Abstract
KIT mutations are the most common secondary mutations in inv(16) acute myeloid leukemia (AML) patients and are associated with poor prognosis. It is therefore important to verify that KIT mutations cooperate with CBFB-MYH11, the fusion gene generated by inv(16), for leukemogenesis. Here, we transduced wild-type and conditional Cbfb-MYH11 knockin (KI) mouse bone marrow (BM) cells with KIT D816V/Y mutations. KIT transduction caused massive BM Lin− cell death and fewer colonies in culture that were less severe in the KI cells. D816Y KIT but not wild-type KIT enhanced proliferation in Lin− cells and led to more mixed lineage colonies from transduced KI BM cells. Importantly, 60% and 80% of mice transplanted with KI BM cells expressing D816V or D816Y KIT, respectively, died from leukemia within 9 months, whereas no control mice died. Results from limiting dilution transplantations indicate higher frequencies of leukemia-initiating cells in the leukemia expressing mutated KIT. Signaling pathway analysis revealed that p44/42 MAPK and Stat3, but not AKT and Stat5, were strongly phosphorylated in the leukemia cells. Finally, leukemia cells carrying KIT D816 mutations were sensitive to the kinase inhibitor PKC412. Our data provide clear evidence for cooperation between mutated KIT and CBFB-MYH11 during leukemogenesis.
Introduction
Chromosome 16 inversion is one of the most frequent chromosomal translocations found in acute myeloid leukemia (AML), occurring in 10% to 15% of AML cases, especially those in subtype M4Eo.1 This translocation fuses the CBFB gene with the MYH11 gene, resulting in a protein product that fuses the first 165 amino acids of core binding factor β (CBFβ) to the coiled-coil region of the smooth muscle myosin heavy chain.2 In mouse models, the CBFB-MYH11 fusion gene predisposes mice to AML but requires cooperating mutations for leukemogenesis.3,4 In humans, mutations in receptor tyrosine kinase genes KIT and FLT3 and in proto-oncogenes N-RAS and K-RAS have been observed in up to 70% of inv(16) AML patients,5–7 and probably serve as cooperating factors during leukemogenesis.8–10
KIT (CD117) is a member of the type 3 subclass of transmembrane receptor tyrosine kinases, with 5 immunoglobulin-like domains in the extracellular region. It has a negative regulatory juxtamembrane domain and a split adenosine triphosphate–binding and phosphotransferase tyrosine kinase domain.11 KIT is a receptor that is specific for stem cell factor (SCF). The interaction between KIT and SCF is crucial for the development of hematopoietic, gonadal, and pigment stem cells.11–13 Genetic mutations that disrupt the expression of KIT are associated with piebaldism, a disorder featured by loosing pigmentation of the skin,13 whereas overexpression or constitutive activation of KIT is associated with tumorigenesis.11,14 KIT is expressed in almost 80% of human AML cases.15–17
Mutations in the KIT gene are especially common in inv(16) AML, compared with other AML subtypes,18 occurring in 10% to 45% of the cases.6,7,19 The most common KIT mutations are deletion and insertion mutations in exon 820,21 and the point mutations in exon 17.21,22 Exon 8 mutations are in the extracellular domain and cause spontaneous receptor dimerization and activation of the downstream signaling pathways without SCF.23 The codon 816 in exon 17 is a mutation hot spot in inv(16) AML,18,21,22 and the most common substitutions, D816V and D816Y, cause constitutive activation of KIT.24 These KIT mutations have been reported to adversely affect overall survival of inv(16) AML patients21; however, the real prognostic value of KIT mutations for inv(16) AML remains controversial.21,25,26
The D816 mutant forms of KIT are resistant to inhibition by the kinase inhibitor imatinib in vitro.27,28 Similarly, results from clinical studies suggest that the kinase inhibitor imatinib has limited clinical benefit to patients with CBF leukemia carrying KIT D816 mutations.29 Conversely, studies in cell lines and primary neoplastic mast cells show that another kinase inhibitor, PKC412, is a promising candidate for blocking D816 mutation,28,30 with demonstrated activity in a patient with mast cell leukemia with the D816V KIT mutation.31
So far, the potential cooperation between mutant KIT and CBFB-MYH11 has not been verified, and no in vivo or in vitro models exist for such studies. Given the high frequency and the potential prognostic value of the KIT mutations in inv(16) AML, it is important to develop an in vivo model not only to confirm the contribution of mutant KIT to leukemogenesis by CBFB-MYH11 but also to test and develop effective therapeutic approaches for this frequent clinical combination.
Methods
Animals
Conditional Cbfb-MYH11 knockin mice (Cbfb+/56m) and wild-type (WT) littermates were genotyped as described previously32 and crossed with Mx1-Cre [Tg(Mx1-Cre)] transgenic mice (The Jackson Laboratory). Six- to 8-week-old Cbfb+/56m; Tg(Mx1-Cre) mice and their littermate controls were treated with 250 μg of poly-IC (pI:pC; Invivogen) intraperitoneally every other day for 3 times to induce the expression of Cbfb-MYH11. Two weeks later, the mice were given 5-fluorouracil at 150 mg/kg body weight using intraperitoneal injection, and 48 hours later bone marrow (BM) cells were harvested from tibia and femur. C57BL/6 × 129/SvEv F1 hybrid mice were sublethally irradiated (650 cGy) and used as recipients for transplantation. All animals used and the procedures performed in this study were approved by the National Human Genome Research Institute Animal Care and Use Committee.
Production of stable retroviral cell lines
Phenix packaging cells were transfected with vectors carrying MSCV-KIT D816V, KIT D816Y, WT KIT, or green fluorescent protein (GFP) alone28 using a calcium transfection kit from Invitrogen. Two days after transfection, the supernatants were harvested and incubated with either the ecotropic packaging cell line GP+E-8633 that was derived from mouse NIH3T3 cells, or 293T cells for 2 days. Then, single GFP+ GP+E-86 and 293T cells were sorted to a 96-well plate and 2 to 3 weeks later, the cells were transferred to 24-well plates and later 6-well plates for expansion. Viral titers were measured,34 and the clones with titers more than 1 × 106 particles/mL from GP+E-86 cells were expanded for transduction of mouse bone marrow cells.
Retroviral transduction of bone marrow cells
Bone marrow cells were harvested and cultured in DMEM (Invitrogen) supplemented with 20% FBS (HyClone Laboratories) and cytokines (IL-6, FLT3L, SCF, and Thrombopoietin at 100 ng/mL and IL-3 at 10 ng/mL, PeproTech) for 2 days. They were then transduced by retroviral supernatants of KIT D816V, D816Y, WT KIT, and GFP controls together with 8 μg/mL Polybrene (Sigma-Aldrich) or coculture with stable retrovirus-producing lines of WT KIT or the D816 mutants. Two days later, BM cells were harvested and sorted by FACS for GFP+ BM cells.
Transplantation of BM cells to recipient mice
GFP+ BM cells (1 × 106) were transplanted to recipient mice using retro-orbital injection. Two to 4 weeks later, peripheral blood (PB) was taken from mice for FACS analysis of the GFP+ cells in PB. For limiting dilution transplantation of leukemia cells, 1 × 106, 1 × 105, 1 × 104, 1 × 103, 1 × 102, 10, and 1 live leukemia spleen cell(s) from terminal leukemic mice [4 donors of Cbfb+/56m; Tg(Mx1-Cre)/KIT 816Y/V and 2 donors of Cbfb+/56m; Tg(Mx1-Cre)] were transplanted to recipient mice using retro-orbital injection. They were monitored daily for any abnormal behavior or physiologic changes, and FACS analysis of PB cells was done every 2 weeks after transplantation.
Analysis of PB and leukemia samples from transplanted mice
The PB cells were stained with anti–mouse antibodies for CD3, CD4, CD8, B220, CD117 (c-KIT) Gr-1, Mac-1, and Sca-1 (BD Biosciences). Blood smears were performed at the same time. Mice were killed at the end point according to our protocol. Leukemic spleen cells were stained with anti–human CD117 antibody (BD Biosciences). Histology sections of spleen, liver, kidney, lung, brain, and sternum were analyzed to confirm the leukemia filtration by a mouse pathologist at the Diagnostic & Research Service Branch of the National Institutes of Health (L.B.).
Colony-forming assay of BM cells after transduction with retroviral vectors
BM cells were harvested from WT and Cbfb+/56m; Tg(Mx1-Cre) littermates 2 weeks after pI:pC treatment. BM cells were then transduced with the retroviruses and sorted by FACS. GFP+ BM cells were cultured in M3434 methocellulose (StemCell Technologies) for colony formation assay following the manufacturer's protocol. On days 7 and 12, colonies were classified, and colony numbers were counted according to manufacturer's protocol.
Treatment of primary leukemia cells with PKC412
Primary leukemia cells were seeded in 24-well culture plates (1 × 106 live cells/mL), and PKC412 (LC Laboratories) was added to each well at final concentrations of 0.01, 0.1, and 1μM (dissolved in DMSO). The cells were incubated at 37°C for 17 hours and then were harvested and stained with trypan blue. Live cells were counted using a hemacytometer, and the numbers were normalized relative to control DMSO-treated samples.
Western blot analysis of leukemia samples
Spleen tissue from leukemic mice (frozen at −80°C) was homogenized in radioimmunoprecipitation assay lysis buffer (Millipore) with protease inhibitors (Roche Applied Science), and protein concentration was determined (DC Protein Assay; Bio-Rad Laboratories). Proteins were separated by 4% to 12% SDS-PAGE gel (Invitrogen) electrophoresis. The proteins were then transferred to polyvinylidene difluoride membranes (Invitrogen) for Western blot analysis. Antibodies to regular and phosphorylated p44/42 MAPK, Stat3 (S717, Y705), Stat5 (Y694), and AKT (S473; Cell Signaling Technology) were used to probe the membranes. The ECL detection kit and Hyperfilm ECL (GE Healthcare) films were used to detect the bound antibodies.
Mitotic cell counting of spleen sections from leukemia mice
Mitotic cell counts were taken at 3 areas: at each end of the spleen and in the middle. In each of the 3 areas, the mitotic cells were counted in 10 fields at ×600 magnification. For the average mitoses of each area, the numbers were added and divided by 10. For the overall average for the spleen, the 3 areas were added; the number divided by 3, and the result was rounded up or down to the nearest integer.
BrdU and annexin V staining of transduced/leukemia BM cells
Forty-eight hours after retroviral transduction, BM cells in culture were treated with BrdU (10μM; BD Biosciences) for 30 minutes and then harvested and stained with antibodies against CD3ϵ, CD4, CD8, B220, TCRβ, NK1.1, Gr1, Mac1; KIT, and Sca-1 (BD Biosciences). After staining, cells were separated into 2 aliquots and following the staining protocol for annexin V (BD Biosciences) and BrdU. A BD LSRII flow cytometer was used to acquire the cells, and FlowJo Version 9.4.10 software (TreeStar) was used for data analysis. Because the GFP+/lin−/KIT+/Sca-1+ cell population was very small, we gated on lin−/GFP+ (transduced cells) and lin−/GFP− (untransduced cells) cell populations for annexin V and BrdU analysis.
Leukemic mice were given BrdU (1 mg) via the intraperitoneal injection 1 hour before euthanasia. BM cells were harvested from femurs and stained as described in the preceding paragraph.
Clonality analysis
EcoRI-digested genomic DNA from KIT leukemia spleen cells was separated with gel electrophoresis and transferred to Hybond XL nylon membranes (GE Healthcare) following the manufacturer's protocol. A probe of 600-bp GFP sequence was labeled with α-32P-dCTP using Rediprime II random labeling system (GE Healthcare) and hybridized to the membranes overnight. After wash, the membranes were exposed to phosphor screen for 3 to 4 hours and then read by a FLA-5100 phosphorimager/fluorescent scanner using software IR FLA-5000 series Version 1.01 (Fujifilm).
Results
Generation of stable retrovirus-producing lines
Single GFP+ cells transduced with the retroviral vectors were sorted into the wells of 96-well plates, expanded, and then virus titers were measured. Two to 12 lines of each virus were selected with their titers more than 1 × 106 particles/mL. These lines were derived from either PGE86 cells or Ecopack-293 cells. We compared the efficiency of 1 cell line from Ecopack-293 and 1 cell line from PGE86 of D816Y with similar titer (1 × 107/mL), for transducing WT BM cells. The transduction efficiency for Ecopack-293 D816Y was 10% and that for PGE86 D816Y was 40%. Therefore, PGE86-producing cell lines have been used for our subsequent experiments.
Transduction efficiency of BM cells by retrovirus
We found that coculturing of BM cells with stable virus-producing cell lines gave robust and efficient transduction, resulting in 34% to 77% of the transduced BM cells being GFP+ (supplemental Table 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). The transduction efficiency of Cbfb+/56m; Tg(Mx1-Cre) BM cells varied from 8% to 78.6% among different experiments using different KIT vectors (supplemental Table 1; supplemental Figure 1). The efficiency was determined not only by the titers of the retroviruses but also the state of the BM cells after culturing with cytokines for 2 days.
Effects of KIT overexpression on colony-forming ability of bone marrow cells in vitro
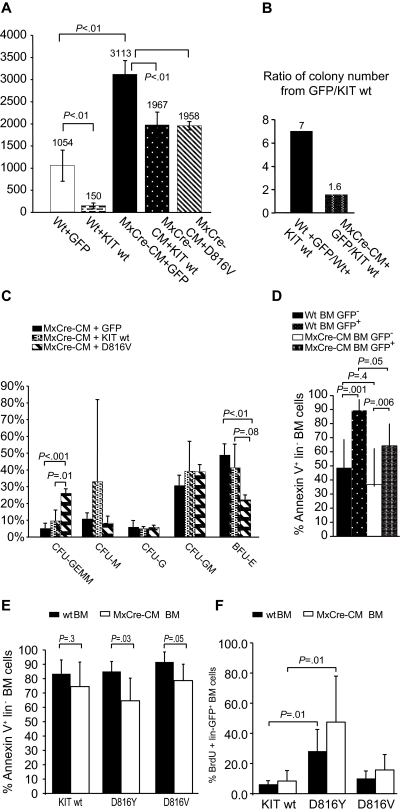
To assess the effect of coexpressing CBFB-MYH11 and mutant KIT on hematopoiesis, BM cells were transduced with retroviral vectors for WT and D816Y/V-mutated KIT and then plated for colony-forming assay. We found that the total colony numbers were decreased (P < .01) in all KIT-transduced BM cells, from both WT and Cbfb+/56m; Tg(Mx1-Cre) mice (Figure 1A). Expression of Cbfb-MYH11 increased the number of total colonies, as we reported previously.32 Significantly, the effect of colony number reduction by KIT was attenuated by Cbfb-MYH11: 7-fold reduction in WT BM cells versus 1.6-fold reduction in Cbfb-MYH11 BM cells (Figure 1B). In addition, there was a decrease in the number of BFU-E (compared with GFP-transduced BM cells; P < .01) and an increase of CFU-GEMM (compared with those transduced with WT KIT; P = .01) in cultures of Cbfb+/56m; Tg(Mx1-Cre) BM cells transduced with D816V/Y (Figure 1C). These results suggest that D816 KIT mutants may cause an accumulation of early myeloid progenitors.
Figure 1.
Differentiation potential of BM cells transduced with D816Y/V KIT and control vectors. (A) Total numbers of colonies from 105 cultured WT (wt) or Cbfb+/56m; Tg(Mx1-Cre) BM cells transduced with retroviral vectors containing WT KIT, KITD816V or GFP and sorted for GFP expression. Wt + GFP indicates WT BM cells transduced with GFP vector alone; Wt + KIT wt, WT BM cells transduced with WT KIT vector; and MxCre-CM + GFP, KIT wt, or D816Y, Cbfb+/56m; Tg(Mx1-Cre) BM cells transduced with GFP, Wt KIT, or KITD816V vector, respectively. The numbers above the bars are average colony numbers for the group. (B) Ratio of total colony numbers from 105 cultured WT or Cbfb+/56m; Tg(Mx1-Cre) (MxCre-CM) BM cells transduced with GFP versus those transduced with WT KIT. (C) Types of colonies formed from transduced Cbfb+/56m; Tg(Mx1-Cre) BM cells. CFU indicates colony-forming unit; G, granulocyte; E, erythrocyte; MM, monocyte and megakaryocyte; and GM, granulocyte and monocyte. All BM cells were sorted for GFP+ cells before plating. (D) Comparison of annexin V+ cell numbers (percentage) between the KIT (both WT and 816 mutants)–transduced (GFP+) and untransduced (GFP−) BM lin− cell populations, for both WT and Cbfb+/56m; Tg(Mx1-Cre). (E) Comparison of annexin V+ cell numbers (percentage) between the WT and Cbfb+/56m; Tg(Mx1-Cre) BM lin− cell populations that were transduced with WT KIT or KIT816Y/V. (F) BrdU+ cells in KIT (WT and 816 mutants)–transduced BM cells.
KIT expression is toxic to lin− BM cells
Both WT and Cbfb+/56m; Tg(Mx1-Cre) lin− BM cells exhibited greatly increased cell death after transduction with human KIT (both WT and 816 mutants; Figure 1D). Consistent with the data in Figure 1B, KIT-transduced Cbfb+/56m; Tg(Mx1-Cre) lin− BM cells showed less cell death compared with WT lin− BM cells (P = .05; Figure 1D), especially with the 816 mutants (Figure 1E). However, there is no significant difference in cell death between WT KIT and 816 mutant KIT, for either Cbfb+/56m; Tg(Mx1-Cre) or WT lin− BM cells. Overall, the data suggest that KIT is toxic to lin− BM cells, and this effect is more severe against WT than against Cbfb+/56m; Tg(Mx1-Cre) cells.
D816Y mutants enhance the proliferation of lin− BM cells
We assessed the proliferation rate of KIT-transduced lin− BM cells by BrdU staining. For both WT and Cbfb+/56m; Tg(Mx1-Cre) cells, there is a significant increase of proliferation by D816Y KIT transduction than by WT KIT transduction (Figure 1F). In contrast, the D816V mutation did not affect the proliferation rate of the transduced cells.
816-KIT transduced BM cells contribute to all lineages in PB
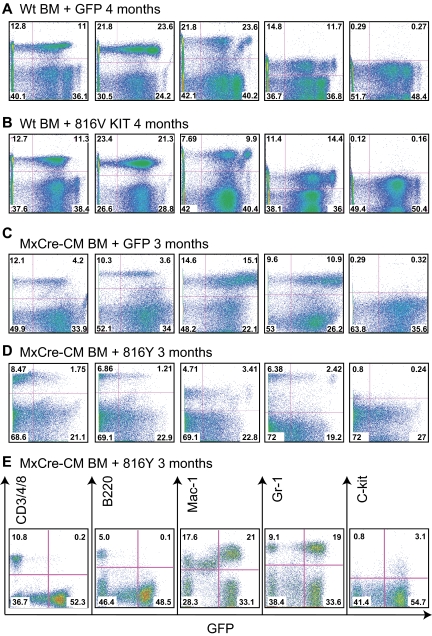
WT BM cells transduced with WT, KIT D816 mutations, or GFP vector alone were transplanted to irradiated congenic C57BL/6 × 129/SvEv F1 hybrid mice. GFP+ cells were found in the PB as early as 1 week after transplantation, which remained detectable 12 months after transplantation (the end point of the experiments). This result indicates that the retroviral vectors transduced not only progenitor cells but also hematopoietic stem cells. T (CD3+ or CD4+ or CD8+), B (B220+), and myeloid cells (Mac1+ or Gr1+) derived from the transplanted cells were all detectable in the recipient mice (Figure 2A-B), suggesting that the KIT D816 mutations do not impair the differentiation potential of WT BM cells.
Figure 2.
Lineage contribution of KIT-transduced bone marrow cells in transplanted mice. PB cells were collected at the indicated times after transplantation from the recipient mice, stained for the indicated markers, and analyzed by FACS. (A) WT BM cells transduced with GFP. (B) WT BM cells transduced with KITD816. (C) Cbfb+/56m; Tg(Mx1-Cre) BM cells transduced with GFP. (D) Cbfb+/56m; Tg(Mx1-Cre) BM cells transduced with KITD816Y. (E) PB from a mouse that was starting to develop leukemia that was transplanted with Cbfb+/56m; Tg(Mx1-Cre) BM cells expressing KITD816Y. CD3/4/8 indicates combination staining of CD3, CD4, and CD8.
The Cbfb+/56m; Tg(Mx1-Cre) BM cells also were transduced with WT or D816 Y/V KIT retroviruses and then transplanted into recipient mice. In the PB of the transplanted mice, GFP+ cells also contributed to all lineages (Figure 2C-D and supplemental Figure 2). However, when leukemia started to develop (Figure 2E), GFP+ cells were mostly Mac1+, Gr-1+, or Mac1+Gr-1+ myeloid cells with few or no B and T cells (Figure 2E), suggesting blockage of the lymphoid differentiation during leukemic transformation. This result correlates with previously published studies demonstrating blockage of T- and B-cell development by CBFB-MYH1132,35 and suggests that the KIT D816 mutations do not impair the contribution of WT BM cells to mature blood cells in the PB in the early stages. However, we cannot rule out hidden differentiation defects in the bone marrow.
KIT D816Y/V mutants cooperate with Cbfb-MYH11 for leukemia initiation
We monitored the disease progression with FACS analysis of GFP+ cells in the PB. In mice that received WT BM cells transduced with KIT, the percentages of GFP+ cells dropped greatly 3 months after transplantation (Table 1). In mice that received the transduced Cbfb+/56m; Tg(Mx1-Cre) BM cells, the decreasing of GFP+ cells was not as dramatic as in the WT BM cells at 12 weeks (Table 2). However, for those mice that did not eventually develop leukemia, the percentages of GFP+ cells in PB also dropped to very low levels. These in vivo findings are consistent with the in vitro data demonstrating higher toxicity of human KIT expression to WT BM cells (Figure 1D-E).
Table 1.
GFP level at different time points after transplantation with WT BM cells
| Vector | Animal sample size, N | Mean ± SD GFP, % |
|||
|---|---|---|---|---|---|
| 3 wk | 12 wk | 6 mo | 12 mo | ||
| GFP | 4 | 61 ± 30 | 50 ± 29 | 58 ± 36 | 47 ± 25 (N = 3) |
| KIT wt | 5 | 24 ± 8 | 3 ± 2 | 5 ± 5 | 4 ± 3.7 (N = 3) |
| 816Y | 3 | 21 ± 7 | 0.7 ± 0.2 | 0.4 ± 0.6 | 1.5 ± 0.8 |
| 816V | 6 | 35 ± 10 | 1.6 ± 0.9 | 0.8 ± 0.6 | 2.5 ± 1.0 |
Table 2.
GFP level at different time points after transplantation with MxCre-CM BM cells (excluding mice that developed leukemia)
| Vector | Animal sample size, N | Mean ± SD GFP, % |
||||
|---|---|---|---|---|---|---|
| 3 wk | 8 wk | 12 wk | 6 mo | 12 mo | ||
| GFP | 4 | 36 ± 7 | 30 ± 12 | 2 ± 2 | 3 ± 0.4 (N = 2) | |
| KIT wt | 5 | 24 ± 7 | 33 ± 8 | 0.8 ± 0.8 | 2.4 ± 0.7 (N = 4) | |
| 816Y | 9 | 48 ± 14 | 37 ± 29 (N = 7) | 3 ± 2 (N = 4) | 0.8 ± 0.1 (N = 2) | |
| D816V | 9 | 38 ± 7 | 42 ± 10 | 4 ± 6 | 3.5 ± 2.9 (N = 5) | |
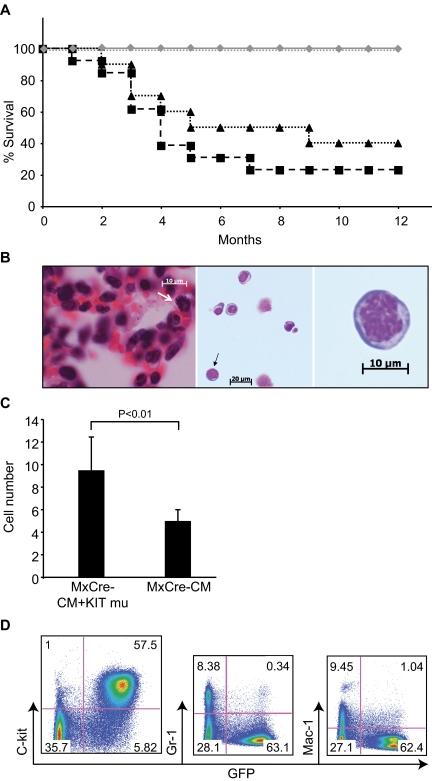
The transplanted mice were observed for up to 12 months after transplantation. By 5 months after transplantation, 70% mice transplanted with Cbfb+/56m; Tg(Mx1-Cre) /KITD816Y (N = 13) and 50% mice transplanted with Cbfb+/56m; Tg(Mx1-Cre)/KITD816V (N = 10) BM cells died from leukemia (Figure 3A). Sixty to 80% of mice died from leukemia in these 2 groups within a year (Figure 3A). Alternatively, no mice transplanted with Cbfb+/56m; Tg(Mx1-Cre)/KIT or Cbfb+/56m; Tg(Mx1-Cre)/GFP BM cells developed leukemia during the 1 year period (N = 18; Figure 3A). In addition, WT BM cells transduced with KITD816Y/V did not cause leukemia within the 12 months after transplantation (N = 15; Figure 3A). These results suggest that KITD816Y/V does not cause leukemia by itself but cooperates with Cbfb-MYH11 for leukemogenesis in this mouse model.
Figure 3.
Leukemia development in mice transplanted with Cbfb+/56m; Tg(Mx1-Cre)/KITD816Y/V BM cells. (A) Survival curves of transplanted mice. Black dashed line with squares indicate mice transplanted with Cbfb+/56m; Tg(Mx1-Cre)/KITD816Y BM cells (N = 13); black dotted line with triangles, mice transplanted with Cbfb+/56m; Tg(Mx1-Cre)/KITD816V BM cells (N = 10); gray line with diamonds, mice transplanted with Cbfb+/56m; Tg(Mx1-Cre)/KIT or Cbfb+/56m; Tg(Mx1-Cre)/GFP BM cells (N = 18); and gray dotted line, mice transplanted with KIT (both WT and D816 mutants)–transduced WT BM cells (N = 15). (B) H&E stained lung tissue (left) and Wright Giemsa–stained leukemia cells in PB (middle and right, respectively) from a Cbfb+/56m; Tg(Mx1-Cre)/KITD816Y leukemic mouse. White arrow in the left panel indicates a cell in mitosis. Black arrow in the middle panel indicates a cell that has been enlarged in the right panel. (Imager D2, Zeiss; 20×, 40×, and 63× plan-Apochromat objective lenses; AxioVision 4.8 acquision software; AxioCam HRC, Zeiss). (C) Mitotic cell count from leukemic spleen sections of Cbfb+/56m; Tg(Mx1-Cre) mice (MxCre-CM; N = 3) and Cbfb+/56m; Tg(Mx1-Cre)/KITD816Y (MxCre-CM + KIT mu; N = 3) mice. (D) FACS analysis of PB cells from a leukemic mouse transplanted with Cbfb+/56m; Tg(Mx1-Cre)/KITD816Y BM cells.
The morphology of the leukemia cells showed a progenitor cell appearance with large nuclei and scant cytoplasm, and mitotic cells were easily detectable, suggesting active proliferation of these cells (Figure 3B). The mitotic cell count from the spleen sections of leukemia mice showed a significant increase in the Cbfb+/56m; Tg(Mx1-Cre)/KIT816Y mice (N = 5) than the Cbfb+/56m; Tg(Mx1-Cre) mice (N = 5; Figure 3C). There were significantly higher percentages (P = .05) of lin− BM cells stained positive for BrdU from leukemic mice carried KIT mutations (48% of the GFP+/lin− cells; N = 5) than from leukemic mice with only Cbfb-MYH11 (28% of the c-kit+/lin− cells; N = 5). FACS staining showed that they were mainly c-kit+ and lineage-negative cells, with minimal myeloid differentiation (Figure 3D). The leukemia cells infiltrated BM, spleen, liver, kidneys, lungs, heart, and meninges of the spinal cord in all mice examined (supplemental Figure 3). In addition, leukemia cells infiltrated the brain tissue in 2 of the Cbfb+/56m; Tg(Mx1-Cre)/KIT816Y mice.
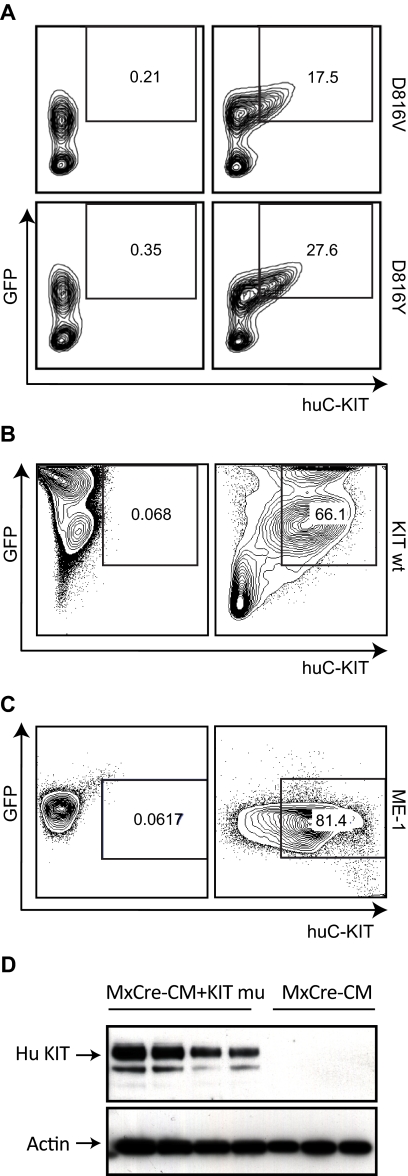
To confirm that the transplanted GFP+ cells expressed the human KIT proteins encoded by the retroviral vectors, we stained the cells for the human KIT protein by FACS analysis. Human KIT protein was detected on the surface of some of the GFP+ leukemic spleen cells (Figure 4A) from mice transplanted with Cbfb+/56m; Tg(Mx1-Cre)/KITD816Y/V bone marrow cells. Human KIT also was detected on the surface of nonleukemic spleen cells from mice transplanted with Cbfb+/56m; Tg(Mx1-Cre)/KIT bone marrow cells (Figure 4B). The human leukemia cell line ME-1 was used as a positive control for human KIT staining (Figure 4C). However, human KIT was not readily detectable on the PB cells before or after leukemia development.
Figure 4.
Expression of human KIT protein in the transplanted mice. (A) Leukemic cells from spleens of mice transplanted with Cbfb+/56m; Tg(Mx1-Cre)/KITD816V or Cbfb+/56m; Tg(Mx1-Cre)/KITD816Y BM cells. (B) Splenocytes from a mouse (nonleukemic) transplanted with Cbfb+/56m; Tg(Mx1-Cre)/KIT wt BM cells. (C) Leukemia cell line ME-1. The cells in panels A-C were stained with an anti–human KIT antibody and analyzed by FACS. In panels A through C, the cells in the left-hand panels were unstained and those in the right-hand panels were stained with the anti–human KIT. Cells in the boxes are GFP+ and human KIT+. (D) Western blot of leukemic spleen cells from Cbfb+/56m; Tg(Mx1-Cre)/KITD816Y (MxCre-CM + KIT mu) and Cbfb+/56m; Tg(Mx1-Cre) (MxCre-CM) mice using the anti–human KIT antibody. Actin was probed as the loading control.
We also performed Western blot using leukemia tissue from different mice and confirmed that the human KIT protein was expressed abundantly in the Cbfb+/56m; Tg(Mx1-Cre)/KITD816Y but not the Cbfb+/56m; Tg(Mx1-Cre) leukemic cells (Figure 4D).
Accelerated leukemia progression and enrichment of leukemia-initiating cells by the KITD816Y/V mutants
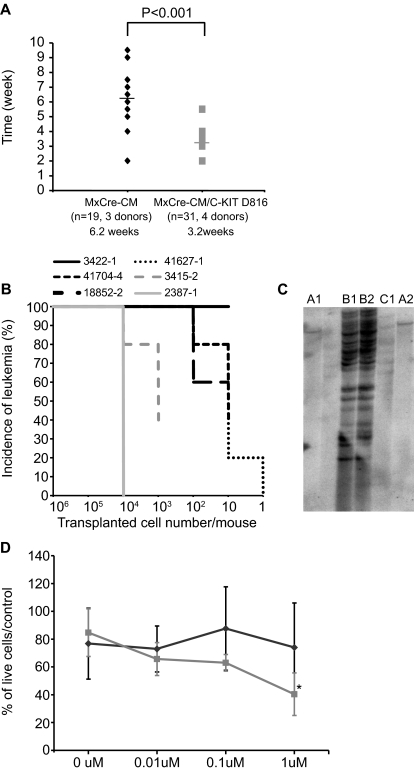
To see the effect of KITD816Y/V mutants on disease progression, the life span of mice transplanted with leukemia cells carrying Cbfb-MYH11 and KIT D816 mutants [Cbfb+/56m; Tg(Mx1-Cre)/KITD816Y/V] or Cbfb-MYH11 alone [Cbfb+/56m; Tg(Mx1-Cre)] was measured. As shown in Figure 5A, mice transplanted with leukemia cells carrying Cbfb-MYH11 and KIT D816 mutants had a much shorter average life span of 3.2 weeks (N = 31, from 4 donors) compared with 6.2 weeks (N = 19, from 3 donors) for mice transplanted with Cbfb-MYH11 leukemia cells.
Figure 5.
Accelerated leukemia development and increased LICs by KITD816V/Y. (A) Disease latency in secondarily transplanted mice with leukemic cells of Cbfb+/56m; Tg(Mx1-Cre) (black diamonds) and Cbfb+/56m; Tg(Mx1-Cre)/KITD816V/Y (gray squares). Each recipient mouse was given 106 leukemia cells. (B) Leukemia incidences from limiting dilution transplantation with Cbfb+/56m; Tg(Mx1-Cre)/KITD816Y leukemia cells (N = 4; black lines) and Cbfb+/56m; Tg(Mx1-Cre) leukemia cells (N = 2; gray lines). Numbers of cells injected per mouse are shown on x-axis. N = 5 in each dose group. (C) Genomic Southern blot hybridization for clonality analysis. Each lane is from one secondarily transplanted mouse. Panels A, B, and C are 3 different donors (lanes not labeled had degraded DNA). (D) Leukemia cell viability in culture after PKC412 treatment. Gray line indicates Cbfb+/56m; Tg(Mx1-Cre)/KITD816Y mice (N = 4); and black line, Cbfb+/56m; Tg(Mx1-Cre) mice (N = 5; *P < .01).
We then did limiting-dilution transplantation using leukemia cells (total live splenocytes with > 80% of GFP+ leukemia cells) that carried Cbfb+/56m; Tg(Mx1-Cre)/KITD816Y/V from 4 different donor mice (Figure 5B). For 1 donor (3422-1), all recipient mice died from leukemia with only 10 cells transplanted. For the other 3 donors, all mice that received 1000 cells died from leukemia as well. In contrast, 105 cells were needed for the Cbfb+/56m; Tg(Mx1-Cre) leukemia cells (from 2 donors) to achieve 100% leukemia development in the recipient mice (Figure 5B). Moreover, the latencies were much longer for mice transplanted with Cbfb+/56m; Tg(Mx1-Cre) leukemia cells (7-15 weeks) than those with the Cbfb+/56m; Tg(Mx1-Cre)/KITD816Y/V leukemic cells (2-10 weeks).
Overall, the data showed that leukemia development in the recipient mice transplanted with Cbfb+/56m; Tg(Mx1-Cre)/KITD816Y/V leukemia cells was faster and required fewer starting cells. This suggests a higher leukemia-initiating cell (LIC) frequency in the Cbfb+/56m; Tg(Mx1-Cre)/KITD816Y/V leukemic cells.
Clonality analysis of leukemia cells
We performed Southern blot hybridization using DNA from secondarily transplanted mice and found that every tested founder was a specific clone and that the secondarily transplanted mice derived from each founder carried the same retroviral insertions (Figure 5C). The copy number of the insertions did not correlate with the latency of leukemia development. One mouse with only 1 copy of the mutant KIT transgene developed leukemia within 3 months, whereas another mouse with multiple copies developed leukemia ∼ 5 months after transplantation. Furthermore, the monoclonal nature of the first mouse suggests that additional events play important roles in transformation, whereas the polyclonal leukemia emergence in the second mouse suggests that coexpression of KIT D816 mutations and CBFb-MYH11 may be sufficient to initiate leukemogenesis.
Cbfb-MYH11/KIT D816 mutant but not Cbfb-MYH11 leukemia cells are sensitive to PKC412
Primary leukemia cells with or without KIT D816 mutations [Cbfb+/56m; Tg(Mx1-Cre)/KITD816Y, N = 4; Cbfb+/56m; Tg(Mx1-Cre), N = 5] were treated with protein kinase inhibitor PKC412 at various concentrations in culture. Cbfb+/56m; Tg(Mx1-Cre)/KITD816Y leukemia cells showed a dose-dependent decrease in cell viability after PKC412 treatment for 17 hours, reaching more than 50% reduction at 1μM (Figure 5D). Alternatively, the viability of Cbfb+/56m; Tg(Mx1-Cre) leukemia cells did not change significantly after PKC412 treatment. The data suggest that KIT D816 mutations sensitize the Cbfb-MYH11 leukemia cells to this protein kinase inhibitor.
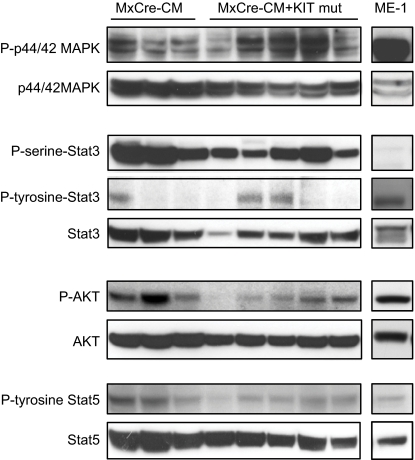
MAPK and Stat3 pathways are activated in leukemic cells
KITD816 V/Y mutations cause constitutive activation of KIT that can then activate several downstream signaling pathways that are potentially involved in the transformation of Cbfb-MYH11 BM cells.28,36,37 Therefore, we checked the downstream signaling pathways of KIT using leukemia cells from our mouse models. We found that p44/42 MAPK was strongly phosphorylated in the Cbfb+/56m; Tg(Mx1-Cre)/KITD816Y leukemic samples that we examined (Figure 6). Interestingly, p44/42 MAPK also was phosphorylated in Cbfb-MYH11 leukemia mouse tissue without the transduced KIT mutants. In addition, serine but not tyrosine phosphorylation of Stat3 was strongly activated in all leukemia cells tested. In contrast, the phosphorylation level of Stat5 and AKT was low in most of the leukemia samples, especially those with Cbfb+/56m; Tg(Mx1-Cre)/KITD816Y (Figure 6).
Figure 6.
Phosphorylation status of MAPK/Stat3/Stat5/Akt in leukemia cells. Western blot analysis of leukemia cells from Cbfb+/56m; Tg(Mx1-Cre)/KITD816Y (MxCre-CM + KIT mut) mice and Cbfb+/56m; Tg(Mx1-Cre) mice (MxCre-CM) and human ME-1 cells for the indicated proteins and their phosphorylated products.
Discussion
KIT mutations have been found in leukemia5,38–40 and solid tumors, such as gastrointestinal stromal tumor,41 breast cancer,42 and lung cancer.43 Two specific mutations affecting the aspartic acid residue 816 of KIT, D816Y and D816V, have been reported in CBF AML cases and are potential indicators of poor clinical outcomes.6,21,25,44 Here, we report a mouse leukemia model that coexpresses the human D816V/Y-KIT and a knockin allele of Cbfb-MYH11 (Cbfb+/56m). Similar to what we reported previously, Cbfb-MYH11 by itself or with WT KIT does not cause leukemia within 12 months after transplantation. In contrast, coexpressing Cbfb-MYH11 and D816Y or D816V KIT led to leukemia development in 60% to 80% of the mice in 9 months (Figure 3). Therefore, our data demonstrate cooperation between Cbfb-MYH11 and mutant KIT during leukemogenesis. Moreover, in the presence of the mutant KIT, the disease progression was more aggressive, and the number of LICs was increased, as shown by shortened life span of secondarily transplanted mice and by limiting dilution transplantation.
KIT is a potent growth factor for hematopoietic progenitor cells, and its expression is normally decreased in more differentiated cells.45 The reduced colony numbers observed in our study (Figure 1A) would have resulted from massive cell death caused by the expression of KIT, either WT or mutant, from the retroviral vectors (Figure 1D). This demonstrates that KIT is toxic to BM cells. However, Cbfb-MYH11–expressing BM cells were more resistant to the KIT toxicity (Figure 1D-E), allowing the cells coexpressing both to survive and undergo leukemic transformation. Expressing the D816Y mutant in BM cells also led to an increased proliferation (Figure 1F) that also may have contributed to leukemia development in these mice. In addition, BM lin− leukemia cells coexpressing D816Y/V KIT and Cbfb-MYH11 proliferated faster than leukemia cells with only Cbfb-MYH11 (48% vs 28%), which correlated with more aggressive disease and higher LIC frequency for the Cbfb+/56m; Tg(Mx1-Cre)/KITD816V/Y leukemia.
Leukemia did take 2 to 9 months to develop in the recipient mice transplanted with Cbfb+/56m; Tg(Mx1-Cre) BM cells expressing mutant KIT transgenes. Such protracted and prolonged latency would suggest that additional mutations are required for full leukemic transformation. Clonality analysis showed that some leukemia samples had as few as 1 copy of KIT integration, whereas others had multiple copies of KIT insertion. These observations also suggest that additional events might contribute to leukemogenesis, especially those with only 1 copy of KIT. Moreover, we used pI:pC to induce the expression of Cbfb-MYH11, which is not 100%. This also may have contributed to the long latency and incomplete penetrance of leukemia.
Interestingly, overexpression of mutant KIT, but not the WT KIT, in the BM cells derived from Cbfb+/56m; Tg(Mx1-Cre) mice led to significant increases of CFU-GEMM, which contains early myeloid progenitor cells (Figure 1C). This effect may lead to an expansion of immature myeloid progenitors in the bone marrow, the potential target cells for transformation by Cbfb-MYH11. Although we did not investigate accumulation of myeloid cells in the BM of transplanted mice, we did observe Mac1+/Gr1+/GFP+ myeloid cells in the PB in early leukemic mice (Figure 2E). Therefore, this expansion might have contributed to the accelerated leukemia development and increased numbers of LICs in the mice coexpressing Cbfb-MYH11 and mutant KIT (Figures 3A and 5).
It has been proposed that SCF and KIT play important roles for the survival and proliferation of cancer stem cells.46 Our data support the notion that activated KIT proteins increase the proliferation of transduced BM cells and leukemia cells (Figures 1F and 3C) and could be a mechanism for the observed higher frequencies of LICs (Figure 5). KIT activation may provide a proliferative or survival signal to the LICs in our model, through signaling pathways such as MAPK and Stat3 that were strongly activated in the Cbfb-MYH11 leukemia cells carrying KIT D816Y/V mutations. Interestingly, MAPK and Stat3 also were phosphorylated in Cbfb-MYH11 leukemia cells without mutant KIT. These data suggest that the MAPK and Stat3 pathways play important roles in Cbfb-MYH11 leukemia cells, with or without KIT mutations.
PKC412 is a broad protein kinase inhibitor that has been reported to be effective for KITD816 mutations.28,30 PKC412 reduced viability of Cbfb+/56m; Tg(Mx1-Cre)/KITD816Y leukemia cells, whereas it did not affect Cbfb+/56m; Tg(Mx1-Cre) leukemia cells (Figure 5D). This suggested that even though the leukemia cells from both genotypes had activated MAPK and Stat3, the causative signal(s) in the KITD816Y leukemia cells were not activating mutations in tyrosine kinase genes. Inhibitors of MAPK and Stat3 pathways might be useful for treating CBF leukemia and should be tested in the future.
Here, we used human KIT cDNA constructs that have been considered problematic in mouse models in previous reports.47,48 Xiang et al reported that human KITD816Vdid not transduce mouse cells well, probably because the protein was blocked at the endoplasmic reticulum in a species-specific manner in mouse cells and did not induce disease in mice.47 Conversely, mouse KITD814V or a human-mouse hybrid KITD816V protein with mouse extracellular or transmembrane domains functioned properly and could induce rapid fatal myeloproliferative diseases.47 In a more recent study, HyC-KITN822K and D816V, as well as juxtamembrane mutants HyC-KIT571 + 14 and 557-558Del, could transform murine 32D cells to cytokine-independent growth, and coexpressing human–mouse hybrid KITN822K and the AML1-ETO fusion gene led to the development of fatal AML.48 In several other reports, however, human KITD816V was found to be functional in mouse cells in culture,28,49,50 and induced factor-independent growth of Ba/F3 in 2 of the studies.28,49
During our experiments, we also encountered significant problems with low transduction efficiency with human KIT constructs. However, we found that producing viral particles using the mouse packaging cell line GP+E-8633 rather than using human cells, such as 293, significantly improved the efficiency of transducing mouse BM cells. In contrast, our observation that the human KIT proteins could be easily detected by Western blot, but only in a small fraction of splenic leukemia cells by FACS (Figure 4), suggests a similar trafficking defect of the human KIT protein in mouse cells as described by Xiang et al.47 Consistent with their results, human KIT D816 mutations alone could not induce leukemia or other blood-related diseases in our mouse model. Nonetheless, our data unequivocally demonstrated cooperation between human KITD816V/Y and CBFB-MYH11 for leukemogenesis. Our data suggest that even though these human KIT D816 mutants do not translocate to the plasma membrane efficiently, they do have oncogenic activity. We found that the mutant KIT proteins not only were less toxic to Cbfb-MYH11 BM cells but also enhanced the proliferation of these BM cells and must have contributed to leukemogenesis. It is possible that murine versions of the KIT mutants or human–mouse hybrid KIT mutants have higher oncogenic activities and may have worked more efficiently. A comparison between them, which is beyond the scope of this manuscript, will be interesting to perform.
In this mouse model, we have proved that leukemic cells with Cbfb-MYH11 and KIT D816Y/V mutations cause more aggressive disease compared with leukemic cells carry only Cbfb-MYH11, which is likely because of increased LICs in the Cbfb+/56m; Tg(Mx1-Cre)/KITD816V/Y leukemia samples. Cell surface staining indicated that GFP+ leukemia cells are mostly CD117+Sca-1−lin− cells, suggesting that future treatment should aim at this early progenitor or stem-like cell population.
Supplementary Material
Acknowledgments
The authors thank David Bodine and Fabio Candotti for helpful discussion for the retroviral transduction of mouse BM cells, Christopher Klug for critical reading of the manuscript, and Julia Fekecs for preparation of the figures.
This work was supported by the Intramural Research Program, National Human Genome Research Institute, National Institutes of Health.
Footnotes
The online version of the article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: L.Z., D.G.G., and P.P.L. designed the research; L.Z., J.J.M., L.A., M. Kirby, S.A., M. Kench, S.H.-M., L.B., and Y.K. performed the research; L.Z., J.J.M., and Y.K. collected the data; L.Z. and P.P.L. analyzed the data; D.G.G. contributed the KIT vectors; and L.Z. and P.P.L. wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
The current affiliation for Y.K. is Department of Hematology and Oncology, University of Tokyo, Tokyo, Japan. The current affiliation for D.G.G. is Merck Research Labs, Whitehouse Station, NJ.
Correspondence: P. Paul Liu, 49 Convent Dr, 49/3A26, Bethesda, MD 20892; e-mail: pliu@mail.nih.gov.
References
- 1.Dunphy CH. Comprehensive review of adult acute myelogenous leukemia: cytomorphological, enzyme cytochemical, flow cytometric immunophenotypic, and cytogenetic findings. J Clin Lab Anal. 1999;13(1):19–26. doi: 10.1002/(SICI)1098-2825(1999)13:1<19::AID-JCLA4>3.0.CO;2-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liu P, Tarle SA, Hajra A, et al. Fusion between transcription factor CBF beta/PEBP2 beta and a myosin heavy chain in acute myeloid leukemia. Science. 1993;261(5124):1041–1044. doi: 10.1126/science.8351518. [DOI] [PubMed] [Google Scholar]
- 3.Castilla LH, Garrett L, Adya N, et al. The fusion gene Cbfb-MYH11 blocks myeloid differentiation and predisposes mice to acute myelomonocytic leukaemia. Nat Genet. 1999;23(2):144–146. doi: 10.1038/13776. [DOI] [PubMed] [Google Scholar]
- 4.Castilla LH, Perrat P, Martinez NJ, et al. Identification of genes that synergize with Cbfb-MYH11 in the pathogenesis of acute myeloid leukemia. Proc Natl Acad Sci U S A. 2004;101(14):4924–4929. doi: 10.1073/pnas.0400930101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Valk PJ, Bowen DT, Frew ME, Goodeve AC, Lowenberg B, Reilly JT. Second hit mutations in the RTK/RAS signaling pathway in acute myeloid leukemia with inv(16). Haematologica. 2004;89(1):106. [PubMed] [Google Scholar]
- 6.Boissel N, Leroy H, Brethon B, et al. Incidence and prognostic impact of c-KIT, FLT3, and Ras gene mutations in core binding factor acute myeloid leukemia (CBF-AML). Leukemia. 2006;20(6):965–970. doi: 10.1038/sj.leu.2404188. [DOI] [PubMed] [Google Scholar]
- 7.Care RS, Valk PJ, Goodeve AC, et al. Incidence and prognosis of c-KIT and FLT3 mutations in core binding factor (CBF) acute myeloid leukaemias. Br J Haematol. 2003;121(5):775–777. doi: 10.1046/j.1365-2141.2003.04362.x. [DOI] [PubMed] [Google Scholar]
- 8.Kim HG, Kojima K, Swindle CS, et al. FLT3-ITD cooperates with inv(16) to promote progression to acute myeloid leukemia. Blood. 2008;111(3):1567–1574. doi: 10.1182/blood-2006-06-030312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grisolano JL, O'Neal J, Cain J, Tomasson MH. An activated receptor tyrosine kinase, TEL/PDGFbetaR, cooperates with AML1/ETO to induce acute myeloid leukemia in mice. Proc Natl Acad Sci U S A. 2003;100(16):9506–9511. doi: 10.1073/pnas.1531730100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shih LY, Liang DC, Huang CF, et al. Cooperating mutations of receptor tyrosine kinases and Ras genes in childhood core-binding factor acute myeloid leukemia and a comparative analysis on paired diagnosis and relapse samples. Leukemia. 2008;22(2):303–307. doi: 10.1038/sj.leu.2404995. [DOI] [PubMed] [Google Scholar]
- 11.Edling CE, Hallberg B. c-KIT–a hematopoietic cell essential receptor tyrosine kinase. Int J Biochem Cell Biol. 2007;39(11):1995–1998. doi: 10.1016/j.biocel.2006.12.005. [DOI] [PubMed] [Google Scholar]
- 12.Kissel H, Timokhina I, Hardy MP, et al. Point mutation in KIT receptor tyrosine kinase reveals essential roles for KIT signaling in spermatogenesis and oogenesis without affecting other KIT responses. EMBO J. 2000;19(6):1312–1326. doi: 10.1093/emboj/19.6.1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Spritz RA, Giebel LB, Holmes SA. Dominant negative and loss of function mutations of the c-KIT (mast/stem cell growth factor receptor) proto-oncogene in human piebaldism. Am J Hum Genet. 1992;50(2):261–269. [PMC free article] [PubMed] [Google Scholar]
- 14.Hirota S, Isozaki K, Moriyama Y, et al. Gain-of-function mutations of c-KIT in human gastrointestinal stromal tumors. Science. 1998;279(5350):577–580. doi: 10.1126/science.279.5350.577. [DOI] [PubMed] [Google Scholar]
- 15.Ikeda H, Kanakura Y, Tamaki T, et al. Expression and functional role of the proto-oncogene c-KIT in acute myeloblastic leukemia cells. Blood. 1991;78(11):2962–2968. [PubMed] [Google Scholar]
- 16.Kita K, Shirakawa S, Kamada N. Cellular characteristics of acute myeloblastic leukemia associated with t(8;21)(q22;q22). The Japanese Cooperative Group of Leukemia/Lymphoma. Leuk Lymphoma. 1994;13(3-4):229–234. doi: 10.3109/10428199409056286. [DOI] [PubMed] [Google Scholar]
- 17.Osato M, Asou N, Okubo T, et al. Myelomonoblastic leukaemia cells carrying the PEBP2beta/MYH11 fusion gene are CD34, c-KIT+ immature cells. Br J Haematol. 1997;97(3):656–658. doi: 10.1046/j.1365-2141.1997.1012917.x. [DOI] [PubMed] [Google Scholar]
- 18.Müller AM, Duque J, Shizuru JA, Lubbert M. Complementing mutations in core binding factor leukemias: from mouse models to clinical applications. Oncogene. 2008;27(44):5759–5773. doi: 10.1038/onc.2008.196. [DOI] [PubMed] [Google Scholar]
- 19.Beghini A, Ripamonti CB, Cairoli R, et al. KIT activating mutations: incidence in adult and pediatric acute myeloid leukemia, and identification of an internal tandem duplication. Haematologica. 2004;89(8):920–925. [PubMed] [Google Scholar]
- 20.Gari M, Goodeve A, Wilson G, et al. c-KIT proto-oncogene exon 8 in-frame deletion plus insertion mutations in acute myeloid leukaemia. Br J Haematol. 1999;105(4):894–900. doi: 10.1046/j.1365-2141.1999.01449.x. [DOI] [PubMed] [Google Scholar]
- 21.Paschka P, Marcucci G, Ruppert AS, et al. Adverse prognostic significance of KIT mutations in adult acute myeloid leukemia with inv(16) and t(8;21): a Cancer and Leukemia Group B Study. J Clin Oncol. 2006;24(24):3904–3911. doi: 10.1200/JCO.2006.06.9500. [DOI] [PubMed] [Google Scholar]
- 22.Beghini A, Peterlongo P, Ripamonti CB, et al. C-KIT mutations in core binding factor leukemias. Blood. 2000;95(2):726–727. [PubMed] [Google Scholar]
- 23.Kohl TM, Schnittger S, Ellwart JW, Hiddemann W, Spiekermann K. KIT exon 8 mutations associated with core-binding factor (CBF)-acute myeloid leukemia (AML) cause hyperactivation of the receptor in response to stem cell factor. Blood. 2005;105(8):3319–3321. doi: 10.1182/blood-2004-06-2068. [DOI] [PubMed] [Google Scholar]
- 24.Furitsu T, Tsujimura T, Tono T, et al. Identification of mutations in the coding sequence of the proto-oncogene c-KIT in a human mast cell leukemia cell line causing ligand-independent activation of c-KIT product. J Clin Invest. 1993;92(4):1736–1744. doi: 10.1172/JCI116761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schnittger S, Kohl TM, Haferlach T, et al. KIT-D816 mutations in AML1-ETO-positive AML are associated with impaired event-free and overall survival. Blood. 2006;107(5):1791–1799. doi: 10.1182/blood-2005-04-1466. [DOI] [PubMed] [Google Scholar]
- 26.Marková J, Trnková Z, Michková P, et al. Monitoring of minimal residual disease in patients with core binding factor acute myeloid leukemia and the impact of C-KIT, FLT3, and JAK2 mutations on clinical outcome. Leuk Lymphoma. 2009;50(9):1448–1460. doi: 10.1080/10428190903085951. [DOI] [PubMed] [Google Scholar]
- 27.Ma Y, Zeng S, Metcalfe DD, et al. The c-KIT mutation causing human mastocytosis is resistant to STI571 and other KIT kinase inhibitors; kinases with enzymatic site mutations show different inhibitor sensitivity profiles than wild-type kinases and those with regulatory-type mutations. Blood. 2002;99(5):1741–1744. doi: 10.1182/blood.v99.5.1741. [DOI] [PubMed] [Google Scholar]
- 28.Growney JD, Clark JJ, Adelsperger J, et al. Activation mutations of human c-KIT resistant to imatinib mesylate are sensitive to the tyrosine kinase inhibitor PKC412. Blood. 2005;106(2):721–724. doi: 10.1182/blood-2004-12-4617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cairoli R, Beghini A, Morello E, et al. Imatinib mesylate in the treatment of Core Binding Factor leukemias with KIT mutations. A report of three cases. Leuk Res. 2005;29(4):397–400. doi: 10.1016/j.leukres.2004.10.005. [DOI] [PubMed] [Google Scholar]
- 30.Gleixner KV, Mayerhofer M, Aichberger KJ, et al. PKC412 inhibits in vitro growth of neoplastic human mast cells expressing the D816V-mutated variant of KIT: comparison with AMN107, imatinib, and cladribine (2CdA) and evaluation of cooperative drug effects. Blood. 2006;107(2):752–759. doi: 10.1182/blood-2005-07-3022. [DOI] [PubMed] [Google Scholar]
- 31.Gotlib J, Berube C, Growney JD, et al. Activity of the tyrosine kinase inhibitor PKC412 in a patient with mast cell leukemia with the D816V KIT mutation. Blood. 2005;106(8):2865–2870. doi: 10.1182/blood-2005-04-1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kuo YH, Landrette SF, Heilman SA, et al. Cbf beta-SMMHC induces distinct abnormal myeloid progenitors able to develop acute myeloid leukemia. Cancer Cell. 2006;9(1):57–68. doi: 10.1016/j.ccr.2005.12.014. [DOI] [PubMed] [Google Scholar]
- 33.Markowitz DG, Goff SP, Bank A. Safe and efficient ecotropic and amphotropic packaging lines for use in gene transfer experiments. Trans Assoc Am Physicians. 1988;101:212–218. [PubMed] [Google Scholar]
- 34.Gallardo HF, Tan C, Ory D, Sadelain M. Recombinant retroviruses pseudotyped with the vesicular stomatitis virus G glycoprotein mediate both stable gene transfer and pseudotransduction in human peripheral blood lymphocytes. Blood. 1997;90(3):952–957. [PubMed] [Google Scholar]
- 35.Zhao L, Cannons JL, Anderson S, et al. CBFB-MYH11 hinders early T-cell development and induces massive cell death in the thymus. Blood. 2007;109(8):3432–3440. doi: 10.1182/blood-2006-10-051508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kent D, Copley M, Benz C, Dykstra B, Bowie M, Eaves C. Regulation of hematopoietic stem cells by the steel factor/KIT signaling pathway. Clin Cancer Res. 2008;14(7):1926–1930. doi: 10.1158/1078-0432.CCR-07-5134. [DOI] [PubMed] [Google Scholar]
- 37.Masson K, Ronnstrand L. Oncogenic signaling from the hematopoietic growth factor receptors c-KIT and Flt3. Cell Signal. 2009;21(12):1717–1726. doi: 10.1016/j.cellsig.2009.06.002. [DOI] [PubMed] [Google Scholar]
- 38.Cairoli R, Grillo G, Beghini A, et al. C-KIT point mutations in core binding factor leukemias: correlation with white blood cell count and the white blood cell index. Leukemia. 2003;17(2):471–472. doi: 10.1038/sj.leu.2402795. [DOI] [PubMed] [Google Scholar]
- 39.Goemans BF, Zwaan CM, Cloos J, et al. FLT3 and KIT mutated pediatric acute myeloid leukemia (AML) samples are sensitive in vitro to the tyrosine kinase inhibitor SU11657. Leuk Res. 2010;34(10):1302–1307. doi: 10.1016/j.leukres.2010.04.004. [DOI] [PubMed] [Google Scholar]
- 40.Jiao B, Wu CF, Liang Y, et al. AML1-ETO9a is correlated with C-KIT overexpression/mutations and indicates poor disease outcome in t(8;21) acute myeloid leukemia-M2. Leukemia. 2009;23(9):1598–1604. doi: 10.1038/leu.2009.104. [DOI] [PubMed] [Google Scholar]
- 41.Agaimy A, Dirnhofer S, Wunsch PH, Terracciano LM, Tornillo L, Bihl MP. Multiple sporadic gastrointestinal stromal tumors (GISTs) of the proximal stomach are caused by different somatic KIT mutations suggesting a field effect. Am J Surg Pathol. 2008;32(10):1553–1559. doi: 10.1097/PAS.0b013e31817587ea. [DOI] [PubMed] [Google Scholar]
- 42.Eroğlu A, Sari A. Expression of c-KIT proto-oncogene product in breast cancer tissues. Med Oncol. 2007;24(2):169–174. doi: 10.1007/BF02698036. [DOI] [PubMed] [Google Scholar]
- 43.Yaren A, Oztop I, Kargi A, et al. Bax, bcl-2 and c-KIT expression in non-small-cell lung cancer and their effects on prognosis. Int J Clin Pract. 2006;60(6):675–682. doi: 10.1111/j.1368-5031.2006.00742.x. [DOI] [PubMed] [Google Scholar]
- 44.Cairoli R, Beghini A, Grillo G, et al. Prognostic impact of c-KIT mutations in core binding factor leukemias: an Italian retrospective study. Blood. 2006;107(9):3463–3468. doi: 10.1182/blood-2005-09-3640. [DOI] [PubMed] [Google Scholar]
- 45.Smith MA, Pallister CJ, Smith JG. Stem cell factor: biology and relevance to clinical practice. Acta Haematol. 2001;105(3):143–150. doi: 10.1159/000046556. [DOI] [PubMed] [Google Scholar]
- 46.Levina V, Marrangoni A, Wang T, et al. Elimination of human lung cancer stem cells through targeting of the stem cell factor-c-KIT autocrine signaling loop. Cancer Res. 2010;70(1):338–346. doi: 10.1158/0008-5472.CAN-09-1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xiang Z, Kreisel F, Cain J, Colson A, Tomasson MH. Neoplasia driven by mutant c-KIT is mediated by intracellular, not plasma membrane, receptor signaling. Mol Cell Biol. 2007;27(1):267–282. doi: 10.1128/MCB.01153-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang YY, Zhao LJ, Wu CF, et al. C-KIT mutation cooperates with full-length AML1-ETO to induce acute myeloid leukemia in mice. Proc Natl Acad Sci U S A. 2011;108(6):2450–2455. doi: 10.1073/pnas.1019625108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pedersen M, Ronnstrand L, Sun J. The c-KIT/D816V mutation eliminates the differences in signal transduction and biological responses between two isoforms of c-KIT. Cell Signal. 2009;21(3):413–418. doi: 10.1016/j.cellsig.2008.11.008. [DOI] [PubMed] [Google Scholar]
- 50.Mayerhofer M, Gleixner KV, Hoelbl A, et al. Unique effects of KIT D816V in BaF3 cells: induction of cluster formation, histamine synthesis, and early mast cell differentiation antigens. J Immunol. 2008;180(8):5466–5476. doi: 10.4049/jimmunol.180.8.5466. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.