Abstract
Idiopathic CD4 lymphopenia (ICL) is an immunodeficiency disorder of unclear etiology. Here we describe a heterozygous dominant-negative missense mutation (codon 22 GGC→GTC; V22G) of the signaling adaptor protein Uncoordinated 119 (Unc119) in an ICL patient. The patient is a 32-year-old female with < 300 CD4 T cells/μL and with a history of recurrent sinusitis/otitis media, frequent episodes of shingles, a widespread fungal nail infection, fungal dermatitis, oral herpetic lesions, and bronchiolitis obliterans organizing pneumonia after 2 episodes of bacterial pneumonia. The patient's cells have reduced response to TCR stimulation, with impairment in both localization and enzymatic activation of the lymphocyte-specific kinase (Lck) resulting in decreased cell proliferation. Transduction of the mutant Unc119 but not wild-type Unc119 into normal T cells reproduces the signaling and proliferation defects. The mutation disrupts the Unc119-Lck interaction which is normally needed for stimulation of the Lck catalytic activity by TCR. The mutant protein also causes mislocalization of Lck to Rab11+ perinuclear endosomes. The mutation is not present in 2 other patients with ICL, patients with secondary CD4 lymphopenia or 60 healthy subjects. The V22G mutation of Unc119 represents a novel genetic defect in ICL.
Introduction
The idiopathic CD4 lymphopenia (ICL) is a rare and heterogeneous syndrome defined by a reproducible reduction in the CD4 T-lymphocyte count (< 300 cells/μL or < 20% of total T cells) in the absence of HIV infection or other known causes of immunodeficiency.1 The ICL predisposes to infections and malignancy. Several studies attempted to characterize the T-cell defect. There are reports showing reduced T-cell proliferation, impaired cytokine production, decreased numbers of circulating naive CD45RA+ T cells, reduced clonogenic capacity of BM progenitors, impaired chemotaxis, and augmented apoptosis.2–6 The etiology of the syndrome is likely heterogeneous and in most cases unclear. ICL can be hereditary.2,3 Two very recent articles began to uncover the genetic basis of ICL. The described patients had defects in the magnesium transporter gene (MAGT1) or the recombination activating gene 1 (RAG1).7,8 Here we characterize a mutation in the Unc119 gene as a novel genetic defect in ICL. Unc119 is a signaling adaptor protein which is essential for activation of the key T-cell tyrosine kinase Lck. Lck initiates signaling from the TCR and is critical for T-cell development and for mature T-cell activation.9–11 Activation of Lck involves several orderly steps, that is, dephosphorylation of the C-terminal tyrosine (Y505), binding of an SH3 ligand and phosphorylation of the tyrosine residue in the activation loop (Y394).12 We previously showed that Unc119 functioned as an SH3 ligand and activated Lck.13 Subsequently, we demonstrated that Unc119 participated in Lck translocation to the plasma membrane. The mechanism involved activation of the GTPase–ras gene from rat brain 11 (Rab11) and transport via recycling endosomes.14 Through Lck, Unc119 controlled immunologic synapse formation, T-cell proliferation, and Th cell differentiation.13–15 Here we demonstrate that an Unc119 mutation causes impaired activation and localization of Lck in an ICL patient, which is likely to underlie the pathogenesis of this disease.
Methods
Research subjects
Three groups of subjects were enrolled in the study: 3 patients with ICL, 8 patients with secondary CD4 lymphopenia, and 60 healthy subjects. The secondary CD4 lymphopenia group comprised 1 patient with SLE, 3 patients with Mycobacterium avium complex infection, and 4 patients with common variable immunodeficiency (CVID). The diagnosis of ICL and SLE was made in accordance with the guidelines of the US Centers for Disease Control and Prevention (CDC) and the American College of Rheumatology, respectively.1,16 CVID was diagnosed based on low IgG and IgA, impaired Ab response to bacterial vaccines, and a history of frequent bacterial infections. All subjects gave a written informed consent. The study was approved by the Institutional Review Board at National Jewish Health and was conducted in accordance with Declaration of Helsinki principles.
Isolation of mononuclear cells and CD4 T cells and short-term stimulation with an anti-CD3 Ab
Mononuclear cells were isolated from the peripheral blood using Histopaque-1077 (Sigma-Aldrich) as described by the manufacturer. CD4 T cells were purified by magnetic depletion of non-CD4 cells using the human CD4+ T Cell Isolation Kit II (Miltenyi Biotec). For measurements of Lck activation and ERK1/2 phosphorylation, primary CD4 T cells or Jurkat CD4 T cells in RPMI medium were briefly (1 minute) spun in 12-well plates coated with an anti-CD3 Ab (clone OKT3; eBioscience) or in noncoated plates. Cells in plates were then incubated at 37°C for 4 minutes. Next, the medium was removed and cells which adhered to a well bottom were lysed in the modified radioimmunoprecipitation assay (RIPA) buffer.13
Sequence analysis of the Unc119 gene and mRNA
RNA was isolated from patients' PBMCs using the TRIzol method. RNA was reverse transcribed with oligo-dT primers and the Superscript III enzyme (Invitrogen). Two sets of specific primers (supplemental Table 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article) and the Pfu Ultra high-fidelity polymerase (Stratagene) were used to amplify Unc119 cDNA. After A-tailing, PCR products were ligated into pGEM-T Easy Vector (Promega) and cloned into competent Escherichia coli cells. Eight to 10 clones per single PCR were isolated and sequenced. A cDNA amplification, cloning, and sequencing procedure was done twice for each patient. Genomic DNA was extracted from patients' PBMCs and the P1 buccal swab sample using the DNA Isolation Kit from Roche. The putative Unc119 promoter region (759-bp region between the Unc119 gene and the upstream gene for phosphatidylinositol-glycan biosynthesis class S protein [PIGS]), all 5 exons of the Unc119 gene (including its 5′ and 3′ untranslated regions) as well as all 12 protein-coding exons (exon 3-exon 14) and the adjacent 5′ untranslated exon 2 of the Lck gene were amplified by PCR using specific primers and the Pfu Ultra polymerase. Primers were placed outside of the promoter and exons to permit analysis of the promoter/gene and intron/exon boundaries. Primer sequences are in supplemental Tables 2 (the Unc119 gene) and 3 (the Lck gene). PCR products were gel-purified and sequenced.
The 5′-nuclease allelic discrimination assay
The genomic DNA was analyzed using the Custom TaqMan SNP Genotyping Assay (Applied Biosystems). The region flanking the potential mutation site (G22V) in exon 1 of the Unc119 gene was amplified using TaqMan Universal PCR Master Mix No AmpErase UNG (Applied Biosystems), specific primers (supplemental Table 4) in the presence of 2 allele-specific probes (supplemental Table 4) each conjugated to a single fluorescent reporter dye (VIC or FAM), minor groove binder (MGB), and a nonfluorescent quencher. The genotype was determined from the fluorescence emission at the end of the PCR using the ABI Prism 7000 Sequence Detection System (Applied Biosystems).
The generation of Unc119 constructs and retroviral infection
The cloning of the wild-type Unc119 cDNA into retroviral vectors pMSCVneo and GFP-RV and an addition of the FLAG tag (GFP-RV constructs) was described previously.13 In a GFP-RV construct, cDNA for a molecule of interest (eg, Unc119) and cDNA for GFP are under control of a common promoter but are separated by the internal ribosomal entry site (IRES) sequence. This design leads to the generation of a single transcript which is translated into 2 separate proteins (GFP and the molecule of interest). The G22V substitution in pMSCVneo-based and GFP-RV–based constructs was generated using the QuikChange (Stratagene) mutagenesis approach. Production of retroviruses, CD4 T-cell infection, Jurkat CD4 T-clone generation (limiting dilution), and primary CD4 T-cell sorting (flow cytometry) were done as described.13–15
Western blotting
PAGE, protein transfer to PVDF membranes, and Western blotting were performed as explained elsewhere.13 The following Abs were used: mouse monoclonal anti-Lck (clone 3A5; Santa Cruz Biotechnology), mouse monoclonal anti-ERK1 (clone MK12; BD Biosciences), goat polyclonal anti-actin (Santa Cruz Biotechnology), rabbit monoclonal anti-phospho-ERK1/2 (T202/Y204; clone 197G2; Cell Signaling Technology), and rabbit polyclonal anti-Unc119 Ab. The generation of the anti-Unc119 Ab was described.13,17
Coimmunoprecipitation
Lysates of stimulated CD4 T cells were subjected to immunoprecipitation with an anti-FLAG Ab (clone M2; Sigma-Aldrich) using a previously described protocol.13 Immunoprecipitates were run on polyacrylamide gels, transferred to PVDF membranes, and Western blotted with anti-FLAG and anti-Lck Abs.
Immune complex kinase assay
Lck was isolated from cell lysates by immunoprecipitation using a rabbit polyclonal anti-Lck Ab (Santa Cruz Biotechnology). The enzymatic activity of the immunoprecipitated kinase was studied as explained elsewhere.13
Immunofluorescent staining, image acquisition, and analysis
Staining of paraformaldehyde-fixed CD4 T cells using a mouse monoclonal anti-Lck Ab plus or minus a rabbit polyclonal anti-Rab11 Ab (Invitrogen) and labeled (Alexa Fluor 594 or Alexa Fluor 350) secondary Abs (Invitrogen) was performed as described.14 Images of stained cells were acquired using a Nikon TE 2000 inverted microscope with a 100× objective. The microscope is equipped with a mercury lamp (Chiu Technical Corporation), z-motor (Prior Scientific), excitation and emission filter wheels, and a CoolSnap HQ camera (Roper Scientific). The data acquisition and analysis was done by Metamorph Version 7.0 (Molecular Devices). Z-stacks of 8-12 fluorescent images were acquired at 1-μm increments and deconvolved using the measured point spread function (PSF) of the Metamorph program. For quantification of Lck fluorescence 3 deconvolved planes at 2-μm increments were analyzed. Results from 3 planes were averaged. Images were thresholded to eliminate the contribution from the background fluorescence. Thresholded areas within selected regions (a single cell or its plasma membrane) were used for integrated fluorescence intensity measurement. The integrated intensity was defined as a sum of intensities of all selected pixels.
Thymidine incorporation assay
PBMCs from P1 and a healthy control subject were stimulated in 96-well plates at 105 cells/well with mitogens or Ags specified in Table 3 for 3 or 6 days, respectively. Retrovirally infected and sorted CD4 T cells were stimulated as indicated in Figure 6 for 3 days. [3H] thymidine (1μCi/well; PerkinElmer) was added to all cultures 6 hours before cell harvesting. [3H] thymidine incorporation was measured by liquid scintillation counter.
Table 3.
Proliferation of the patient 1's lymphocytes
| P1, CPM | C, CPM | Reference value, CPM | P1, SI | C, SI | Reference value, SI | |
|---|---|---|---|---|---|---|
| Media, control for PHA | 218 | 170 | ||||
| PHA, μg/mL | ||||||
| 5 | 242 | 49 431 | > 27 000 | 1 | 291 | |
| 2.5 | 269 | 32 403 | > 14 000 | 1 | 191 | |
| 1 | 139 | 12 142 | > 2000 | 1 | 71 | |
| Media, control for ConA | 309 | 170 | ||||
| ConA, μg/mL | ||||||
| 5 | 296 | 15 402 | > 4500 | 1 | 91 | |
| 1.7 | 336 | 2663 | > 1400 | 1 | 16 | |
| 0.6 | 259 | 410 | > 350 | 1 | 2 | |
| Media, control for PWM | 275 | 170 | ||||
| PWM, μg/mL | ||||||
| 25 | 362 | 7174 | > 3800 | 1 | 42 | |
| 8.3 | 322 | 13 795 | > 3500 | 1 | 81 | |
| 2.8 | 223 | 12 467 | > 3000 | 1 | 73 | |
| Media, control for Candida | 282 | 1054 | ||||
| Candida, 6.5 K PNU | 242 | 54 902 | 0.9 | 52.1 | > 5 | |
| Media, control for TET | 229 | 894 | ||||
| TET, LFU | ||||||
| 0.2 | 353 | 10 436 | 1.5 | 11.7 | > 5 | |
| 0.07 | 339 | 16 664 | 1.5 | 18.6 | > 5 | |
| 0.02 | 462 | 14 407 | 2 | 16.1 | > 5 |
PBMCs from P1 and a healthy control subject were incubated in the media alone or with ConA, PHA, PWM, Candida Ag, and TET Ag. [3H]Thymidine was added to cultures 6 hours before cell harvesting into filters. Filters were read by a liquid scintillation counter. SI is a ratio of [3H] thymidine counts for simulated cells divided by counts for unstimulated cells. Reference value represents counts or SI which are considered normal. Note that the Clinical Immunology Laboratory at National Jewish Health uses the CPM reference for PHA, ConA, and PWM, and the SI reference for Candida and TET.
P indicates patient; C, healthy control subject; CPM, counts per minute; SI, stimulation index; ConA, concanavalin A; PWM, pokeweed mitogen; PNU, protein nitrogen unit; TET, tetanus; and LFU, lytic forming unit.
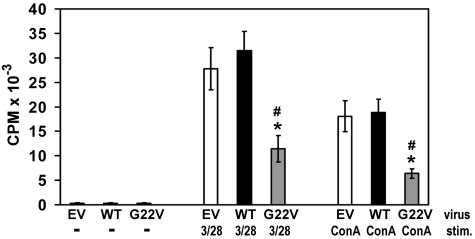
Figure 6.
Effect of Unc119 G22V on CD4 T-cell proliferation. CD4 T cells were infected and sorted as in Figure 3A. Cells were then stimulated (stim.) with plastic-bound anti-CD3 and anti-CD28 Abs (3 of 28) or Concanavalin A (ConA) or left unstimulated (−) for 3 days. Cultures were pulsed with [3H] thymidine 6 hours before cell harvesting. [3H] thymidine incorporation was measured by liquid scintillation counter. N = 4 experiments using CD4 T cells from 4 different healthy donors, P < .05 (Mann-Whitney U test) was found for comparison of G22V and EV cells (*) and comparison of G22V and WT cells (#).
Flow cytometry
PBMCs were stained with CFSE as described.18 Cells were stimulated as indicated in Figure 5 and supplemental Figure 4, stained either with allophycocyanin-labeled anti-CD4 (clone OKT4; eBioscience) and PE-labeled anti-CD8 (clone RPA-T8; eBioscience) Abs or with PE-labeled anti-CD3 (clone OKT3; eBioscience) and allophycocyanin-labeled anti-CD56 (clone CMSSB; eBioscience) Abs, and analyzed using the CyAn ADP cytometer (Beckman Coulter).
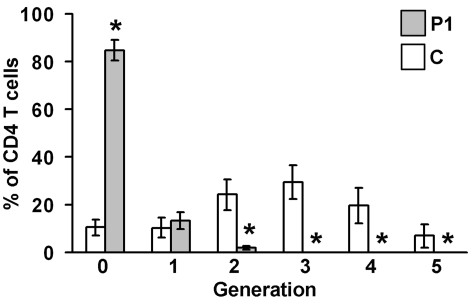
Figure 5.
Proliferation of patient's (P1) CD4 T cells. PBMCs from the patient P1 and healthy controls (C) were labeled with CFSE and stimulated with plastic-bound anti-CD3 and anti-CD28 Abs for 72 hours. Cells were then stained with an allophycocyanin-labeled anti-CD4 Ab. The progressive dilution of the CFSE dye in daughter CD4 T-cell generations (“0” denotes undivided cells, “1” denotes cells after first division, etc) was analyzed by flow cytometry. The graph shows percentage (mean ± SD) of CD4 T cells in each generation. N = 4 experiments; *P < .05 (Mann-Whitney U test, pairwise comparison).
Statistical analysis
Results are shown as mean ± SD. Statistical analyses were done with the Mann-Whitney U test. P < .05 was considered statistically significant.
Results
Clinical and immunologic features of ICL patients
We studied 3 ICL patients (P1, P2, and P3) who were recruited from the Allergy and Immunology Clinic at National Jewish Health. The diagnosis of ICL was made based on the CDC guidelines.1 All 3 patients had reduced CD4 counts (< 300 cells/μL) at the first and follow-up visits. We excluded known causes of CD4 lymphopenia, that is, an infection with HIV or mycobacteria, systemic lupus erythematosus (SLE), Sjogren syndrome, lymphoma, myelodysplastic syndrome, aplastic anemia, CVID, therapy with corticosteroids and other immunosuppressants.19 P1 had a history of corticosteroid therapy (for bronchiolitis obliterans organizing pneumonia [BOOP]) but the described immunologic defect was noted before the therapy had begun. These 3 patients also presented with lymphopenia and low CD3 T-cell counts (Table 1). P2 and P3 had an inverted CD4:CD8 ratio and a low CD19 B-cell count. The patients had normal levels of IgM, IgA, IgG, and its subclasses. The patients' Ab response to bacterial (pneumococcal, tetanus, hemophilus influenza B) Ags was unimpaired. The neutrophil count and function (nitroblue tetrazolium test [NBT] test) were normal. The ICL patients presented with various infections of viral, fungal, and bacterial origin (Table 2).
Table 1.
Lymphocyte subsets in the blood of ICL patients
| ICL patients |
Healthy subjects | |||
|---|---|---|---|---|
| P1 | P2 | P3 | ||
| Lymphocytes/μL | ||||
| Total | 962 | 738 | 794 | 1000-4800 |
| CD3+ | 523 | 533 | 538 | 678-2504 |
| CD4+ | 262 | 186 | 254 | 414-1679 |
| CD8+ | 235 | 310 | 257 | 162-1038 |
| CD19+ | 231 | 7 | 77 | 96-515 |
| CD4/CD8 ratio | 1.11 | 0.60 | 0.99 | 1-3.60 |
| NK cells/μL | ||||
| CD16/56+ | 187 | 148 | 213 | 45-523 |
ICL indicates idiopathic CD4 lymphopenia; and P, patient.
Table 2.
Demographics and clinical characteristics of ICL patients
| Demographics and clinical characteristics | ||
|---|---|---|
| P1 | P2 | P3 |
| White female, age 32 y | White female, age 30 y | Asian female, age 35 y |
| 1. Recurrent sinusitis and otitis media | 1. Recurrent upper respiratory track infections (acute sinusitis, pharyngitis) | 1. Recurrent upper respiratory infections |
| 2. Recurrent shingles | 2. Nasal polyposis | 2. An episode of bacterial pneumonia followed by bronchiectasis with recurrent infections, chronic cough and purulent sputum production |
| 3. Persistent severe fungal infections of fingernails and toenails with partial loss of nails | 3. Aspergillosis of the sinuses | |
| 4. Fungal dermatitis | 4. Two episodes of bacterial pneumonia | |
| 5. Two episodes of bacterial pneumonia followed by BOOP | 5. A single episode of viral meningitis | |
| 6. Oral herpetic lesions | 6. Allergic rhinitis | |
| 7. Moderate persistent asthma | ||
ICL indicates idiopathic CD4 lymphopenia; and BOOP, bronchiolitis obliterans organizing pneumonia.
Lck activation in ICL patients is impaired
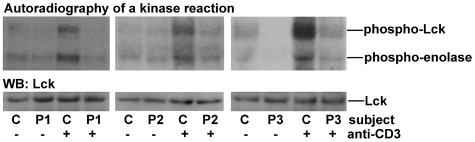
A previous report showed reduced Lck activity in ICL patients.20 We set out to determine whether this biochemical defect was also present in our patients. Indeed, TCR stimulation triggered only a marginal increase in Lck activity in CD4 T cells from 3 ICL patients under study (Figure 1). We also examined 3 representative secondary lymphopenic patients (supplemental Table 5: P4-P6). Lck activation was reduced in a patient with secondary CD4 lymphopenia in the course of M avium complex infection (P4; supplemental Figure 1). Lck activation was normal in patients with CD4 lymphopenia secondary to SLE (P5) and CVID (P6) (supplemental Figure 1). The level of Lck protein was normal in all 6 lymphopenic patients (Figure 2A, supplemental Figure 2).
Figure 1.
Lck activity in CD4 T cells from ICL patients. Peripheral blood CD4 T cells from healthy control subjects (C) and ICL patients (P1, P2, P3) were incubated with (+) or without (−) a plate-bound anti-CD3 Ab for 5 minutes and lysed. Lysates were immunoprecipitated with a rabbit polyclonal anti-Lck Ab. Lck immunoprecipitates were incubated with [32P]γ-ATP, cold ATP and the tyrosine kinase substrate–enolase. Reactions were run on polyacrylamide gels, transferred to PVDF membranes, and exposed to radiosensitive films. Bands on radiography images represent autophosphorylated Lck (phospho-Lck) and phosphorylated enolase (phospho-enolase). Membranes were subsequently Western blotted (WB) with a mouse monoclonal anti-Lck Ab. Each radiography image and a corresponding blot is representative of 3 experiments.
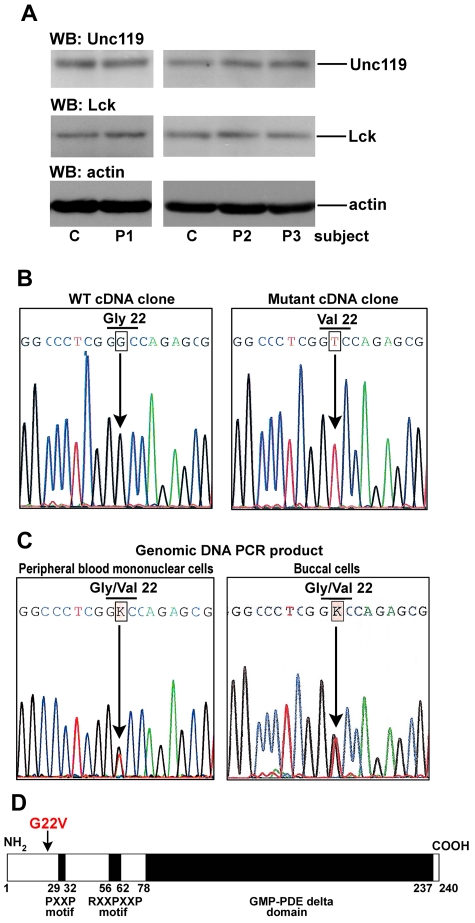
Figure 2.
Heterozygous missense mutation of Unc119 at glycine 22 in an ICL patient 1 (P1). (A) Western blot analysis of CD4 T-cell lysates. CD4 T cells from subjects as in Figure 1 were lysed and blotted with anti-Unc119, anti-Lck, and anti-actin Abs. Each blot represents one of 3 independent experiments. (B) Sequence of cDNA clones. RNA from PBMCs obtained from P1 was reverse transcribed. Unc119 cDNA was amplified by PCR, cloned, and sequenced. Wild-type (WT) and mutant clones were identified. The G to T transversion in codon 22 resulted in a glycine (Gly) to valine (Val) substitution. (C) Sequence of the genomic DNA PCR products. Genomic DNA was isolated from P1 PBMCs (left panel) and from the P1 buccal swab sample (right panel). The Unc119 exons were amplified and the PCR products were sequenced. The figure shows select sequence from the exon 1 PCR product. In the sequence, a single peak of G overlapped with that of T at codon 22. The sequencer read these overlapping peaks as an unidentified base K. (D) Schematic representation of the Unc119 protein structure. The position of the G22V mutation in P1 is marked with an arrow. GMP-PDE δ domain, the cyclic guanosine monophosphate phosphodiesterase δ domain
Unc119 gene is mutated in patient 1
We hypothesized that reduced Lck activation in foregoing lymphopenic patients was because of a defect in its activator—Unc119. The approximate molecular weight of the Unc119 protein and its expression level were normal in patients 1 through 6 (P1-P6; Figure 2A, supplemental Figure 2). The next step was to analyze the sequence of the Unc119 gene. Genomic DNA and RNA were isolated from PBMCs of patients with reduced Lck activity (P1-P4). We sequenced all 5 exons, the intron/exon boundaries, the putative promoter region, and the cDNA. P1 had a heterozygous G to T transversion in exon 1 at the middle position of codon 22 (G136T; base numbering according to the NCBI Reference Sequence of the human Unc119 transcript: NM_005148.3; Figure 2B-C). The mutation led to glycine to valine substitution (G22V). Sequencing of genomic DNA from the buccal swab sample of P1 revealed the presence of the same mutation (Figure 2C). Patients P2, P3, and P4 did not have any mutations in the analyzed genomic regions and cDNA.
We wanted to make sure that reduced Lck activity in P1 was not because of a defect in the Lck gene. Although expressed at a normal level (Figure 2A), the enzyme could be inactive because of a mutation in its coding sequence. We sequenced all 12 protein-coding exons (exon 3–exon 14) and the closely located 5′ untranslated exon 2 of the Lck gene from P1 but did not find any mutations. Therefore, a change in the Lck sequence and/or level do not account for impaired activation of this enzyme in P1.
The identified nucleotide substitution is not found in healthy subjects
The identified Unc119 mutation was not an annotated single nucleotide polymorphism (SNP) in the dbSNP Build 132 and HapMap databases. We screened genomic DNA from PBMCs obtained from 60 healthy subjects for the G/T transversion in the Unc119 gene using a 5′-nuclease allelic discrimination assay. We did not detect the mutation in these subjects. The assay confirmed the presence of the heterozygous mutation in P1 and the absence of this mutation in P2, P3, and 8 secondary lymphopenic patients including P4, P5, and P6. Patients P7 and P8 had disseminated refractory infection with M avium complex, patients P9, P10, and P11 had CVID. Supplemental Table 5 contains information about lymphocyte subsets in these patients.
The genetic screen of P1 family members was impossible because the patient was adopted and she did not know her biologic parents. Furthermore, she did not have any children.
Expression of the Unc119 mutant in normal CD4 T cells prevents Lck activation and blocks downstream signaling
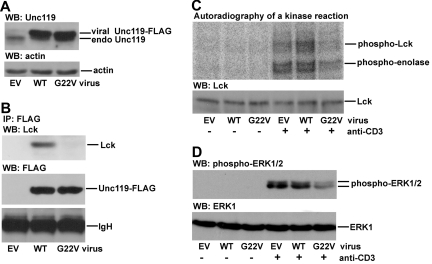
The G22V mutation in P1 is situated near the PXXP motif of Unc119, which is responsible for Lck activation (Figure 2D).13 The PXXP motif of Unc119 binds to the SH3 domain of Lck and this interaction is predicted to relax the conformation of Lck leading to enzymatic activation.13,21 A mutant form of the PXXP motif of Unc119 (Unc119 P29, 32A) does not bind to Lck. The expression of this mutant in normal T cells blocks Lck activation.13 Therefore, the PXXP motif mutant has a dominant-negative effect on Lck. The G22V mutation may interfere with the function of the PXXP motif and have similar dominant-negative properties as the aforementioned P29, 32A mutant. The rationale is that glycine and valine tend to play opposing roles in a protein structure, thus, the G→V substitution may change the conformation of the adjacent regions. We investigated the contribution of the Unc119 G22V mutation to the impairment of Lck activity. We expressed the G22V mutant of Unc119 in normal primary CD4 T cells and the Jurkat CD4 T-cell line by retroviral infection. The virus-derived mutant was expressed at a level comparable with that of the virus-derived wild-type Unc119 (Figure 3A, supplemental Figure 3A) which was in line with the results obtained with P1 T cells (Figure 2A). The wild-type Unc119 protein but not the G22V mutant coprecipitated with Lck (Figure 3B). The mutant blocked Lck activation in spite of the endogenous Unc119 protein (Figure 3C, supplemental Figure 3B). Therefore, the G22V mutant functioned in a similar fashion as the P29, 32A mutant and exerted a dominant-negative effect. We studied the effect of the mutant on propagation of signals downstream of Lck. The MAPKs ERK1 and ERK2 are critical for biologic effects of Lck, for example, for T-cell proliferation.22 The G22V mutant inhibited ERK1/2 phosphorylation (Figure 3D).
Figure 3.
Effect of Unc119 G22V on activation of Lck and its downstream target ERK1/2. (A) CD4 T cells were isolated from PBMCs obtained from a healthy donor. To facilitate retroviral infection, cells were stimulated with plate-bound anti-CD3 and anti-CD28 Abs for 48 hours. Cells were then infected with bicistronic retroviruses encoding GFP alone (EV, empty virus) or GFP and wild-type Unc119 (WT) or the G22V mutant of Unc119 (G22V). GFP+ cells were sorted, lysed, and blotted with anti-Unc119 and anti-actin Abs. The viral Unc119 and its mutant had a FLAG tag attached to their C terminus. Therefore, corresponding bands have a higher molecular weight compared with the endogenous Unc119 (endo Unc119) band and are shifted upward. (B) Infected/sorted primary CD4 T cells were stimulated as in Figure 1 and lysed. Lysates were subjected to immunoprecipitation with an anti-FLAG Ab. Immunoprecipitates were run on polyacrylamide gels and sequentially blotted with anti-Lck and anti-FLAG Abs. IgH indicates Ig heavy chain. (C) Infected/sorted primary CD4 T cells were analyzed for Lck activity as in Figure 1. (D) Infected/sorted CD4 T cells were stimulated as in Figure 1 and lysed. Lysates were blotted with anti-phospho-ERK1/2 (T202/Y204) and anti-ERK1 Abs. Panels A through D are each representative of 3 independent experiments performed using CD4 T cells from 3 different healthy donors.
The G22V mutant reduces the amount of Lck at the T-cell plasma membrane
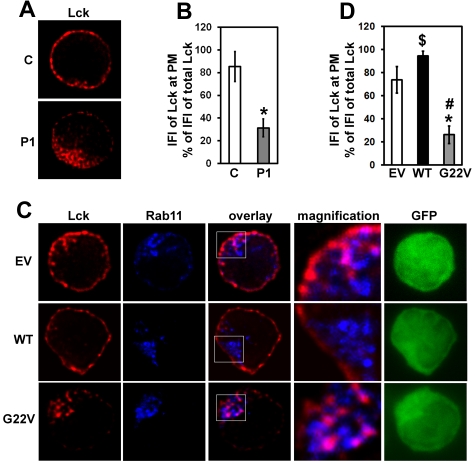
We previously reported on impaired membrane localization and endosomal sequestration of Lck in Unc119-deficient CD4 T cells.14 We observed a similar redistribution of Lck in CD4 T cells from P1 (Figure 4A-B) as well as in G22V mutant-transfected CD4 T cells from a healthy donor (Figure 4C-D). Lck accumulated in the Rab11+ endosomal compartment (Figure 4C). The amount of Lck at the plasma membrane of P1 CD4 T cells and mutant-transfected normal CD4 T cells was, on average, reduced 2.7-fold (P < .05) and 2.8-fold (P < .05), compared with that of nontransfected and empty vector-transfected normal CD4 T cells, respectively. The results suggested that the G22V mutant of Unc119 interfered with the proper subcellular localization of Lck.
Figure 4.
Effect of Unc119 G22V on Lck localization. (A) Peripheral blood CD4 T cells from P1 and a healthy control subject (C) were immunostained for Lck (red) and inspected with a fluorescent microscope (magnification ×1000). Z-stacks of images were collected and subjected to three-dimensional (3D) deconvolution using the measured point spread function (PSF) method. A representative plane is shown. (B) The relative amount of Lck at the plasma membrane of cells from panel A was quantified. Deconvolved z-stacks from 15 cells obtained from 3 separate experiments were used for fluorescent intensity measurements as described in “Methods.” The integrated fluorescent intensity (IFI) of Lck at the plasma membrane (PM) was expressed as a percentage (mean ± SD) of the IFI of the entire cellular Lck. *P < .05 (Mann-Whitney U test). (C) Normal CD4 T cells were infected and sorted as in Figure 3A. Cells were immunostained for Lck (red) and Rab11 (blue). Z-stacks of green (GFP), red, and blue fluorescence were collected. Z-stacks of red and blue fluorescence were subjected to 3D deconvolution as in panel A. The boxed area in the overlay image is magnified and displayed to the right of the image. Pink areas in cells denote colocalization of red Lck and blue Rab11. (D) Deconvolved Z-stacks of Lck fluorescence from panel C were quantified as in panel B. Images of 15 cells from 3 experiments (each experiment done using blood from a different healthy donor) were measured. P < .05 (Mann-Whitney U test) for difference between G22V and EV cells (*), G22V and WT cells (#), and WT and EV cells ($).
The G22V mutant of Unc119 blocks T-cell proliferation
One of the fundamental functions of Unc119 and Lck is to induce T-cell proliferation on TCR stimulation.10–13 Using the thymidine incorporation assay, we determined that proliferation of P1 lymphocytes in response to activation with mitogen (concanavalin A, PHA, pokeweed mitogen) and Ag (Candida and tetanus toxoid) was profoundly reduced (on average, 20-fold reduction compared with minimal normal [reference] value; Table 3) and was comparable with proliferation of unstimulated cells. Accordingly, P1 showed no reactivity to Candida, Trichophyton, and purified protein derivative (PPD) Ags in the delayed-type hypersensitivity (DTH) skin test. Using the CFSE dilution assay, we studied the proliferative capacity of the CD4 and CD8 T-cell subsets on TCR stimulation. In the P1 and healthy control CD4 T-cell populations, the daughter cells constituted 15.3% ± 4.3% and 89.6% ± 3.5%, respectively (Figure 5). In the P1 and healthy control CD8 T-cell populations, the daughter cells comprised 33.4.3% ± 9.5% and 93.1% ± 2.0%, respectively (supplemental Figure 4A). The P1 CD4 T cells and P1 CD8 T cells divided up to 3 times while their healthy control counterparts divided up to 5 times. Using the CFSE dilution assay, we also demonstrated that the IL-15–induced proliferation of P1 NK cells was not different from the proliferation of control NK cells (supplemental Figure 4B). We asked whether ectopic expression of the G22V mutant altered T-cell function. The introduction of the mutant Unc119 cDNA to normal primary CD4 T cells via retroviral infection inhibited their proliferation in response to stimulation with concanavalin A and an anti-CD3 Ab (2.8-fold [P < .05] and 2.4-fold [P < .05] reduction, respectively, compared with empty virus-infected cells; Figure 6). Therefore, the G22V mutation of Unc119 impaired the Lck-dependent T-cell function.
Developmental regulation of Unc119 expression
Lck is critically important for T-cell development in the thymus. Therefore, one would expect a severe phenotype in a patient with a dominant-negative Unc119 mutation. Yet, the patient P1 had a relatively moderate phenotype. For this reason, we wanted to check the Unc119 level in thymocytes. Because of the difficulties of obtaining human thymocytes, we examined Unc119 expression in mouse thymocytes. The expression of the Unc119 protein in mouse thymocytes was substantially lower (borderline detectable) than that in splenic T cells (supplemental Figure 5). If the expression level has any predictive value for a protein function, the result would suggest that Unc119 plays a lesser role in Lck activation during T-cell development in the thymus.
Discussion
Here we describe for the first time an ICL patient with a heterozygous dominant-negative missense mutation (codon 22 GGC→GTC; V22G) in the Unc119 gene. The patient suffered from multiple infections of viral, fungal, and bacterial origin. The patient's T-cell defect (impaired plasma membrane anchoring of Lck, its reduced activation, and decreased T-cell proliferation) was reproduced in normal T cells on transfection with mutant cDNA. The pathogenic mechanism of the mutation involved a disruption of the Unc119-Lck interaction resulting in impaired Lck activation and its mislocalization to the Rab11+ perinuclear endosomal compartment.
Lck is essential for T-cell development and function. Lck regulates both CD4 and CD8 T-cell compartment. There is ample genetic evidence that a reduction in Lck expression and/or activity in humans and in mice leads to immunodeficiency.10,20,23,24 A reduction in Lck expression to < 10% of the normal level in 1 infant was associated with severe combined immunodeficiency (SCID).23 The patient expressed aberrant Lck transcripts, which lacked the exon 7. The child demonstrated a pronounced reduction in peripheral blood T cells. An identical genetic abnormality of the Lck transcript but with a less pronounced expression defect was subsequently demonstrated in an adult patient, who manifested CVID but not SCID.24 A moderate impairment of Lck enzymatic activity (∼ 50% reduction) with no defect in the Lck gene was described in an adult patient with ICL.20 The authors of the report suspected a defect in an activator of Lck in this patient. Collectively, mouse and human data point to a critical role of Lck in T-cell development. Lck also regulates mature T-cell function. Very few naive T cells that manage to mature and exit the thymus of Lck-deficient mice have a reduced proliferative response to TCR stimulation.10 Peripheral blood T cells from the Lck-low SCID infant proliferated poorly as well.23 The patient P1 demonstrated reduced proliferative capacity of CD4 and CD8 T cells. This result reflects the importance of Lck for TCR signaling in both T-cell compartments. The result is in agreement with other studies which showed that a CD4 T-cell defect was accompanied by a CD8 T-cell defect in a subgroup of ICL patients.25 The P1 NK cells expanded normally in response to IL-15 stimulation. Lck does not play a major role in signaling from the IL-15 receptor nor in NK-cell activation.26,27
A question may be raised as to why the G22V mutation in the Unc119 gene is not associated with a more severe form of immunodeficiency such as SCID. Below we provide several explanations. We believe that Unc119 plays a lesser role in Lck activation during T-cell development in the thymus. This is because the Unc119 expression is very low in thymocytes compared with splenic T cells. Therefore, we predict that Lck activity in P1 thymocytes, and consequently, their maturation are not significantly impaired. The requirement for Lck in T-cell function varies among humans. As discussed in the previous paragraph, a single genetic alteration (lack of exon 7 in the Lck transcript), although deleterious to Lck, gives rise to 2 quite different clinical presentations, that is, CVID and SCID.23,24 A mutated allele may produce different phenotypes depending on other concurrent genetic and environmental factors. This is best illustrated by RAG (recombination activating gene) mutations, which produce variable immune defects.8,28,29 Complete RAG deficiency with no (< 1% of wild type) recombination activity is associated with classic SCID and absence of T and B cells. In RAG deficiency with minimal residual activity (> 1% recombination activity of wild type), several clinical and immunologic phenotypes have been described: (1) classic Omenn syndrome with skin inflammation and α-β T-cell expansion; (2) incomplete Omenn syndrome with skin inflammation and without T-cell expansion; (3) RAG deficiency with γ-δ T-cell expansion; (4) RAG deficiency with granulomas; and (5) ICL. Finally, the Lck activation defect in patient P1 may be partially compensated by hyperactive Abl family kinases. Abl kinases participate in signal generation by TCR and promote T-cell development/activation.30,31 Unc119 suppresses these enzymes through the action of its PXXP motif.32
A mutation in a different region of the Unc119 gene was reported in a single patient with late-onset retinal dystrophy.33 The patient had a heterozygous A to T transition in codon 57, changing lysine to a premature termination codon. As a result, the RXXPXXP motif and the cyclic guanosine monophosphate phosphodiesterase (GMP-PDE) δ domain were missing in the mutant protein but the N-terminal region flanking glycine 22 and the PXXP motif remained intact. The expression of a truncated Unc119 cDNA under the rhodopsin promoter caused retinal degeneration in mice. It is unknown whether the patient suffered from any immunodeficiency as the report focused exclusively on the eye defect. The absence of overt immunodeficiency in this patient would suggest that in contrast to the N terminus, the C terminus of Unc119 is redundant for the immune system function. So far, the ICL patient P1 did not have problems with vision. We cannot exclude the possibility of future retinopathy in this patient. The patient is 32 years old. The reported Unc119 mutation-associated retinal degeneration33 had a late onset with symptoms starting at the age of 40. No retinopathy development in patient P1 would suggest that the N terminus of Unc119 is redundant for retina.
Our studies indicate that the G22V mutant has a dominant-negative effect as its expression in normal T cells blocks Unc119-regulated T-cell functions, that is, Lck activation and T-cell proliferation. A similar dominant-negative effect was seen on expression of the PXXP mutant of Unc119 (Unc119 P29, 32A).13 Glycine 22 is located in close proximity of the PXXP motif. We speculate that the G22V mutation impairs the PXXP motif function. The mutant may sequester proteins, which are upstream of Lck and Rab11 activation and therefore diminish their interactions with wild-type Unc119. The mutant may sequester the wild-type Unc119 protein through formation of a wild-type protein-mutant protein heterodimer. Finally, the mutation may lead to the acquisition of a new abnormal function by Unc119, for example, activation and/or recruitment of inhibitory molecules for Lck (eg, Csk) and Rab11 (eg, GTPase activating proteins [GAPs]).
In summary, our study reveals a novel mutation associated with the ICL syndrome. A further understanding of the molecular basis of this immunodeficiency disorder will improve diagnosis and disease classification, and pave the way for development of new therapies.
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health (NIH) grants R01 AI0059719 and PPG HL36577 (R.A. and M.M.G.), and by the Colorado Clinical and Translational Sciences Institute (CCTSI) Junior Faculty Pilot Award (NIH/NCRR Colorado CTSI UL1 RR025780) and the CCTSI KL2 Research Scholar Award (NIH/NCRR Colorado CTSI KL2 RR025779) to M.M.G.
Footnotes
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: M.M.G. designed and performed research, analyzed data, and wrote the manuscript; and R.A. designed research, recruited research subjects, analyzed data, and wrote the manuscript.
Conflict-of-interest disclosures: The authors declare no competing financial interests.
Correspondence: Rafeul Alam, MD, PhD, Department of Medicine, Division of Allergy & Immunology, National Jewish Health, 1400 Jackson St, Denver, CO 80206; e-mail: alamr@njhealth.org.
References
- 1.Smith DK, Neal JJ, Holmberg SD. Unexplained opportunistic infections and CD4+ T-lymphocytopenia without HIV infection. An investigation of cases in the United States. The Centers for Disease Control Idiopathic CD4+ T-lymphocytopenia Task Force. N Engl J Med. 1993;328(6):373–379. doi: 10.1056/NEJM199302113280601. [DOI] [PubMed] [Google Scholar]
- 2.Lin SJ, Chao HC, Yan DC, Kuo ML. Idiopathic CD4+ T lymphocytopenia in two siblings. Pediatr Hematol Oncol. 2001;18(2):153–156. doi: 10.1080/088800101300003008. [DOI] [PubMed] [Google Scholar]
- 3.Freier S, Kerem E, Dranitzki Z, et al. Hereditary CD4+ T lymphocytopenia. Arch Dis Child. 1998;78(4):371–372. doi: 10.1136/adc.78.4.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Isgrò A, Sirianni MC, Gramiccioni C, Mezzaroma I, Fantauzzi A, Aiuti F. Idiopathic CD4+ lymphocytopenia may be due to decreased bone marrow clonogenic capability. Int Arch Allergy Immunol. 2005;136(4):379–384. doi: 10.1159/000084258. [DOI] [PubMed] [Google Scholar]
- 5.Laurence J, Mitra D, Steiner M, Lynch DH, Siegal FP, Staiano-Coico L. Apoptotic depletion of CD4+ T cells in idiopathic CD4+ T lymphocytopenia. J Clin Invest. 1996;97(3):672–680. doi: 10.1172/JCI118464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Scott-Algara D, Balabanian K, Chakrabarti LA, et al. Idiopathic CD4+ T-cell lymphocytopenia is associated with impaired membrane expression of the chemokine receptor CXCR4. Blood. 2010;115(18):3708–3717. doi: 10.1182/blood-2009-02-202796. [DOI] [PubMed] [Google Scholar]
- 7.Li FY, Chaigne-Delalande B, Kanellopoulou C, et al. Second messenger role for Mg2+ revealed by human T-cell immunodeficiency. Nature. 2011;475(7357):471–476. doi: 10.1038/nature10246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kuijpers TW, Ijspeert H, van Leeuwen EM, et al. Idiopathic CD4+ T lymphopenia without autoimmunity or granulomatous disease in the slipstream of RAG mutations. Blood. 2011;117(22):5892–5896. doi: 10.1182/blood-2011-01-329052. [DOI] [PubMed] [Google Scholar]
- 9.Straus DB, Weiss A. Genetic evidence for the involvement of the lck tyrosine kinase in signal transduction through the T cell antigen receptor. Cell. 1992;70(4):585–593. doi: 10.1016/0092-8674(92)90428-f. [DOI] [PubMed] [Google Scholar]
- 10.Molina TJ, Kishihara K, Siderovski DP, et al. Profound block in thymocyte development in mice lacking p56lck. Nature. 1992;357(6374):161–164. doi: 10.1038/357161a0. [DOI] [PubMed] [Google Scholar]
- 11.Lovatt M, Filby A, Parravicini V, Werlen G, Palmer E, Zamoyska R. Lck regulates the threshold of activation in primary T cells, while both Lck and Fyn contribute to the magnitude of the extracellular signal-related kinase response. Mol Cell Biol. 2006;26(22):8655–8665. doi: 10.1128/MCB.00168-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Salmond RJ, Filby A, Qureshi I, Caserta S, Zamoyska R. T-cell receptor proximal signaling via the Src-family kinases, Lck and Fyn, influences T-cell activation, differentiation, and tolerance. Immunol Rev. 2009;228(1):9–22. doi: 10.1111/j.1600-065X.2008.00745.x. [DOI] [PubMed] [Google Scholar]
- 13.Gorska MM, Stafford SJ, Cen O, Sur S, Alam R. Unc119, a novel activator of Lck/Fyn, is essential for T cell activation. J Exp Med. 2004;199(3):369–379. doi: 10.1084/jem.20030589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gorska MM, Liang Q, Karim Z, Alam R. Uncoordinated 119 protein controls trafficking of Lck via the Rab11 endosome and is critical for immunological synapse formation. J Immunol. 2009;183(3):1675–1684. doi: 10.4049/jimmunol.0900792. [DOI] [PubMed] [Google Scholar]
- 15.Gorska MM, Goplen N, Liang Q, Alam R. Uncoordinated 119 preferentially induces Th2 differentiation and promotes the development of asthma. J Immunol. 2010;184(8):4488–4496. doi: 10.4049/jimmunol.0903115. [DOI] [PubMed] [Google Scholar]
- 16.Klippel JH. Primer on the Rheumatic Diseases. Atlanta, GA: Arthritis Foundation; 2001. [Google Scholar]
- 17.Cen O, Gorska MM, Stafford SJ, Sur S, Alam R. Identification of UNC119 as a novel activator of SRC-type tyrosine kinases. J Biol Chem. 2003;278(10):8837–8845. doi: 10.1074/jbc.M208261200. [DOI] [PubMed] [Google Scholar]
- 18.Hasbold J, Gett AV, Rush JS, et al. Quantitative analysis of lymphocyte differentiation and proliferation in vitro using carboxyfluorescein diacetate succinimidyl ester. Immunol Cell Biol. 1999;77(6):516–522. doi: 10.1046/j.1440-1711.1999.00874.x. [DOI] [PubMed] [Google Scholar]
- 19.Walker UA, Warnatz K. Idiopathic CD4 lymphocytopenia. Curr Opin Rheumatol. 2006;18(4):389–395. doi: 10.1097/01.bor.0000231908.57913.2f. [DOI] [PubMed] [Google Scholar]
- 20.Hubert P, Bergeron F, Ferreira V, et al. Defective p56Lck activity in T cells from an adult patient with idiopathic CD4+ lymphocytopenia. Int Immunol. 2000;12(4):449–457. doi: 10.1093/intimm/12.4.449. [DOI] [PubMed] [Google Scholar]
- 21.Moarefi I, LaFevre-Bernt M, Sicheri F, et al. Activation of the Src-family tyrosine kinase Hck by SH3 domain displacement. Nature. 1997;385(6617):650–653. doi: 10.1038/385650a0. [DOI] [PubMed] [Google Scholar]
- 22.Gupta S, Weiss A, Kumar G, Wang S, Nel A. The T-cell antigen receptor utilizes Lck, Raf-1, and MEK-1 for activating mitogen-activated protein kinase. Evidence for the existence of a second protein kinase C-dependent pathway in an Lck-negative Jurkat cell mutant. J Biol Chem. 1994;269(25):17349–17357. [PubMed] [Google Scholar]
- 23.Goldman FD, Ballas ZK, Schutte BC, et al. Defective expression of p56lck in an infant with severe combined immunodeficiency. J Clin Invest. 1998;102(2):421–429. doi: 10.1172/JCI3205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sawabe T, Horiuchi T, Nakamura M, et al. Defect of lck in a patient with common variable immunodeficiency. Int J Mol Med. 2001;7(6):609–614. doi: 10.3892/ijmm.7.6.609. [DOI] [PubMed] [Google Scholar]
- 25.Zonios DI, Falloon J, Bennett JE, et al. Idiopathic CD4+ lymphocytopenia: natural history and prognostic factors. Blood. 2008;112(2):287–294. doi: 10.1182/blood-2007-12-127878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rochman Y, Spolski R, Leonard WJ. New insights into the regulation of T cells by gamma(c) family cytokines. Nat Rev Immunol. 2009;9(7):480–490. doi: 10.1038/nri2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wen T, Zhang L, Kung SK, Molina TJ, Miller RG, Mak TW. Allo-skin graft rejection, tumor rejection and natural killer activity in mice lacking p56lck. Eur J Immunol. 1995;25(11):3155–3159. doi: 10.1002/eji.1830251125. [DOI] [PubMed] [Google Scholar]
- 28.Sobacchi C, Marrella V, Rucci F, Vezzoni P, Villa A. RAG-dependent primary immunodeficiencies. Hum Mutat. 2006;27(12):1174–1184. doi: 10.1002/humu.20408. [DOI] [PubMed] [Google Scholar]
- 29.Niehues T, Perez-Becker R, Schuetz C. More than just SCID–the phenotypic range of combined immunodeficiencies associated with mutations in the recombinase activating genes (RAG) 1 and 2. Clin Immunol. 2010;135:183–192. doi: 10.1016/j.clim.2010.01.013. [DOI] [PubMed] [Google Scholar]
- 30.Zipfel PA, Zhang W, Quiroz M, Pendergast AM. Requirement for Abl kinases in T cell receptor signaling. Curr Biol. 2004;14(14):1222–1231. doi: 10.1016/j.cub.2004.07.021. [DOI] [PubMed] [Google Scholar]
- 31.Gu JJ, Zhang N, He YW, Koleske AJ, Pendergast AM. Defective T cell development and function in the absence of Abelson kinases. J Immunol. 2007;179(11):7334–7343. doi: 10.4049/jimmunol.179.11.7334. [DOI] [PubMed] [Google Scholar]
- 32.Vepachedu R, Karim Z, Patel O, Goplen N, Alam R. Unc119 protects from Shigella infection by inhibiting the Abl family kinases. PLoS One. 2009;4(4):e5211. doi: 10.1371/journal.pone.0005211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kobayashi A, Higashide T, Hamasaki D, et al. HRG4 (UNC119) mutation found in cone-rod dystrophy causes retinal degeneration in a transgenic model. Invest Ophthalmol Vis Sci. 2000;41(11):3268–3277. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.