Abstract
Itraconazole (ITZ) is a mixture of four cis-stereoisomers that inhibit CYP3A4 potently and coordinate CYP3A4 heme via the triazole nitrogen. However, (2R,4S,2′R)-ITZ and (2R,4S,2′S)-ITZ also undergo stereoselective sequential metabolism by CYP3A4 at a site distant from the triazole ring to 3′-OH-ITZ, keto-ITZ, and N-desalkyl-ITZ. This stereoselective metabolism demonstrates specific interactions of ITZ within the CYP3A4 active site. To further investigate this process, the binding and metabolism of the four trans-ITZ stereoisomers by CYP3A4 were characterized. All four trans-ITZ stereoisomers were tight binding inhibitors of CYP3A4-mediated midazolam hydroxylation (IC50 16–26 nM), and each gave a type II spectrum upon binding to CYP3A4. However, instead of formation of 3′-OH-ITZ, they were oxidized at the dioxolane ring, leading to ring scission and formation of two new metabolites of ITZ. These two metabolites were also formed from the four cis-ITZ stereoisomers, although not as efficiently. The catalytic rates of dioxolane ring scission were similar to the dissociation rates of ITZ stereoisomers from CYP3A4, suggesting that the heme iron is reduced while the triazole moiety coordinates to it and no dissociation of ITZ is necessary before catalysis. The triazole containing metabolite [1-(2,4-dichlorophenyl)-2-(1H-1,2,4-triazol-1-yl)ethanone] also inhibited CYP3A4 (IC50 >15 μM) and showed type II binding with CYP3A4. The dioxolane ring scission appears to be clinically relevant because this metabolite was detected in urine samples from subjects that had been administered the mixture of cis-ITZ isomers. These data suggest that the dioxolane ring scission is a metabolic pathway for drugs that contain this moiety.
Introduction
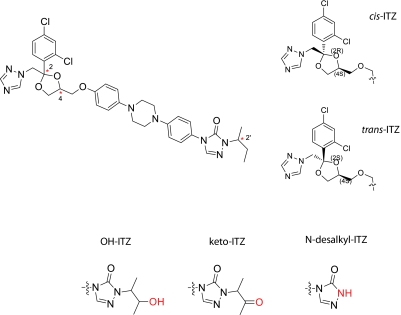
Itraconazole (ITZ) is a broad-spectrum antifungal agent that was designed to inhibit the fungal cytochrome P450 (P450) enzyme lanosterol demethylase by coordinating the heme of the P450 via the triazole nitrogen (Vanden Bossche et al., 1989; Haria et al., 1996). Itraconazole is also a potent inhibitor of human P450 enzymes, specifically CYP3A4, resulting in potent drug-drug interactions (DDIs) with CYP3A4 substrates (Isoherranen et al., 2004; Templeton et al., 2008). In addition to CYP3A4 inhibition, the clinical use of ITZ is limited by its rare hepatotoxicity (Wang et al., 2010). Although ITZ contains three chiral centers (at C-2 and C-4 in the dioxolane ring and at C-2′ on the sec-butyl side chain), resulting in a total of eight possible ITZ stereoisomers (Scheme 1), a mixture of four of them with cis configuration in the dioxolane ring is marketed. Representative cis- and trans-ITZ are shown in Scheme 1.
Scheme 1.
The structure of ITZ with three stereocenters (2, 4, and 2′) indicated with an asterisks in the top left. One example of cis-ITZ and trans-ITZ is shown in the top right. The three metabolites of cis-ITZ are shown below the ITZ structure.
All four cis-ITZ stereoisomers are high-affinity ligands of CYP3A4 and produce type II binding spectra with CYP3A4 indicating that the triazole nitrogen coordinates the heme iron (Kunze et al., 2006). However, two of the four marketed ITZ stereoisomers, (2R,4S,2′R)-ITZ and (2R,4S,2′S)-ITZ, also undergo metabolism by CYP3A4 to form 3′-OH-ITZ, keto-ITZ, and N-desalkyl-ITZ (Scheme 1). The formation of 3′-OH-ITZ, keto-ITZ, and N-desalkyl-ITZ results from oxidation of ITZ at the opposite end of the molecule from the triazole ring that coordinates to the heme (Kunze et al., 2006). Hence, metabolism to the 3′-OH-ITZ requires either reorientation of ITZ within the CYP3A4 active site or dissociation of the type II complex and rebinding of ITZ in the catalytically productive orientation. The latter process was previously shown to be likely for (2R,4S,2′R)-ITZ based on surface plasmon resonance (SPR) experiments (Pearson et al., 2006). However, the reasons for the observed stereoselectivity in ITZ metabolism have not been elucidated.
Recently, all eight stereoisomers of ITZ were reported with evaluation of their antifungal and antiangiogenic activity (Shi et al., 2010). In cell culture models, the trans-ITZ stereoisomers (2R,4R,2′R)-ITZ, (2R,4R,2′S)-ITZ, (2S,4S,2′R)-ITZ, and (2S,4S,2′S)-ITZ were less potent than cis-ITZ stereoisomers in antiangiogenesis activity, but two of the trans-ITZs (2S,4S pair) were as potent as the cis-diastereomers in their antifungal activity (Shi et al., 2010). Whether these trans-ITZs inhibit and/or are metabolized by CYP3A4 is unknown. On the basis of the observed stereoselectivity in cis-ITZ metabolism by CYP3A4, it was hypothesized that the metabolism of trans-ITZ stereoisomers would be stereoselective, and that all four stereoisomers would inhibit CYP3A4 and coordinate the heme iron via the triazole nitrogen. The aims of this study were to determine whether 1) trans-ITZ stereoisomers are potent, stereoselective inhibitors of CYP3A4; 2) trans-ITZ stereoisomers bind to CYP3A4 with affinity similar to that of cis-ITZs in type II fashion; and 3) trans-ITZs are metabolized by CYP3A4 at similar sites as the cis-ITZ isomers. A novel metabolite resulting from dioxolane ring scission was identified from both cis-ITZs and trans-ITZs, and the kinetics of the formation of this metabolite was characterized. The inhibition of CYP3A4 by the 1-(2,4-dichlorophenyl)-2-(1H-1,2,4-triazol-1-yl)ethanone metabolite (M2) was also examined, and the formation of this metabolite in vivo in humans was established. This metabolic pathway may be important for other antifungal drugs that have the dioxolane ring in their structures such as ketoconazole.
Materials and Methods
Chemicals and Reagents.
Trans-itraconazole stereoisomers were synthesized as previously published (Shi et al., 2010), cis-ITZ stereoisomers were gifts from Sepracor (Marlborough, MA). 3′-OH-ITZ was purchased from Research Diagnostics (Flanders, NJ), keto-ITZ was provided by Janssen Pharmaceutica (New Brunswick, NJ), and N-desalkyl-ITZ was prepared as previously reported (Kunze et al., 2006). Midazolam, 1′-hydroxymidazolam (OH-MDZ), and 1′-hydroxymidazolam-d4 were purchased from Cerilliant Corporation (Round Rock, TX). 1-(2,4-Dichlorophenyl)-2-(1H-1,2,4-triazol-1-yl)ethanone was purchased from ChemBridge Screening Library (San Diego, CA). Ammonium chloride was obtained from Mallinckrodt Baker, Inc. (Phillipsburg, NJ), and high-performance liquid chromatography (HPLC)-grade methanol, acetonitrile (ACN), and ethyl acetate were obtained from Thermo Fisher Scientific (Waltham, MA). NADPH was from Sigma-Aldrich (St. Louis, MO). CYP3A4 Supersomes coexpressed with P450 reductase and cytochrome-b5 were purchased from BD Biosciences (San Jose, CA). Pooled human liver microsomes (HLM) were prepared from three livers (all CYP3A5*3/*3 genotype) obtained from the human liver bank of the University of Washington, using equal microsomal protein amounts from each liver to make the pooled stock HLM. The CYP3A4 NF-14 construct was expressed and purified from Escherichia coli as described previously (Gillam et al., 1993). The CYP3A4 concentration was determined by CO difference spectrum using the ε 450 of 91 · mM−1 · cm−1 (Omura and Sato, 1964). CYP3A4 was stored at −80°C in 20% glycerol 80% 100 mM potassium phosphate (KPi) buffer (pH 7.4).
IC50 Determination of Trans-Itraconazole Stereoisomers Toward CYP3A4.
Pooled HLM (0.025 mg/ml, CYP3A5*3/*3) and midazolam (1 μM, MDZ) were incubated with varying concentrations of trans-ITZ isomers (5–1000 nM) in 0.5 ml of KPi buffer to determine IC50. The 1 μM concentration of MDZ, which is ≪Km for MDZ (4 μM) was used to ensure that the IC50 value determined was not dependent on the mechanism (competitive or noncompetitive) of inhibition or was elevated because of competition of CYP3A4 between the substrate and inhibitor. The reaction mixtures were preincubated for 5 min at 37°C and were initiated by adding NADPH (final concentration 1 mM). After 2 min, the reaction was stopped by adding 1 ml of ethyl acetate. Internal standard (20 μl of 0.1 μg/ml 1′-OH-MDZ-d4) was added to each sample. The organic phase was evaporated to dryness under nitrogen stream, reconstituted in 100 μl of 1:1 methanol/water (v/v), transferred to liquid chromatography/mass spectrometry (LC/MS) vials, and analyzed as described previously (Isoherranen et al., 2004) using a Micromass Platform LCZ: Quadrupole Mass Spectrometer (Waters, Milford, MA) operated in the positive ion electrospray mode, monitoring ions m/z 342.2 for 1′-OH-MDZ and m/z 346.2 for 1′-OH-MDZ-d4 using Micromass MassLynx data analysis software (Waters). The metabolites were separated by Waters Alliance 2690 HPLC and an Agilent Zorbax XDB-C8 5-μm column (2.1 i.d. × 50 mm) (Agilent Technologies, Palo Alto, CA). Gradient elution with 0.3 ml/min flow was used starting from 55% aqueous (0.1% acetic acid in water) and 45% methanol (0.1% acetic acid in methanol) to 60% methanol over 1.5 min, maintained for 0.5 min, followed by an increase to 90% methanol over 0.5 min, maintained for one min and then returned to initial conditions. The percentage remaining activity was plotted against the trans-ITZ concentrations, and the IC50 values were determined by nonlinear regression using GraphPad Prism (GraphPad Software Inc., San Diego, CA), according to eq. 1:
in which 100% × (vi/v) is the percentage of activity remaining, (vi/v)max ×100% is the fitted percentage of maximum activity remaining, and (vi/v)min × 100% is the percentage of minimum activity remaining. Terms v and vi refer to the reaction velocity under control and inhibited conditions, I refers to the inhibitor concentrations, and IC50 to the concentration of inhibitor that causes 50% inhibition of enzyme activity.
Determination of Ligand-Induced Binding Spectra.
The binding mode and affinity of the trans-ITZ isomers were determined by spectral titration with purified CYP3A4. The ligand-induced binding spectra (difference spectra) were recorded with Varian Cary 3E UV-Vis spectrophotometer (Varian, Inc., Palo Alto, CA). Matched cuvettes containing purified CYP3A4 (500 nM) in KPi buffer (pH 7.4) with 20% glycerol at 22°C were used. The trans-ITZ stereoisomers (0.005–3.96 μM) or M2 [1-(2,4-dichlorophenyl)-2-(1H-1,2,4-triazol-1-yl)ethanone] (1–127.2 μM) were added in 1-μl increments to the sample cuvette, and the same volume of solvent was added to the reference cuvette. Ligand-induced difference spectra were recorded, and because of the tight binding of the ITZ stereoisomers with CYP3A4, the binding constant (Ks) values for ITZ stereoisomers were obtained by fitting the “Morrison” equation (eq. 2) to the spectral titration data. The enzyme-inhibitor (EI) complex concentration was determined by Lambert-Beer law using the extinction coefficient determined from maximum absorbance detected when CYP3A4 protein was saturated as described previously (Kunze et al., 2006). In eq. 2,
Ks is the affinity constant of the ligand, [I] is the concentration of ligand, [E] is the concentration of enzyme, and [EI] is the concentration of the enzyme-ligand complex. For M2, the Ks was determined by fitting the Michaelis-Menten equation to the data.
Identification of Itraconazole Metabolites.
Trans-ITZ stereoisomers (1 μM) were incubated with CYP3A4 Supersomes (100 pmol) in 0.5 ml of KPi buffer at 37°C for 1 h with and without NADPH. A control incubation with no Supersomes was also included for each substrate. To determine whether the same metabolites were formed from cis-ITZs and from the 3′-OH-ITZ, keto-ITZ, and N-desalkyl-ITZ, these compounds were also incubated with CYP3A4 Supersomes using similar conditions. The reactions were quenched with 1 ml of ethyl acetate. The extracted organic layer was dried under nitrogen gas in a water bath, and the residues were reconstituted in 100 μl of ACN/H2O (1:4). The metabolites were separated using a Shimadzu LC-10AD HPLC system (Shimadzu, Kyoto, Japan) equipped with an Agilent Zorbax XDB-C8 5-μm column (2.1 i.d. × 50 mm) by gradient elution (0.3 ml/min) from 90% aqueous (5 mM ammonium acetate) and 10% ACN to 40% ACN over 3 min, to 70% ACN over 3 min, then to 85% ACN over 3 min and to 95% ACN over 1 min, followed by a 2-min hold before returning to initial conditions. The metabolites were detected with a Synapt High-Definition Quadrupole/Triwave/Orthogonal acceleration time-of-flight tandem hybrid mass spectrometer (QToa-TOF) (Waters) operated on the positive ion electrospray mode monitoring ions m/z 100-1500 using cone voltage of 35 V with two modes: the low-energy mode (collision energy 6 eV) for detection of the parent compounds and the high-energy mode (ramped collision energy) for the fragmentation of the parent compounds. For the tandem mass spectrometry (MS/MS) analysis, the collision energy ramped from 11 to 30 eV for [M + 1]+ ion of m/z 466 and from 11 to 25 eV for [M + 1]+ ion of m/z 256. The resolving quadrupole (Q1) low mass resolution was 4.7 Da to collect fragmentation of all chlorine isotopes. The data were analyzed using Micromass MassLynx data analysis software (Waters).
The formation of M1 and M3 and the reduction of M3 (m/z 466) to M1 (m/z 468) was monitored by Applied Biosystems API 4000 MS/MS (AB Sciex, Foster City, CA) operated on the positive ion electrospray mode. The ions monitored were m/z 466.0 and m/z 468.0. The declustering potential was 60 V, and the gas temperature was 450°C. The metabolites were separated as described above with a gradient from 80% aqueous (5 mM ammonium acetate) and 20% ACN to 50% ACN over 3 min, then to 60% ACN over 2 min and to 95% ACN over 0.1 min, followed by a 1.5-min hold at 95% ACN before returning to initial conditions.
Characterization of Catalytic Rates of Itraconazole Metabolism.
The catalytic rates (kcat) for M2 formation were determined at saturating concentrations of the four trans-ITZs (500 nM) in incubations with CYP3A4 Supersomes (1 pmol) in 0.5 ml of KPi buffer at 37°C for 1 min after addition of NADPH (final concentration of 1 mM). The reaction mixtures were preincubated for 5 min at 37°C before NADPH addition. The kcat was also determined on the basis of results from incubations with reconstituted purified CYP3A4. CYP3A4 reconstitution was modified from the previous report (Shaw et al., 1997) to include 5 × CYP3A4 protein premix (0.5 μM CYP3A4, 1.0 μM NADPH-P450 reductase, 0.5 μM cytochrome-b5, 0.5 mg/ml CHAPS (3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonic acid), 0.1 mg/ml liposomes [l-α-dilauroyl-sn-glycero-3-phosphocholine, l-α-dioleoyl-sn-glycero-3-phosphocholine, l-α-dilauroyl-sn-glycero-3-phosphoserine (1:1:1, w/w/w)], 3.0 mM GSH, in 100 mM KPi buffer, pH 7.4), and a 5× buffer mix (100 mM KPi buffer, pH 7.4, 12 mM GSH, and 150 mM MgCl2). For reconstitution, 60 μl of 5 × CYP3A4 protein premix and equal volume of 5× buffer mix were mixed and left on ice for 10 min. Trans-ITZ (1.67 μM) and KPi buffer were added to bring the volume to 0.294 ml. The mixture was preincubated at 37°C for 3 min, followed by the addition of 6 μl of an NADPH solution (2 mM final concentration) to start the reaction, and the reaction was allowed to proceed for 1 min. The reactions were quenched with 1 ml of ethyl acetate, and internal standard (10 μl of 710 nM ITZ-d5) was added. The organic layer was evaporated under a nitrogen stream, the dry residue was reconstituted in 100 μl of ACN/water (v/v, 1:4) and transferred to a LC/MS vial for analysis using an Applied Biosystems API 4000 MS/MS (AB Sciex) as described above. The mass spectrometer was operated in the positive ion electrospray mode, and multiple reaction monitoring transitions of m/z 256.0 > 158.8 for M2 and m/z 712.32 > 399.23 for ITZ-d5 were monitored. The declustering potential was 60 V, collision energy was 50 V, and gas temperature was 450°C. Gradient elution (0.3 ml/min) from 80% aqueous (5 mM ammonium acetate) and 20% ACN to 50% ACN over 3 min, to 60% ACN over 2 min and to 95% ACN over 0.1 min, followed by 1.5-min hold at 95% ACN before returning to initial conditions was used. The standard curve was constructed on the basis of extractions of M2 at concentrations from 0.039 to 30 nM. Data were analyzed using Analyst software (AB Sciex). The kcat (pmol · min−1 · pmol P450−1) was calculated from the amount of product formed divided by incubation time and amount of CYP3A4 in the incubation.
Human Urine Sample Extraction.
To establish the formation of M2 in vivo in humans, urine samples of six healthy volunteers before and after single oral cis-ITZ administration (400 mg) were analyzed. Urine samples were from previously conducted clinical ITZ-MDZ DDI study (Templeton et al., 2010). The University of Washington Institutional Review Board approved this protocol. Six subjects were enrolled in the study (five males and one female). Urine samples were obtained for 24 h before and 0 to 12 and 12 to 24 h after ITZ dosing (400 mg), and the volume of the urine was recorded. In addition, urine was collected for three additional 24-h intervals, 24 to 48, 72 to 96, and 120 to 144 h after the 400-mg ITZ dose. The urine samples were stored at −20°C until analysis. Urine samples (0.5 ml) were extracted with 1 ml of ethyl acetate, and the residue was reconstituted in 100 μl of ACN/water (1:4). Samples were analyzed for M2 by liquid chromatography/tandem mass spectrometry (LC/MS/MS) using the Applied Biosystems API 4000 MS as described above.
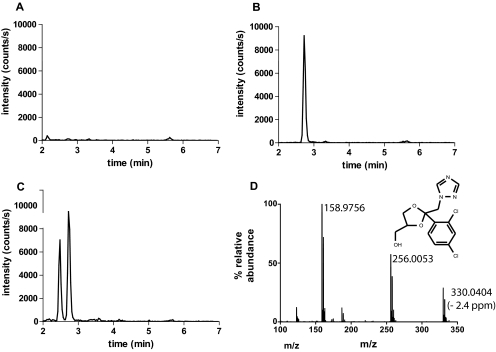
An additional metabolite was detected in the urine samples in the same MS/MS channel as M2. To identify this metabolite, the same urine samples described above from two of the volunteers were reanalyzed. These urine samples (5 ml) were extracted with 25 ml of ethyl acetate, and the residue was reconstituted in 100 μl of ACN/water (1:4). The samples were analyzed using the Synapt High Definition MS system, QToa-TOF, as described above for incubation samples using the same chromatographic conditions. The parent mass was first determined via monitoring ions m/z 100 to 1000 using cone voltage of 35 V with two modes: a low-energy mode (collision energy 6 eV) and a high-energy mode (ramped collision energy from 10 to 40 eV). After identification of a metabolite with a mass of m/z 330, an MS/MS fragmentation pattern was collected for the metabolite. For the MS/MS analysis, the collision energy ramped from 10 to 40 eV for [M + 1]+ ion of m/z 330. All other detection and separation methods were as described above.
Surface Plasmon Resonance.
The SPR experiments were performed as described previously for (2R,4S,2′R)-ITZ and CYP3A4 (Pearson et al., 2006). The sensor surface was pretreated as described previously and then was normalized by standard Biacore protocols. Binding of the drug (2S,4S,2′S-ITZ; 2, 4, and 8 μM) to CYP3A4 in 3% methanol in 100 mM KPi (pH 7.4) was monitored. For coupling of CYP3A4 to CM5 sensor surfaces, the surface was activated with a 5-min pulse of 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide/N-hydroxysuccinimide [5 μl/min in standard Biacore buffer containing 10 mM Hepes (pH 7.4), 150 mM NaCl, 3 mM EDTA, and 0.005% v/v P20 surfactant], followed by injection of the protein solution (400 nM in 10 mM sodium acetate buffer, pH 5.5), and then was deactivated by a 5-min pulse of ethanolamine HCl (pH 8.5, 5 μl/min). Protein injections were done for variable lengths of time to produce surfaces of variable protein densities, yielding responses that reached targeted 8000 relative response units (RU). Blank control surfaces were activated and deactivated as the CYP3A4 immobilization process. SPR data were double-referenced and analyzed with Biacore T100 evaluation software (GE Healthcare, Chalfont St. Giles, Buckinghamshire, UK). Dissociation phase data were fitted by a one phase decay model to obtain a koff rate constant using GraphPad Prism.
Results
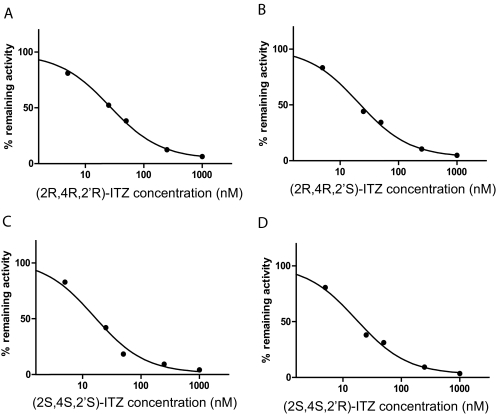
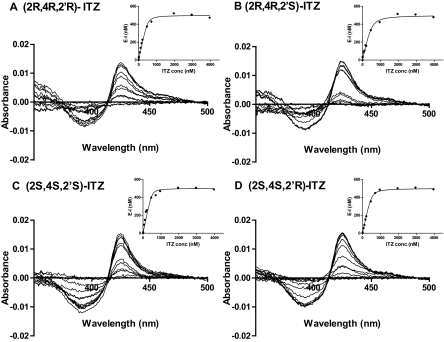
To determine whether the four trans-ITZ stereoisomers bind to CYP3A4, their inhibitory potency was determined in HLM using MDZ as a probe substrate. The trans-ITZ concentration-dependent inhibition of CYP3A4 is shown in Fig. 1. The IC50 values for the four trans-ITZs were approximately 20 nM (Table 1), demonstrating that trans-ITZs are tight binding inhibitors of CYP3A4. To determine whether trans-ITZs also bind to CYP3A4 via triazole nitrogen coordinating to the heme iron, the ligand-induced difference spectra (binding spectra) of trans-ITZs were determined with purified CYP3A4 (Fig. 2). All four trans-ITZ stereoisomers showed typical type II binding spectra with CYP3A4, characteristic of the triazole nitrogen coordinating to the heme and causing a high to low spin shift of the heme iron. The spectral dissociation constant, Ks, of the four trans-ITZ stereoisomers was determined from spectral titration (Fig. 2), and the Ks values (obtained by fitting eq. 2 to the data) ranged from 16 to 62 nM (Table 1), confirming the high-affinity binding of all four trans-ITZ stereoisomers with CYP3A4.
Fig. 1.
Inhibition of CYP3A4 by trans-ITZ stereoisomers. IC50 inhibition of CYP3A4 by trans-ITZs (2R,4R,2′R in A; 2R,4R,2′S in B; 2S,4S,2′S in C; and 2S,4S,2′R in D) in human liver microsomes using midazolam as the probe substrate. IC50 was determined by fitting eq. 1 to the data.
TABLE 1.
Inhibitory potency (IC50) of trans-ITZ stereoisomers toward CYP3A4 determined in HLMs (IC50) and spectroscopic dissociation constants (Ks) determined using purified CYP3A4
| Trans- | (2R,4R,2′R)-ITZ | (2R,4R,2′S)-ITZ | (2S,4S,2′S)-ITZ | (2S,4S,2′R)-ITZ |
|---|---|---|---|---|
| IC50, nM | 26 ± 1 | 20 ± 1 | 16 ± 1 | 17 ± 1 |
| Ks, nM | 23.8 ± 11.2 | 61.7 ± 17.4 | 16.1 ± 12.2 | 31.9 ± 4.1 |
Fig. 2.
Binding of trans-ITZ (2R,4R,2′R in A; 2R,4R,2′S in B; 2S,4S,2′S in C; and 2S,4S,2′R in D) in CYP3A4 active site. UV-visible difference spectra of CYP3A4 with trans-ITZs. The spectral titration curves are shown in the insets. The Ks value for each trans-ITZ was determined from fitting to the quadratic equation (Morrison, eq. 2) to the titration data.
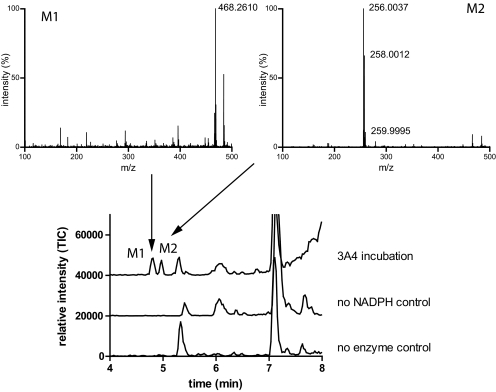
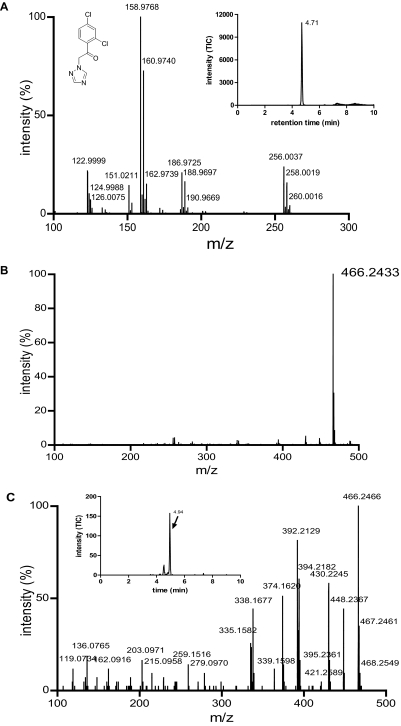
Because cis-ITZ stereoisomers have previously been shown to undergo stereoselective metabolism by CYP3A4 despite their high-affinity type II binding (Kunze et al., 2006), the metabolism of trans-ITZs by CYP3A4 was examined using CYP3A4 Supersomes. The four trans-ITZs were not metabolized to the same metabolites as two of the cis-ITZs. No 3′-OH-ITZ, keto-ITZ, or N-desalkyl-ITZ metabolites were found in trans-ITZ incubations, but considerable substrate depletion by recombinant CYP3A4 was observed. Because trans-ITZs were efficiently depleted by CYP3A4, the products from the incubations were analyzed using a Waters Synapt Q-TOF LC/MS/MS instrument to identify the metabolites formed. Two new metabolites, not observed in the control incubations (no NADPH and no enzyme), were detected from the incubations with trans-ITZs (Fig. 3). The two metabolites had [M + 1]+ ions of m/z 468.261 (M1) and m/z 256.004 (M2) (Fig. 3). The mass spectrum of M2 also showed a typical chlorine isotope pattern with an abundant isotope peak at m/z 258.0012, suggesting that this metabolite retained the two Cl atoms in ITZ. A similar isotope pattern was observed for ITZ as well. The MS/MS spectrum of M2 is shown in Fig. 4. On the basis of the chlorine isotope pattern and accurate mass (<5 ppm accuracy), M2 was tentatively identified as 1-(2,4-dichlorophenyl)-2-(1H-1,2,4-triazol-1-yl)ethanone (M2; Scheme 2). A reference material for M2 was commercially available, and the MS/MS fragmentation pattern and retention time of the reference material were found to be identical to those of the enzymatically formed M2. Therefore, M2 was confirmed as 1-(2,4-dichlorophenyl)-2-(1H-1,2,4-triazol-1-yl)ethanone (Scheme 2; Fig. 4A).
Fig. 3.
Identification of metabolites generated from trans-ITZs by CYP3A4. A representative chromatogram of an incubation of cis-ITZ and trans-ITZ with CYP3A4. When incubated in the presence of CYP3A4 Supersomes, the new ITZ metabolites (M1 and M2) are formed from both cis-ITZ and trans-ITZ. The new metabolites were not found in the absence of CYP3A4 or NADPH control samples, as shown in the control traces. The MS data were collected by full scan using Synapt Q-TOF for high-accuracy MS. Ions monitored are m/z 100-1500 in low-energy mode, the cone voltage was 35 V, and the collision energy was 6 eV, as described under Materials and Methods.
Fig. 4.
Characterization of M2 and M3 as metabolites of ITZ. A, the MS/MS spectrum of M2 (parent [M + 1]+ m/z 256) formed from ITZ in incubation with CYP3A4 is shown. The MS/MS spectrum was identical to that obtained from the reference standard of M2. B, the molecular ion of M3. C, the MS/MS spectrum of M3 (parent ion [M + 1]+ m/z 466). The insets show the selected ion chromatograms for the parent ion. The spectra were collected using Synapt Q-TOF LC/MS as described under Materials and Methods.
Scheme 2.
Proposed mechanism for the formation of the new ITZ metabolites.
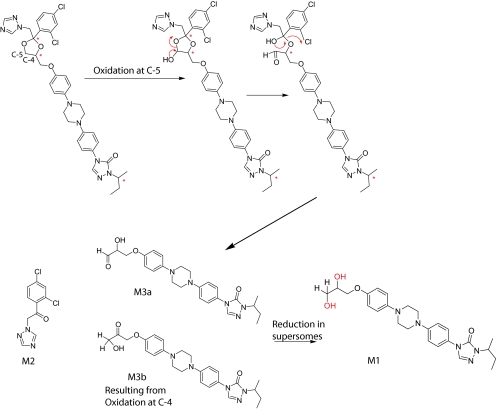
A tentative mechanism for the formation of M2 from ITZ by CYP3A4 was proposed (Scheme 2). On the basis of this proposed mechanism, the dioxolane ring is cleaved by CYP3A4, resulting in formation of M2 and a second metabolite with [M + 1]+ ion of m/z 466. However, the second metabolite M1, which was detected in the 1-h incubations with CYP3A4 Supersomes, had an accurate mass that was 2 Da higher, [M + 1]+ of m/z 468.2610 (Fig. 3). This mass difference suggested that M1 is a product from reduction of the aldehyde (M3a) or ketone (M3b) to an alcohol, resulting in the [M + 1]+ ion at m/z 468 (Scheme 2). It was hypothesized that M3a and/or M3b was reduced to M1 in the CYP3A4 Supersomes in a non-P450-mediated manner. To test this hypothesis, the incubation time in Supersomes was shortened, and samples from seven different time points were analyzed. Indeed, when trans-(2S,4S,2′S)-ITZ was incubated for 5 min with CYP3A4 Supersomes, a metabolite with [M + 1]+ ion of m/z 466.2466 was detected (Fig. 4B). Its formation was NADPH and enzyme-dependent, and the accurate mass of this metabolite was within 5 ppm of the calculated molecular ion of M3a and M3b (Scheme 2), which is consistent with the proposed mechanism for CYP3A4-mediated oxidative cleavage of the dioxolane ring. The MS/MS spectrum of this metabolite is shown in Fig. 4C. As shown in the inset to Fig. 4C, two peaks were detected in this m/z channel, suggesting that both M3a and M3b were formed by CYP3A4.
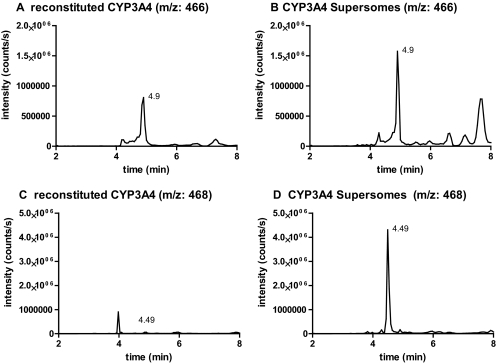
To test whether the reduction of M3a and/or M3b to M1 was mediated by an endogenous reductase in the CYP3A4 Supersomes rather than CYP3A4, trans-(2S,4S,2′S)-ITZ was incubated with reconstituted purified CYP3A4 and CYP3A4 Supersomes, and the formation of the metabolites with [M + 1]+ m/z 466 (M3a and/or M3b) and [M + 1]+ m/z 468 (M1) was monitored. Formation of M3a/M3b was detected in both systems with similar abundance (2-min incubation time for reconstituted system; 3-min incubation time for Supersomes) (Fig. 5, A and B). However, no formation of M1 was observed in incubations with purified reconstituted CYP3A4 (Fig. 5C), whereas M1 was the dominant metabolite observed in incubations with CYP3A4 Supersomes (Fig. 5D). On the basis of this information, the mechanism shown in Scheme 2 for the CYP3A4-mediated dioxolane ring cleavage is proposed. Unfortunately no reference material for M3a or M3b is available; therefore, it is not possible to confirm whether the observed metabolite has the structure of M3a and/or M3b and whether both of these metabolites are formed (Scheme 2). Methylene carbon 5 (Scheme 2) is sterically less hindered than carbon 4; thus, formation of M3a appears more likely. However, the MS/MS spectrum of the enzymatically formed product (Fig. 4C) does not show any losses of CO (loss of m/z 28) or formaldehyde (loss of m/z 30), which would be expected from an aldehyde metabolite (M3a), suggesting that oxidation may occur at carbon 4. At present, it is not possible to differentiate between the two products.
Fig. 5.
Characterization of the formation of M1 and M3 by CYP3A4 microsomes and reconstituted CYP3A4. Selected ion chromatograms of m/z 466 and m/z 468 recorded with Applied Biosystems API 4000 MS/MS, as described under Materials and Methods, are shown. Trans-(2S,4S,2′S)-ITZ was incubated with purified CYP3A4 reconstituted with P450 reductase (A and C) and CYP3A4 Supersomes (B and D), and the formation of M1 and M3 was followed at m/z 468 and m/z 466. The metabolite M3, m/z 466, was found in both enzyme systems; however, reduction to M1, m/z 468, occurred only in the Supersome incubation.
To investigate whether the dioxolane ring scission is important for therapeutically administered ITZ (cis-ITZ), the four cis-ITZ stereoisomers and the CYP3A4 formed metabolites 3′-OH-ITZ, keto-ITZ, and N-desalkyl-ITZ were incubated with CYP3A4, and the formation of M2 and M3a/M3b was monitored. Indeed, both M2 and M3a/M3b were detected in incubations with all four cis-ITZs, and M2 was detected in the incubations with 3′-OH-ITZ, keto-ITZ, and N-desalkyl-ITZ (data not shown). These results demonstrate that dioxolane ring scission is a common metabolic pathway for all ITZ stereoisomers and their metabolites. The kcat of M2 formation found for trans-ITZ was approximately 10-fold faster than the kcat found for cis-ITZ. However, the rapid substrate depletion of cis-ITZs by CYP3A4 and the fact that M2 is formed from cis-ITZs as well as from the sequentially oxidized metabolites make it impossible to accurately characterize the kinetics of M2 formation from cis-ITZ.
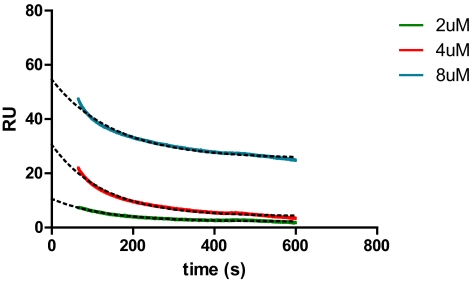
The trans-ITZs do not undergo metabolism to 3′-OH-ITZ, keto-ITZ, or N-desalkyl-ITZ. Therefore, these ITZ stereoisomers offer a useful molecular probe for studying the interactions of ITZ with CYP3A4. It has been proposed previously that cis-ITZs have two binding orientations with CYP3A4 (Pearson et al., 2006), one of which allows the triazole ring to coordinate to the heme iron and another that brings the isobutyl side chain close to the heme for oxidation. Dissociation of cis-ITZ from CYP3A4 was proposed to be necessary for reorientation of ITZ within the CYP3A4 active site to allow metabolism to 3′-OH-ITZ. It was also found that the binding orientation in which the triazole ring coordinates to the heme iron has a slow dissociation rate that results in a “burst” in the metabolite formation kinetics from cis-ITZ. Because the dissociation rate for cis-ITZ is reportedly slow (0.0023 s−1) and the dioxolane ring is adjacent to the triazole ring in ITZ, it was hypothesized that the dioxolane ring scission and CYP3A4-mediated oxidation of trans-ITZ occur without substrate dissociation from type II complex of the drug and CYP3A4. To test this hypothesis, the kcat for M2 formation was measured for the four trans-ITZs at 500 nM substrate concentration. At this substrate concentration, product formation was linear with time and protein concentration. The kcat values obtained for M2 formation from incubations of trans-ITZs with CYP3A4 Supersomes and reconstituted CYP3A4 are summarized in Table 2. To compare the catalytic rate to the dissociation rate of trans-ITZs from CYP3A4, the koff for trans-(2S,4S,2′S)-ITZ was measured using SPR. A koff of 0.0072 s−1 was obtained by fitting the one phase decay model to the data (Fig. 6). The fact that the off-rate is much slower than the measured kcat in CYP3A4 Supersomes suggests that the dioxolane ring scission occurs without dissociation and rebinding of trans-ITZ.
TABLE 2.
The catalytic rate constants (kcat values) for formation of M2 from trans-ITZ stereoisomers in incubations with reconstituted CYP3A4 and with CYP3A4 Supersomes
Catalytic rates could not be measured for cis-ITZ stereoisomers because of high substrate depletion and formation of M2 from multiple substrates simultaneously.
| Trans- | (2R,4R,2′R)-ITZ | (2R,4R,2′S)-ITZ | (2S,4S,2′S)-ITZ | (2S,4S,2′R)-ITZ |
|---|---|---|---|---|
| kcat, s−1, reconstituted CYP3A4 | 0.0032 ± 0.0005 | 0.0038 ± 0.0001 | 0.0041 ± 0.0007 | 0.0042 ± 0.0004 |
| kcat, s−1, CYP3A4 Supersomes | 0.0159 ± 0.003 | 0.0214 ± 0.004 | 0.0183 ± 0.003 | 0.0198 ± 0.001 |
Fig. 6.
Determination of the dissociation rate for (2S,4S,2′S)-ITZ from CYP3A4 using SPR. The dissociation rate (koff) of (2S,4S,2′S)-ITZ was determined by one phase decay model, and the koff was found to be 0.0072 ± 0.0006 s−1. Only the dissociation phase of the SPR run is shown.
To determine whether the dioxolane ring scission occurs in vivo in humans, urine and plasma samples of six individuals after oral administration of 400 mg of ITZ were analyzed for presence of M2. M2 was found in all of the tested urine samples (Fig. 7) but not in any of the plasma samples from the six subjects. The average amount of M2 found in the urine over 24 h after 400 mg of ITZ administration to the six subjects was 1114 ± 494 ng. As expected on the basis of the long half-life of ITZ, M2 was detected in the urine 2, 4, and 7 days after ITZ administration, and the average amount over 24-h intervals was 429 ± 137 ng on day 2, 319 ± 192 ng on day 4, and 87 ± 31 ng on day 7. The total amount of M2 recovered in urine during these sampling periods suggests that it is a minor metabolite in urine. However, urine collection was incomplete, and exact recovery of this metabolite cannot be determined. An additional unknown metabolite eluting at a retention time slightly before M2 was detected in the urine samples collected after ITZ administration but not in the samples collected before ITZ administration (Fig. 7). To suggest a logical structure for this metabolite, an accurate mass and MS/MS fragmentation pattern for this metabolite was collected. An MS/MS spectrum and the proposed ketal-carbinol structure [2-(2,4-dichlorophenyl)-2-(1H-1,2,4-triazol-1-yl)-1,3-dioxolane-4-methanol] is shown in Fig. 7D. The mass accuracy for this structure was 2.4 ppm. The fragmentation was similar to that observed from M2, as expected.
Fig. 7.
Detection of M2 in urine samples from healthy volunteers after administration of 400 mg of ITZ and identification of an additional metabolite of ITZ in urine. Samples were analyzed as described under Materials and Methods. M2 was detected by MS/MS using ion transition of m/z 256 > 158. A, a representative chromatogram of a urine sample from a study subject before ITZ administration. B, a chromatogram of M2 standard extracted from blank urine. C, a representative chromatogram of a urine sample after administration of 400 mg of ITZ orally. In C, the peak at 2.75 min is M2, and a second unknown metabolite was detected at an earlier elution time, 2.45 min. To identify this metabolite, the urine samples were reanalyzed as described under Materials and Methods, and accurate mass as well as the fragmentation pattern of this metabolite were obtained (D). The MS/MS spectrum of this additional metabolite shows the fragmentation to the two monitored ions m/z 256 and m/z 159 as well as the chlorine isotope pattern characteristic of two chlorine ions present in the metabolite and fragments.
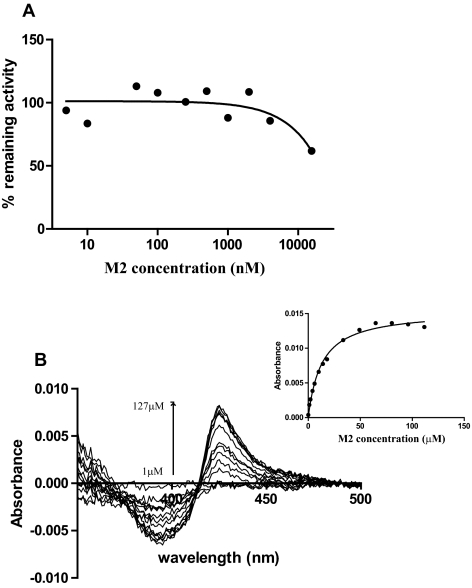
Because M2 was detected in vivo and it possesses a triazole ring in its structure, its inhibitory potency toward CYP3A4 was tested. The IC50 value was found to be >15 μM (Fig. 8A), because of <50% inhibition at 15 μM M2 concentration. The affinity of M2 to CYP3A4 was further determined by spectral titration. A typical type II difference spectrum was obtained with purified CYP3A4, and the Ks value was 12.8 μM (Fig. 8B). Because of the lack of detection of M2 in human plasma after ITZ administration and the high IC50 of M2, it is unlikely that this metabolite contributes to observed CYP3A4 inhibition in vivo after ITZ administration.
Fig. 8.
Determination of the inhibitory potency of M2 toward CYP3A4. A, inhibition of CYP3A4-mediated midazolam hydroxylation in HLM by M2. The percentage of remaining activity versus M2 concentrations is shown. The IC50 of M2 toward CYP3A4 was determined to be >15 μM. B, binding of M2 to purified CYP3A4 shows a type II binding spectrum demonstrating triazole coordination to the heme. The Ks of M2 was determined by spectral titration as described under Materials and Methods (Ks = 12.8 ± 1.1 μM), and the increased absorbance as a function of M2 concentration is shown in the inset. The Michaelis-Menten equation was fitted to the data to obtain the Ks value.
Discussion
ITZ is a strong inhibitor of the CYP3A4-mediated clearance of drugs in humans (Olkkola et al., 1996; Neuvonen et al., 1998) resulting in significant and persistent DDIs. According to some studies, the magnitude and time course of ITZ-mediated DDIs result from inhibition of CYP3A4 by ITZ and its metabolites that are formed by CYP3A4 (Templeton et al., 2008, 2010). In addition to inhibiting CYP3A4, cis-ITZs are also metabolized stereoselectively by CYP3A4 (Kunze et al., 2006). Two of the cis-ITZ isomers, (2R,4S,2′R)-ITZ and (2R,4S,2′S)-ITZ, are metabolized by CYP3A4 to 3′-OH-ITZ, keto-ITZ, and N-desalkyl-ITZ, whereas the other two cis-ITZ stereoisomers do not undergo metabolism to these products (Kunze et al., 2006). Despite this observation, recombinant CYP3A4 depletes over 50% of the mixture of cis-ITZ isomers in vitro (Isoherranen et al., 2004), suggesting that all four cis-ITZ stereoisomers are substrates of CYP3A4. This study shows that dioxolane ring scission is another CYP3A4-mediated metabolic pathway for cis-ITZs and for the circulating cis-ITZ metabolites, 3′-OH-ITZ, keto-ITZ, and N-desalkyl-ITZ.
In early studies in rats and dogs, dioxolane ring scission was proposed as a possible metabolic pathway of ITZ, but the resulting products were not reported (Heykants et al., 1987), and no other metabolites of ITZ except 3′-OH-ITZ, keto-ITZ, and N-desalkyl-ITZ have been quantified in humans. This study shows that the dioxolane ring scission is a clearance pathway of ITZ in vivo. This finding is of interest because the dioxolane ring is a common structural motif in many antifungal agents, including ketoconazole and terconazole, and in related agricultural fungicides. In fact, the metabolite resulting from the dioxolane ring scission of ketoconazole (M + H m/z 255) was also observed in incubations of ketoconazole with CYP3A4 (data not shown). The cleavage of the dioxolane ring in propiconazole and the formation of M2 have been reported previously (Chen et al., 2008), although the enzyme responsible for the reaction was not identified. The effects of M2 as a metabolite of propiconazole on cell viability were also evaluated in hepatic cells, but no significant effects were detected. On the basis of these data, it is likely that dioxolane ring cleavage is a common metabolic pathway for the drugs in this class.
Even though M2 and M1 theoretically can be generated by acid-catalyzed hydrolysis of the dioxolane ring (chiral center carbon, acetal), this mechanism is not likely in the in vitro incubations at pH 7.4 because no product was detected in the absence of NADPH. However, it is possible that after oral administration of ITZ, the acidic conditions of the stomach could catalyze the cleavage. To test whether the hydrolysis occurs under acidic conditions, cis-ITZs (four isomers), OH-ITZ, keto-ITZ, and ND-ITZ, were incubated in pH 2 at 37°C for 1 h, but no hydrolysis products or depletion of ITZ was observed (data not shown), suggesting acid-catalyzed hydrolysis is unlikely.
Whether M2 has any biological activity is not currently known. Although M2 was a weak inhibitor of CYP3A4, it may have higher binding affinity to other drug-metabolizing P450 enzymes or to P450 enzymes relevant for fungal cell survival. It is unlikely that the formation of M2 contributes to inhibition of CYP3A4 after ITZ administration because of the undetectable plasma concentrations and low inhibitory potency toward CYP3A4. However, if this metabolite is formed from other azole antifungals resulting in detectable plasma concentrations of it, further characterization of its inhibitory potency toward other P450s is warranted due to the triazole group. The toxicological importance of the metabolites formed by dioxolane ring scission is not known. Both ITZ and ketoconazole cause rare hepatotoxicity (Lewis et al., 1984; Wang et al., 2010), and it is possible that metabolites contribute to this adverse effect.
This study shows that trans-ITZs are as potent inhibitors of CYP3A4 as the cis-ITZs, suggesting that the coordination of the triazole nitrogen to CYP3A4 heme and type II binding of ITZs in CYP3A4 active site are not sensitive to stereochemistry in the dioxolane ring or in the isobutyl side chain. The IC50 values for the four trans-ITZs were between 16 and 26 nM, which is similar to previously determined IC50 values of cis-ITZ isomers (4–15 nM) (Kunze et al., 2006). Thus, it seems surprising that the oxidation of the isobutyl side chain by CYP3A4 in cis-ITZs and trans-ITZs is very stereoselective and that the formation of M2 from trans-ITZs is significantly more efficient than that from cis-ITZs. It is possible that the lack of 3′-OH-ITZ, keto-ITZ, and N-desalkyl-ITZ formation from trans-ITZs is due to the preferential binding of the trans-ITZs with the triazole ring coordinating the heme, and the oxidation of the dioxolane ring occurs before the type II complex dissociates. Previous SPR studies (Pearson et al., 2006) showed that dissociation of the cis-ITZ must occur from type II binding orientation followed by rebinding in the catalytically accessible orientation (type I) to CYP3A4 in order for the metabolism to happen. For trans-ITZ, the rapid catalysis of the dioxolane ring scission will not allow dissociation and rebinding of trans-ITZ in the alternative binding orientation with the isobutyl side chain close to the heme. This explanation was supported by the SPR data with trans-ITZ that showed that the dissociation rate of trans-ITZ from CYP3A4 (koff = 0.0072 s−1) is slower than the catalysis to M2 in CYP3A4 Supersomes (kcat = 0.019 s−1). These data also suggest that the heme iron is reduced with the type II ligand bound to the heme (low spin complex). This has been suggested for quinoline-4-carboxamide compounds with CYP3A4 (Pearson et al., 2011). Alternatively, the three-dimensional structure of trans-ITZ may prevent the binding of these stereoisomers with the isobutyl side chain close to the heme because of unfavorable interactions between trans-ITZs and CYP3A4 active site residues in this orientation.
In summary, trans-ITZs are as potent inhibitors as cis-ITZs toward CYP3A4. Although the trans-ITZs are also substrates of CYP3A4, they are not metabolized to the corresponding trans-3′-OH-ITZ, keto-ITZ, or N-desalkyl-ITZ. A metabolic pathway was identified in both cis-ITZ and trans-ITZ incubations with CYP3A4, and a new metabolite was formed from a type II binding compound that undergoes metabolism at a carbon close to the coordinating nitrogen. This metabolic pathway is likely to occur for other antifungals that contain a dioxolane ring in their structure, such as ketoconazole. This study increases our overall understanding of how the stereocenters of a substrate affect the metabolism and ligand interactions with CYP3A4 and provides unique insight into the structural determinants of ligand metabolism by P450s.
Acknowledgments
We thank Peter Rademacher for assistance in the high-resolution mass spectrometry work, Kip Conner for help in the SPR studies, and William M. Atkins for helpful discussions during this work. We thank the Analytical Biopharmacy Core in the Department of Medicinal Chemistry at the University of Washington (funded by the Center for Intracellular Delivery of Biologics) for the SPR experiments. We also thank Connie Davis and Angela Hein for their efforts in the itraconazole clinical study.
This work was supported by the National Institutes of Health National Institute of General Medical Sciences [Grant P01-GM32165] (to N.I., K.L.K., and W.L.N.); and the National Institutes of Health National Cancer Institute [Grant CA122814] (to J.O.L.).
Article, publication date, and citation information can be found at http://dmd.aspetjournals.org.
- ITZ
- itraconazole
- P450
- cytochrome P450
- DDI
- drug-drug interactions
- SPR
- surface plasmon resonance
- HPLC
- high-performance liquid chromatography
- ACN
- acetonitrile
- LC/MS
- liquid chromatography/mass spectrometry
- LC/MS/MS
- liquid chromatography/tandem mass spectrometry
- HLM
- human liver microsomes
- KPi
- potassium phosphate
- MDZ
- midazolam
- QToa-TOF
- Quadrupole/Triwave/Orthogonal acceleration time-of-flight tandem hybrid mass spectrometer
- OH-MDZ
- 1′-hydroxymidazolam
- EI
- enzyme inhibitor
- RU
- relative response units.
Authorship Contributions
Participated in research design: Peng, Kunze, Nelson, and Isoherranen.
Conducted experiments: Peng and Lutz.
Contributed new reagents or analytic tools: Shi and Liu.
Performed data analysis: Peng, Shi, Lutz, Kunze, Liu, Nelson, and Isoherranen.
Wrote or contributed to the writing of the manuscript: Peng, Lutz, Shi, Liu, Nelson, and Isoherranen.
References
- Chen PJ, Moore T, Nesnow S. (2008) Cytotoxic effects of propiconazole and its metabolites in mouse and human hepatoma cells and primary mouse hepatocytes. Toxicol in Vitro 22:1476–1483 [DOI] [PubMed] [Google Scholar]
- Gillam EM, Baba T, Kim BR, Ohmori S, Guengerich FP. (1993) Expression of modified human cytochrome P450 3A4 in Escherichia coli and purification and reconstitution of the enzyme. Arch Biochem Biophys 305:123–131 [DOI] [PubMed] [Google Scholar]
- Haria M, Bryson HM, Goa KL. (1996) Itraconazole. A reappraisal of its pharmacological properties and therapeutic use in the management of superficial fungal infections. Drugs 51:585–620 [DOI] [PubMed] [Google Scholar]
- Heykants J, Michiels M, Meuldermans W, Monbaliu J, Lavrijsen K, Van Peer A, Levron J, Woestenborghs R, Cauwenbergh G. (1987) The pharmacokinetics of itraconazole in animals and man: an overview, in Recent Trends in the Discovery, Development, and Evaluation of Antifungal Agents, pp 223–249, J. R. Prous Science Publishers, Barcelona, Spain [Google Scholar]
- Isoherranen N, Kunze KL, Allen KE, Nelson WL, Thummel KE. (2004) Role of itraconazole metabolites in CYP3A4 inhibition. Drug Metab Dispos 32:1121–1131 [DOI] [PubMed] [Google Scholar]
- Kunze KL, Nelson WL, Kharasch ED, Thummel KE, Isoherranen N. (2006) Stereochemical aspects of itraconazole metabolism in vitro and in vivo. Drug Metab Dispos 34:583–590 [DOI] [PubMed] [Google Scholar]
- Lewis JH, Zimmerman HJ, Benson GD, Ishak KG. (1984) Hepatic injury associated with ketoconazole therapy. Analysis of 33 cases. Gastroenterology 86:503–513 [PubMed] [Google Scholar]
- Neuvonen PJ, Kantola T, Kivistö KT. (1998) Simvastatin but not pravastatin is very susceptible to interaction with the CYP3A4 inhibitor itraconazole. Clin Pharmacol Ther 63:332–341 [DOI] [PubMed] [Google Scholar]
- Olkkola KT, Ahonen J, Neuvonen PJ. (1996) The effects of the systemic antimycotics, itraconazole and fluconazole, on the pharmacokinetics and pharmacodynamics of intravenous and oral midazolam. Anesth Analg 82:511–516 [DOI] [PubMed] [Google Scholar]
- Omura T, Sato R. (1964) The carbon monoxide-binding pigment of liver microsomes. II. Solubilization, purification, and properties. J Biol Chem 239:2379–2385 [PubMed] [Google Scholar]
- Pearson J, Dahal UP, Rock D, Peng CC, Schenk JO, Joswig-Jones C, Jones JP. (2011) The kinetic mechanism for cytochrome P450 metabolism of type II binding compounds: evidence supporting direct reduction. Arch Biochem Biophys 511:69–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson JT, Hill JJ, Swank J, Isoherranen N, Kunze KL, Atkins WM. (2006) Surface plasmon resonance analysis of antifungal azoles binding to CYP3A4 with kinetic resolution of multiple binding orientations. Biochemistry 45:6341–6353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw PM, Hosea NA, Thompson DV, Lenius JM, Guengerich FP. (1997) Reconstitution premixes for assays using purified recombinant human cytochrome P450, NADPH-cytochrome P450 reductase, and cytochrome b5. Arch Biochem Biophys 348:107–115 [DOI] [PubMed] [Google Scholar]
- Shi W, Nacev BA, Bhat S, Liu JO. (2010) Impact of absolute stereochemistry on the antiangiogenic and antifungal activities of itraconazole. ACS Med Chem Lett 1:155–159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Templeton I, Peng CC, Thummel KE, Davis C, Kunze KL, Isoherranen N. (2010) Accurate prediction of dose-dependent CYP3A4 inhibition by itraconazole and its metabolites from in vitro inhibition data. Clin Pharmacol Ther 88:499–505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Templeton IE, Thummel KE, Kharasch ED, Kunze KL, Hoffer C, Nelson WL, Isoherranen N. (2008) Contribution of itraconazole metabolites to inhibition of CYP3A4 in vivo. Clin Pharmacol Ther 83:77–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanden Bossche H, Marichal P, Gorrens J, Coene MC, Willemsens G, Bellens D, Roels I, Moereels H, Janssen PA. (1989) Biochemical approaches to selective antifungal activity. Focus on azole antifungals. Mycoses 32 (Suppl 1):35–52 [DOI] [PubMed] [Google Scholar]
- Wang JL, Chang CH, Young-Xu Y, Chan KA. (2010) Systematic review and meta-analysis of the tolerability and hepatotoxicity of antifungals in empirical and definitive therapy for invasive fungal infection. Antimicrob Agents Chemother 54:2409–2419 [DOI] [PMC free article] [PubMed] [Google Scholar]