency and 3′-exon loss
Ribozymes with specific IGSs (Supplemental Table S1) were individually transcribed by T7 RNA polymerase in 20-μL reactions containing α-[32P]-GTP (Perkin Elmer) from DNA templates that were prepared as described above. The reactions were carried out at 30°C for 20 min and were stopped by the addition of formamide loading buffer. The ribozymes were heat-denatured at 80°C for 2 min and separated by denaturing 5% PAGE. The resulting gels were analyzed as described above for the trans-splicing reactions. A total of seven independent reactions per ribozyme were carried out. To calculate the relative transcription efficiencies, the absolute intensity of the band corresponding to each intact ribozyme was divided by the absolute intensity of the band corresponding to ribozyme 97. Error bars are standard deviations of the relative transcription efficiencies. To calculate the fraction of 3′-exons lost during transcription for each ribozyme, the quantitated intensity of the faster migrating band was divided by the sum of the intensities of this band and of the band for the intact ribozyme. Error bars are standard deviations of these fractions. Ribozymes were eluted from the gel slices and individually incubated without substrate under the same trans-splicing conditions described above. Samples taken at times 0 and 4 h reaction time were analyzed by denaturing 5% PAGE and autoradiography as described above for trans-splicing reaction products. No loss of 3′-exon was seen at time 0. Fractions of lost 3′-exon were calculated as for the transcription reactions.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
ACKNOWLEDGMENTS
This work was supported by the National Institutes of Health (T32DK007233 to K.E.O.; Hemoglobin and Blood Protein Chemistry training grant to E. Komives); the ARCS Foundation, San Diego Chapter (Scholarship to D.M.); and U.C. San Diego (to G.A.). We thank the reviewers for their constructive comments, which resulted in several improvements to the manuscript.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.029884.111.
REFERENCES
- Adams PL, Stahley MR, Kosek AB, Wang J, Strobel SA 2004. Crystal structure of a self-splicing group I intron with both exons. Nature 430: 45–50 [DOI] [PubMed] [Google Scholar]
- Ayre BG, Kohler U, Goodman HM, Haseloff J 1999. Design of highly specific cytotoxins by using trans-splicing ribozymes. Proc Natl Acad Sci 96: 3507–3512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Backofen R, Hess WR 2010. Computational prediction of sRNAs and their targets in bacteria. RNA Biol 7: 33–42 [DOI] [PubMed] [Google Scholar]
- Bai Y, Gong H, Li H, Vu GP, Lu S, Liu F 2011. Oral delivery of RNase P ribozymes by Salmonella inhibits viral infection in mice. Proc Natl Acad Sci 108: 3222–3227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Been MD, Cech TR 1986. One binding site determines sequence specificity of Tetrahymena pre-rRNA self-splicing, trans-splicing, and RNA enzyme activity. Cell 47: 207–216 [DOI] [PubMed] [Google Scholar]
- Bell MA, Sinha J, Johnson AK, Testa SM 2004. Enhancing the second step of the trans excision-splicing reaction of a group I ribozyme by exploiting P9.0 and P10 for intermolecular recognition. Biochemistry 43: 4323–4331 [DOI] [PubMed] [Google Scholar]
- Busch A, Richter AS, Backofen R 2008. IntaRNA: Efficient prediction of bacterial sRNA targets incorporating target site accessibility and seed regions. Bioinformatics 24: 2849–2856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustin SA 2000. Absolute quantification of mRNA using real-time reverse transcription polymerase chain reaction assays. J Mol Endocrinol 25: 169–193 [DOI] [PubMed] [Google Scholar]
- Byun J, Lan N, Long M, Sullenger BA 2003. Efficient and specific repair of sickle beta-globin RNA by trans-splicing ribozymes. RNA 9: 1254–1263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell TB, Cech TR 1995. Identification of ribozymes within a ribozyme library that efficiently cleave a long substrate RNA. RNA 1: 598–609 [PMC free article] [PubMed] [Google Scholar]
- Carter JR, Keith JH, Barde PV, Fraser TS, Fraser MJ Jr 2010. Targeting of highly conserved Dengue virus sequences with anti-Dengue virus trans-splicing group I introns. BMC Mol Biol 11: 84 doi: 10.1186/1471-2199-11-84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cech TR, Damberger SH, Gutell RR 1994. Representation of the secondary and tertiary structure of group I introns. Nat Struct Biol 1: 273–280 [DOI] [PubMed] [Google Scholar]
- Chan JH, Lim S, Wong WS 2006. Antisense oligonucleotides: From design to therapeutic application. Clin Exp Pharmacol Physiol 33: 533–540 [DOI] [PubMed] [Google Scholar]
- Condon A, Jabbari H 2009. Computational prediction of nucleic acid secondary structure: Methods, applications, and challenges. Theor Comput Sci 410: 294–301 [Google Scholar]
- Ding Y, Lawrence CE 2001. Statistical prediction of single-stranded regions in RNA secondary structure and application to predicting effective antisense target sites and beyond. Nucleic Acids Res 29: 1034–1046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dotson PP II, Johnson AK, Testa SM 2008. Tetrahymena thermophila and Candida albicans group I intron-derived ribozymes can catalyze the trans-excision-splicing reaction. Nucleic Acids Res 36: 5281–5289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doudna JA, Cormack BP, Szostak JW 1989. RNA structure, not sequence, determines the 5′ splice-site specificity of a group I intron. Proc Natl Acad Sci 86: 7402–7406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Einvik C, Fiskaa T, Lundblad EW, Johansen S 2004. Optimization and application of the group I ribozyme trans-splicing reaction. Methods Mol Biol 252: 359–371 [DOI] [PubMed] [Google Scholar]
- Far RK, Leppert J, Frank K, Sczakiel G 2005. Technical improvements in the computational target search for antisense oligonucleotides. Oligonucleotides 15: 223–233 [DOI] [PubMed] [Google Scholar]
- Fiskaa T, Birgisdottir AB 2010. RNA reprogramming and repair based on trans-splicing group I ribozymes. New Biotechnol 27: 194–203 [DOI] [PubMed] [Google Scholar]
- Fiskaa T, Lundblad EW, Henriksen JR, Johansen SD, Einvik C 2006. RNA reprogramming of alpha-mannosidase mRNA sequences in vitro by myxomycete group IC1 and IE ribozymes. FEBS J 273: 2789–2800 [DOI] [PubMed] [Google Scholar]
- Gao Y, Liu XL, Li XR 2011. Research progress on siRNA delivery with nonviral carriers. Int J Nanomedicine 6: 1017–1025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golden BL, Kim H, Chase E 2005. Crystal structure of a phage Twort group I ribozyme-product complex. Nat Struct Mol Biol 12: 82–89 [DOI] [PubMed] [Google Scholar]
- Guo F, Gooding AR, Cech TR 2004. Structure of the Tetrahymena ribozyme: Base triple sandwich and metal ion at the active site. Mol Cell 16: 351–362 [DOI] [PubMed] [Google Scholar]
- Guo P, Coban O, Snead NM, Trebley J, Hoeprich S, Guo S, Shu Y 2010. Engineering RNA for targeted siRNA delivery and medical application. Adv Drug Deliv Rev 62: 650–666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haugen P, Andreassen M, Birgisdottir AB, Johansen S 2004. Hydrolytic cleavage by a group I intron ribozyme is dependent on RNA structures not important for splicing. Eur J Biochem 271: 1015–1024 [DOI] [PubMed] [Google Scholar]
- Hofacker IL, Fontana W, Stadler PF, Bonhoeffer LS, Tacker M, Schuster P 1994. Fast folding and comparison of RNA secondary structures. Monatsh Chem 125: 167–188 [Google Scholar]
- Inoue T, Sullivan FX, Cech TR 1985. Intermolecular exon ligation of the rRNA precursor of Tetrahymena: Oligonucleotides can function as 5′ exons. Cell 43: 431–437 [DOI] [PubMed] [Google Scholar]
- Inoue T, Sullivan FX, Cech TR 1986. New reactions of the ribosomal RNA precursor of Tetrahymena and the mechanism of self-splicing. J Mol Biol 189: 143–165 [DOI] [PubMed] [Google Scholar]
- Johnson AK, Sinha J, Testa SM 2005. Trans insertion-splicing: Ribozyme-catalyzed insertion of targeted sequences into RNAs. Biochemistry 44: 10702–10710 [DOI] [PubMed] [Google Scholar]
- Jones JT, Lee SW, Sullenger BA 1996. Tagging ribozyme reaction sites to follow trans-splicing in mammalian cells. Nat Med 2: 643–648 [DOI] [PubMed] [Google Scholar]
- Jung HS, Lee SW 2005. Re-engineering of carcinoembryonic antigen RNA with the group I intron of Tetrahymena thermophila by targeted trans-splicing. J Microbiol Biotechnol 15: 1408–1413 [Google Scholar]
- Jung HS, Lee SW 2006. Ribozyme-mediated selective killing of cancer cells expressing carcinoembryonic antigen RNA by targeted trans-splicing. Biochem Biophys Res Commun 349: 556–563 [DOI] [PubMed] [Google Scholar]
- Karbstein K, Carroll KS, Herschlag D 2002. Probing the Tetrahymena group I ribozyme reaction in both directions. Biochemistry 41: 11171–11183 [DOI] [PubMed] [Google Scholar]
- Kay PS, Inoue T 1987. Catalysis of splicing-related reactions between dinucleotides by a ribozyme. Nature 327: 343–346 [DOI] [PubMed] [Google Scholar]
- Kertesz M, Iovino N, Unnerstall U, Gaul U, Segal E 2007. The role of site accessibility in microRNA target recognition. Nat Genet 39: 1278–1284 [DOI] [PubMed] [Google Scholar]
- Kim A, Ban G, Song MS, Bae CD, Park J, Lee SW 2007. Selective regression of cells expressing mouse cytoskeleton-associated protein 2 transcript by trans-splicing ribozyme. Oligonucleotides 17: 95–103 [DOI] [PubMed] [Google Scholar]
- Kohler U, Ayre BG, Goodman HM, Haseloff J 1999. Trans-splicing ribozymes for targeted gene delivery. J Mol Biol 285: 1935–1950 [DOI] [PubMed] [Google Scholar]
- Kruger K, Grabowski PJ, Zaug AJ, Sands J, Gottschling DE, Cech TR 1982. Self-splicing RNA: Autoexcision and autocyclization of the ribosomal RNA intervening sequence of Tetrahymena. Cell 31: 147–157 [DOI] [PubMed] [Google Scholar]
- Kwon BS, Jung HS, Song MS, Cho KS, Kim SC, Kimm K, Jeong JS, Kim IH, Lee SW 2005. Specific regression of human cancer cells by ribozyme-mediated targeted replacement of tumor-specific transcript. Mol Ther 12: 824–834 [DOI] [PubMed] [Google Scholar]
- Lan N, Howrey RP, Lee SW, Smith CA, Sullenger BA 1998. Ribozyme-mediated repair of sickle beta-globin mRNAs in erythrocyte precursors. Science 280: 1593–1596 [DOI] [PubMed] [Google Scholar]
- Lan N, Rooney BL, Lee SW, Howrey RP, Smith CA, Sullenger BA 2000. Enhancing RNA repair efficiency by combining trans-splicing ribozymes that recognize different accessible sites on a target RNA. Mol Ther 2: 245–255 [DOI] [PubMed] [Google Scholar]
- Layton DM, Bundschuh R 2005. A statistical analysis of RNA folding algorithms through thermodynamic parameter perturbation. Nucleic Acids Res 33: 519–524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SJ, Lee SW, Jeong JS, Kim IH 2010. In vivo reprogramming of human telomerase reverse transcriptase (hTERT) by trans-splicing ribozyme to target tumor cells. Methods Mol Biol 629: 307–321 [DOI] [PubMed] [Google Scholar]
- Lehnert V, Jaeger L, Michel F, Westhof E 1996. New loop-loop tertiary interactions in self-splicing introns of subgroup IC and ID: A complete 3D model of the Tetrahymena thermophila ribozyme. Chem Biol 3: 993–1009 [DOI] [PubMed] [Google Scholar]
- Lipchock SV, Strobel SA 2008. A relaxed active site after exon ligation by the group I intron. Proc Natl Acad Sci 105: 5699–5704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long MB, Sullenger BA 1999. Evaluating group I intron catalytic efficiency in mammalian cells. Mol Cell Biol 19: 6479–6487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu ZJ, Mathews DH 2008. Efficient siRNA selection using hybridization thermodynamics. Nucleic Acids Res 36: 640–647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathews DH, Burkard ME, Freier SM, Wyatt JR, Turner DH 1999. Predicting oligonucleotide affinity to nucleic acid targets. RNA 5: 1458–1469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCaskill JS 1990. The equilibrium partition function and base pair binding probabilities for RNA secondary structure. Biopolymers 29: 1105–1119 [DOI] [PubMed] [Google Scholar]
- Milligan JF, Uhlenbeck OC 1989. Synthesis of small RNAs using T7 RNA polymerase. Methods Enzymol 180: 51–62 [DOI] [PubMed] [Google Scholar]
- Milligan JF, Groebe DR, Witherell GW, Uhlenbeck OC 1987. Oligoribonucleotide synthesis using T7 RNA polymerase and synthetic DNA templates. Nucleic Acids Res 15: 8783–8798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitov MI, Greaser ML, Campbell KS 2009. GelBandFitter–A computer program for analysis of closely spaced electrophoretic and immunoblotted bands. Electrophoresis 30: 848–851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan J, Thirumalai D, Woodson SA 1997. Folding of RNA involves parallel pathways. J Mol Biol 273: 7–13 [DOI] [PubMed] [Google Scholar]
- Park Y-H, Jung H-S, Kwon B-S, Lee S-W 2003. Replacement of thymidine phosphorylase RNA with group I intron of Tetrahymena thermophila by targeted trans-splicing. J Microbiol 41: 340–344 [Google Scholar]
- Phylactou LA, Darrah C, Wood MJ 1998. Ribozyme-mediated trans-splicing of a trinucleotide repeat. Nat Genet 18: 378–381 [DOI] [PubMed] [Google Scholar]
- Pichon C, Felden B 2008. Small RNA gene identification and mRNA target predictions in bacteria. Bioinformatics 24: 2807–2813 [DOI] [PubMed] [Google Scholar]
- Rogers CS, Vanoye CG, Sullenger BA, George AL Jr 2002. Functional repair of a mutant chloride channel using a trans-splicing ribozyme. J Clin Invest 110: 1783–1789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu KJ, Lee SW 2003. Identification of the most accessible sites to ribozymes on the hepatitis C virus internal ribosome entry site. J Biochem Mol Biol 36: 538–544 [DOI] [PubMed] [Google Scholar]
- Ryu KJ, Kim JH, Lee SW 2003. Ribozyme-mediated selective induction of new gene activity in hepatitis C virus internal ribosome entry site-expressing cells by targeted trans-splicing. Mol Ther 7: 386–395 [DOI] [PubMed] [Google Scholar]
- Scott WG 2007. Ribozymes. Curr Opin Struct Biol 17: 280–286 [DOI] [PubMed] [Google Scholar]
- Shadle SE, Allen DF, Guo H, Pogozelski WK, Bashkin JS, Tullius TD 1997. Quantitative analysis of electrophoresis data: Novel curve fitting methodology and its application to the determination of a protein-DNA binding constant. Nucleic Acids Res 25: 850–860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao Y, Wu Y, Chan CY, McDonough K, Ding Y 2006. Rational design and rapid screening of antisense oligonucleotides for prokaryotic gene modulation. Nucleic Acids Res 34: 5660–5669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao Y, Chan CY, Maliyekkel A, Lawrence CE, Roninson IB, Ding Y 2007a. Effect of target secondary structure on RNAi efficiency. RNA 13: 1631–1640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao Y, Wu S, Chan CY, Klapper JR, Schneider E, Ding Y 2007b. A structural analysis of in vitro catalytic activities of hammerhead ribozymes. BMC Bioinformatics 8: 469 doi: 10.1186/1471-2105-8-469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi X, Mollova ET, Pljevaljcic G, Millar DP, Herschlag D 2009. Probing the dynamics of the P1 helix within the Tetrahymena group I intron. J Am Chem Soc 131: 9571–9578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullenger BA, Cech TR 1994. Ribozyme-mediated repair of defective mRNA by targeted, trans-splicing. Nature 371: 619–622 [DOI] [PubMed] [Google Scholar]
- Tafer H, Ameres SL, Obernosterer G, Gebeshuber CA, Schroeder R, Martinez J, Hofacker IL 2008. The impact of target site accessibility on the design of effective siRNAs. Nat Biotechnol 26: 578–583 [DOI] [PubMed] [Google Scholar]
- Thomas CE, Ehrhardt A, Kay MA 2003. Progress and problems with the use of viral vectors for gene therapy. Nat Rev Genet 4: 346–358 [DOI] [PubMed] [Google Scholar]
- Thomas M, Lieberman J, Lal A 2010. Desperately seeking microRNA targets. Nat Struct Mol Biol 17: 1169–1174 [DOI] [PubMed] [Google Scholar]
- Thompson AJ, Herrin DL 1991. In vitro self-splicing reactions of the chloroplast group I intron Cr.LSU from Chlamydomonas reinhardtii and in vivo manipulation via gene-replacement. Nucleic Acids Res 19: 6611–6618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Horst G, Inoue T 1993. Requirements of a group I intron for reactions at the 3′ splice site. J Mol Biol 229: 685–694 [DOI] [PubMed] [Google Scholar]
- Walton SP, Stephanopoulos GN, Yarmush ML, Roth CM 1999. Prediction of antisense oligonucleotide binding affinity to a structured RNA target. Biotechnol Bioeng 65: 1–9 [PubMed] [Google Scholar]
- Walton SP, Wu M, Gredell JA, Chan C 2010. Designing highly active siRNAs for therapeutic applications. FEBS J 277: 4806–4813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JY, Drlica K 2004. Computational identification of antisense oligonucleotides that rapidly hybridize to RNA. Oligonucleotides 14: 167–175 [DOI] [PubMed] [Google Scholar]
- Waring RB, Towner P, Minter SJ, Davies RW 1986. Splice-site selection by a self-splicing RNA of Tetrahymena. Nature 321: 133–139 [Google Scholar]
- Watanabe T, Sullenger BA 2000. Induction of wild-type p53 activity in human cancer cells by ribozymes that repair mutant p53 transcripts. Proc Natl Acad Sci 97: 8490–8494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarrinkar PP, Sullenger BA 1998. Probing the interplay between the two steps of group I intron splicing: Competition of exogenous guanosine with omega G. Biochemistry 37: 18056–18063 [DOI] [PubMed] [Google Scholar]