Abstract
Protein kinase D1 (PKD1) is a stress-activated serine/threonine kinase that plays a vital role in various physiologically important biological processes, including cell growth, apoptosis, adhesion, motility, and angiogenesis. Dysregulated PKD1 expression also contributes to the pathogenesis of certain cancers and cardiovascular disorders. Studies to date have focused primarily on the canonical membrane-delimited pathway for PKD1 activation by G protein-coupled receptors or peptide growth factors. Here, agonist-dependent increases in diacylglycerol accumulation lead to the activation of protein kinase C (PKC) and PKC-dependent phosphorylation of PKD1 at two highly conserved serine residues in the activation loop; this modification increases PKD1 catalytic activity, as assessed by PKD1 autophosphorylation at a consensus phosphorylation motif at the extreme C terminus. However, recent studies expose additional controls and consequences for PKD1 activation loop and C-terminal phosphorylation as well as additional autophosphorylation reactions and trans-phosphorylations (by PKC and other cellular enzymes) that contribute to the spatiotemporal control of PKD1 signaling in cells. This review focuses on the multisite phosphorylations that are known or predicted to influence PKD1 catalytic activity and may also influence docking interactions with cellular scaffolds and trafficking to signaling microdomains in various subcellular compartments. These modifications represent novel targets for the development of PKD1-directed pharmaceuticals for the treatment of cancers and cardiovascular disorders.
Introduction
Protein kinase D1 (PKD1) is the founding member of a family of stress-activated enzymes that play multifunctional roles in fundamental biological processes that regulate cell proliferation, differentiation, apoptosis, immune regulation, cardiac contraction, cardiac hypertrophy, angiogenesis, and cancer (Rozengurt et al., 2005; Avkiran et al., 2008; Guha et al., 2010; LaValle et al., 2010; Steiner et al., 2010). PKD1 is structurally characterized by a C-terminal kinase domain and a N-terminal regulatory domain that contains tandem C1A/C1B motifs that anchor full-length PKD1 to diacylglycerol-/phorbol ester-containing membranes and a pleckstrin homology (PH) domain that participates in intramolecular autoinhibitory interactions that limit catalytic activity (Fig. 1) (Iglesias and Rozengurt, 1998; Chen et al., 2008). PKD1 activation is generally attributed to growth factor-dependent mechanisms that promote diacylglycerol accumulation, colocalize PKD1 at lipid membranes with allosterically activated novel PKC isoforms (nPKCs), and promote nPKC-dependent trans-phosphorylation of PKD1 at two highly conserved serine residues in the activation loop (Ser738/Ser742; nomenclature based upon human PKD1; Fig. 2A) (Waldron et al., 1999). The activated form of PKD1 then autophosphorylates at Ser910, a serine at the extreme C terminus that resides in a consensus PKD1 phosphorylation motif (Nishikawa et al., 1997).
Fig. 1.
Domain structure and regulatory phosphorylation sites in PKD1. C1A/C1B, cysteine-rich Zn finger domains; Kinase, kinase domain. Numbering based upon the human PKD1 enzyme.
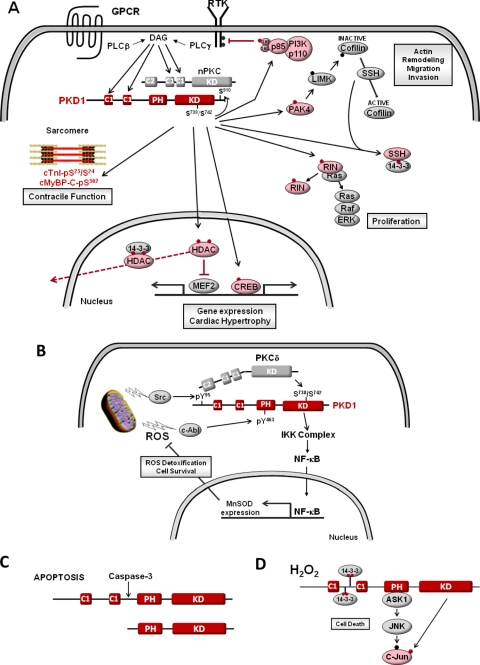
Fig. 2.
PKD1 activation mechanisms. A, G protein-coupled receptors (GPCRs) and receptor tyrosine kinases (RTKs) activate PKD1 via an allosteric mechanism involving lipid cofactors and phosphorylation by nPKC isoforms. PKD1 then phosphorylates a range of cellular substrates, including HDAC5, the sarcomeric proteins cTnI and cardiac myosin binding protein-C (cMyBP-C), CREB, the 27-kDa heat shock protein (HSP27), p21 protein (Cdc42/Rac)-activated kinase 4 (PAK4), c-Jun, Bit1 (Bcl-2 inhibitor of transcription, a mitochondrial protein that induces caspase-independent apoptosis), the F-actin-binding protein cortactin, the cofillin phosphatase slingshot 1, RIN1 (a Ras effector protein that influences ERK and c-Abl pathways), and the p85 regulatory subunit of PI3K (which is inhibited—no longer binds to RTKs—when phosphorylated in the SH2 domain by PKD1); direct substrates of PKD1 are in pink (Hurd et al., 2002; Döppler et al., 2005; Biliran et al., 2008; Eiseler et al., 2009, 2010; Peterburs et al., 2009; Barišić et al., 2011; Lee et al., 2011; Spratley et al., 2011; Ziegler et al., 2011). B and C depict alternative mechanism for PKD1 regulation by reactive oxygen species (ROS) or caspase-3 in the setting of oxidative stress or apoptosis (see Other PKD1 Activation Mechanisms).
Other PKD1 Activation Mechanisms
Recent studies indicate that the common stereotypic PKD1 activation mechanism involving activation loop phosphorylation by nPKCs does not account for PKD1 activation in all cell types (or even by all G protein-coupled receptors). Rather, studies in cardiomyocytes identify stimulus-specific differences in PKD1 activation by α1-adrenergic receptors (α1-ARs) and endothelin-1 receptors, two seemingly similar Gq-coupled receptors. Here, α1-ARs induce a rapid increase in PKD1 activity that is sustained for at least 1 h; the rapid and sustained phases of α1-AR-dependent PKD1 activation both require PKC activity (Guo et al., 2011). In contrast, endothelin-1 receptors induce a transient PKC-dependent increase in PKD1 activity that is followed by a more sustained increase in PKD1 that does not require PKC activity (Guo et al., 2011). This PKC-independent mechanism for PKD1 activation may have evolved to support signaling responses at late time points, when PKC isoforms are down-regulated. Spatiotemporal differences in PKD1 activation also have been detected in adult cardiomyocytes. Here, phenylephrine (α1-AR agonist) and endothelin-1 act in a similar manner to induce rapid PKD1 translocation to the sarcolemma (Bossuyt et al., 2011). However, the activated form of PKD1 remains stably associated with the sarcolemma only in endothelin-1-treated cardiomyocytes. In phenylephrine-treated cardiomyocytes, activated PKD1 shuttles to the nucleus, where it phosphorylates the class IIa histone deacetylase HDAC5 (Haworth et al., 2000; Harrison et al., 2006; Bossuyt et al., 2008, 2011); because HDAC5 phosphorylation creates docking sites for 14-3-3 proteins that escort HDAC5 from the nucleus, this pathway provides a mechanism to derepress pathologic gene programs that promote cardiomyocyte hypertrophy (Fig. 2A). In theory, these subtle differences in PKD1 activation by α1-AR agonists and endothelin-1 also might influence the phosphorylation of cAMP response element-binding protein (CREB), sarcomeric proteins such as cardiac troponin I (cTnI) or cardiac myosin-binding protein C, or other cardiac PKD1 substrates that regulate contraction, influence tissue remodeling, and contribute to the pathogenesis of certain cardiomyopathies (Ozgen et al., 2008; Bardswell et al., 2010).
PKD1 is activated during oxidative stress through a mechanism that requires nonreceptor tyrosine kinases (c-Abl and Src) and PKCδ (and probably not other PKCs; Fig. 2B). Here, c-Abl-dependent PKD1 phosphorylation at Tyr463 (in the PH domain) releases intramolecular autoinhibition, and Src-dependent PKD1 phosphorylation at Tyr95 creates a docking site for the C2 domain of PKCδ; PKCδ then phosphorylates the PKD1 activation loop at Ser738/Ser742 (Storz and Toker, 2003; Storz et al., 2003; Döppler and Storz, 2007). A redox-dependent pathway involving Src and c-Abl also promotes PKD1-PH domain phosphorylation at Tyr432 and Tyr502 (Fig. 1), but the significance of these modifications is uncertain because they do not lead to gross changes in PKD1 activity (Storz et al., 2003). There is evidence that the reactive oxygen species-activated PKD1 enzyme is localized (although not necessarily restricted) to mitochondria and that it recruits a nuclear factor κB (NFκB) pathway that induces expression of antioxidant/antiapoptotic genes (such as manganese superoxide dismutase) and promotes cell survival (Storz et al., 2004; Storz et al., 2005). It is noteworthy that the canonical growth factor-dependent PKD1 signaling pathway does not activate NFκB or induce manganese superoxide dismutase, emphasizing that the signaling repertoire and cellular actions of PKD1 can be highly contextual.
PKD1 also is cleaved by caspase-3; it is a component of the signaling machinery mobilized by proapoptotic stimuli (Fig. 2C). Although there is general consensus that caspase-3 cleaves PKD1 at a site in the C1-PH interdomain, the precise cleavage site remains uncertain (Häussermann et al., 1999; Endo et al., 2000; Vántus et al., 2004). The consequences of this proteolytic event (which removes the C1 domain, but not the “autoinhibitory” PH domain) also have been disputed. Vántus et al. (2004) concluded that PKD1 is a proteolytically activated enzyme on the basis of evidence that the PKD1 cleavage product generated during apoptosis displays a modest increase in basal activity compared with WT-PKD1. However, Häussermann et al. (1999) showed that the C-terminal cleavage product (which lacks a C1 domain) does not respond to lipid cofactors (phosphatidylserine/PMA); as a result, the maximal activity of this catalytic fragment is inconsequential compared with the activity of the phosphatidylserine/PMA-activated full-length PKD1 enzyme. The notion that cleavage limits maximal PKD1 activity also is more consistent with recent results in cardiomyocytes, where the action of PKD1 to regulate lipoprotein lipase-mediated triglyceride accumulation is lost during apoptosis (under conditions associated with the activation of caspase-3 and caspase-3-dependent cleavage of PKD1) (Kim et al., 2009).
Phosphorylation and the Control of PKD1 Activity
The prevailing dogma regarding the structural basis for PKD1 activation is based upon early studies that relied primarily on Ser910 autophosphorylation or PKD1 phosphorylation of syntide-2 (a peptide substrate) as measures of PKD1 activity. Recent studies indicate that these measures do not necessarily provide valid surrogates for PKD1 activity toward more physiologically relevant protein substrates (Rybin et al., 2009). This review summarizes recent studies that use more comprehensive experimental approaches and expose novel controls and consequences of PKD1 phosphorylation at the activation loop, C terminus, and elsewhere in the enzyme.
Mechanisms and Consequences of PKD1-Ser910 Phosphorylation.
The observations that PKD1-Ser910 phosphorylation increases in the context of PKD1 activation by growth factor receptors or phorbol esters and that constitutively active forms of PKD1 (such as the PH domain-deleted or S738E/S742E-substituted mutants) display high levels of basal Ser910 phosphorylation led to the widespread use of PKD1-Ser910 phosphorylation as a surrogate marker of PKD1 activity, in place of more cumbersome direct enzyme activity measurements (Matthews et al., 1999). The assumption inherent in this experimental approach is that PKD1 activation loop phosphorylation is followed by PKD1 autophosphorylation at Ser910 and that Ser910 phosphorylation provides a valid measure of the activity of that particular PKD1 molecule. However, there is ample evidence that these assumptions do not apply to all experimental conditions. First, several laboratories have described agonist-dependent increases in PKD1 activation loop phosphorylation and catalytic activity that are not accompanied by increased PKD1-Ser910 phosphorylation (Brändlin et al., 2002; Storz et al., 2004; Celil and Campbell, 2005). Second, we and others reported that PKD1-K612W (a catalytically inactive form of PKD1 that by definition cannot undergo a intramolecular cis-autophosphorylation) is phosphorylated at Ser910 in trans by endogenous PKD1 or other enzymes with Ser910 kinase activity in several cell types (Sánchez-Ruiloba et al., 2006; Rybin et al., 2009). We also identified major discrepancies between the controls of PKD1-Ser910 autophosphorylation versus PKD1 phosphorylation of target substrates. In particular, we showed that PKD1-Ser910 autophosphorylation is a privileged catalytic reaction that proceeds at exceedingly low ATP concentrations, does not require prior PKD1 phosphorylation at Ser738/Ser742, and is not necessarily accompanied by increased PKD1 activity toward heterologous protein substrates (Rybin et al., 2009). Collectively, these results seriously undermine the assumption that immunoblotting studies that track PKD1-Ser910 phosphorylation provide a reliable measure of PKD1 activity under all experimental conditions.
The distinct mechanisms for PKD1-Ser910 autophosphorylation and PKD1 trans-phosphorylation of target substrates are consistent with findings recently reported for several other protein kinases. For example, the epidermal growth factor receptor family member ErbB3 binds ligands, forms heterodimeric complexes with other epidermal growth factor receptor family members, and possesses some trans-autocatalytic activity, but ErbB3 does not phosphorylate exogenous target substrates (Shi et al., 2010). DYRK and GSK-3 are serine/threonine kinases that autoactivate through an intramolecular autophosphorylation at tyrosine residues in the activation loop (Lochhead et al., 2006; Lochhead, 2009). The mitogen-activated protein kinase p38α phosphorylates target substrates as a proline-directed serine/threonine kinase, but under some stimulatory conditions p38α autophosphorylates at Thr180 and Tyr182 in the activation loop as a dual-specificity kinase (Ge et al., 2002). It is noteworthy that although the p38α inhibitor 4-(4-fluorophenyl)-2-(4-methylsulfinylphenyl)-5-(4-pyridyl)1H-imidazole (SB203580) blocks all p38α activities, the inhibitor sensitivities of the cis-autophosphorylation versus target substrate phosphorylation reactions catalyzed by DYRKs and GSK-3 are quite different. These results emphasize that conventional drug screens—designed to identify compounds that prevent phosphorylation of peptide substrates—may miss compounds that specifically block intramolecular cis-autophosphorylations. Recent studies suggest that this caveat may be pertinent to the development of therapeutics targeted to PKD1, because PKD1-Ser910 autophosphorylation (a reaction that requires only very low concentrations of ATP) is relatively resistant to inhibition by 12-(2-cyanoethyl)-6,7,12,13-tetrahydro-13-methyl-5-oxo-5H-indolo(2,3-a)pyrrolo(3,4-c)-carbazole (Gö6976; an inhibitor that is competitive with ATP) (Rybin et al., 2009). As a result, Gö6976 treatment protocols that effectively block PKD1 phosphorylation of target substrates (such as HDAC5, CREB, or cTnI) do not induce coordinate decreases in PKD1-Ser910 autophosphorylation (Rybin et al., 2009). Similar discrepancies have been identified in studies of BPKDi, another ATP competitive inhibitor recently identified in a high-throughput medicinal chemistry screen (Meredith et al., 2010). Here, typical BPKDi treatment protocols that inhibit HDAC5 phosphorylation do not block PKD1-Ser910 autophosphorylation or confer protection from thoracic aortic constriction-induced cardiac hypertrophy. It is interesting to note that BPKDi exerts a modest growth inhibitory effect only at very high doses that induce a modest decrease in PKD1-Ser910 autophosphorylation. These results raise the intriguing hypothesis that Ser910 phosphorylation may be critical for PKD1 regulation of cardiac growth responses (and that PKD1 exerts some cardiac actions as a scaffold).
PKD1 can act as a signal-regulated scaffold, because sequence flanking the PKD1 autophosphorylation site at Ser910 conforms to a type I PDZ domain-binding motif (S/T-X-φ, where X is any amino acid and φ is a hydrophobic amino acid); Ser910 phosphorylation disrupts this PKD1 docking interaction with PDZ domain-containing scaffolding proteins. Recent studies identify another docking interaction between PKD1 and Na+/H+ exchanger regulatory factor, a PDZ domain-containing protein that colocalizes PKD1 with phosphatases that “fine tune” the local amplitude and tempo of PKD1 responses (relative to PKD1 responses in the bulk cytosol) (Kunkel et al., 2009). Other studies identified an additional role for Ser910 to structure the kinase core for some aspect of catalysis, showing that a S910A substitution abrogates PKD1 autophosphorylation at Ser742 and prolongs in vivo PKD1 signaling responses (Rybin et al., 2009). The S910A substitution does not lead to other gross changes in PKD1 activity; PKD1-S910A autophosphorylates at Ser738 and trans-phosphorylates various protein substrates (Rybin et al., 2009). Collectively, these results suggest that a S910A substitution alters the dynamics of PKD1 cellular responses by disrupting docking interactions with PDZ domain containing scaffolding proteins that regulate PKD1 trafficking or terminate PKD1's cellular actions. Alternatively, a secondary Ser742 autophosphorylation defect induced by the S910A substitution regulates these processes; phosphorylation at the homologous activation loop site in D kinase family-2 (a PKD-related enzyme in Caenorhabditis elegans) enhances enzyme stability (Feng et al., 2007).
PKD1-Ser738/Ser742 Loop Phosphorylation.
Concepts regarding the controls and consequences of PKD1-Ser738/Ser742 (activation loop) phosphorylation are based largely on early studies that used an anti-PKD1-Ser(P)738/Ser(P)742 PSSA (from Cell Signaling Technology, Danvers, MA) and showed that PMA increases PKD1 activation loop phosphorylation in many cell types via a mechanism that requires nPKC isoform activity (PKCδ, PKCε, PKCη, and/or PKCθ). In vitro kinase assays showing direct phosphorylation of the PKD1 activation loop by certain nPKC isoforms also have been published (Brändlin et al., 2002). However, there is recent evidence that the Cell Signaling Technology anti-PKD1-Ser(P)738/Ser(P)742 PSSA primarily recognizes PKD1 phosphorylation at Ser738 and that PKD1 phosphorylation at Ser742 can be tracked with a different PSSA (commercially available from Abcam Inc., Cambridge, MA). Experiments that use a combined approach with these two PSSAs expose differences in the controls and consequences of PKD1 phosphorylation at Ser738 and Ser742 (Jacamo et al., 2008; Rybin et al., 2009). First, there is evidence that the kinase-inactive PKD1-K612W mutant displays a high level of trans-phosphorylation at Ser738 but only a low level of phosphorylation at Ser742. In the context of in vitro studies showing that Ser742 is a target for autocatalytic phosphorylation, these results suggest that PKD1-Ser742 phosphorylation in vivo is mediated primarily by a cis-autophosphorylation reaction that is defective in the catalytically inactive enzyme (Rybin et al., 2009). Second, there is evidence that certain G protein-coupled receptor agonists induce a rapid/coordinate PKC-dependent increase in PKD1 phosphorylation at Ser738/Ser742 that is followed by a more sustained increase in PKD1 phosphorylation at Ser742; Ser742 phosphorylation during the late phase of G protein-coupled receptor activation occurs via an autocatalytic mechanism that does not require PKC activity (Jacamo et al., 2008; Guo et al., 2011). This sustained GPCR-dependent mechanism for PKD1 activation loop phosphorylation (via an autocatalytic mechanism that does not require PKC activity) promotes extracellular signal-regulated kinase activation and mitogenic signaling in some cell types (Sinnett-Smith et al., 2009).
Mutagenesis studies expose a mechanism for PKD1 autophosphorylation at Ser742, showing that PKD1-Ser742 autophosphorylation is abrogated by a S910A substitution (Rybin et al., 2009). The observation that PKD1-Ser742 autophosphorylation is a hierarchical process that requires a prior priming phosphorylation at Ser910 is important for two reasons. First, these results identify a heretofore-unrecognized role for the Ser910-phosphorylated C terminus to structure the kinase core for some aspects of catalysis. The observation that Ser910 is specifically required for PKD1 autophosphorylation at Ser742 (a site that does not conform to a PKD1 consensus phosphorylation motif) but not PKD1 phosphorylation of target substrates is intriguing—given evidence that other autoactivating kinases (such as GSK-3, p38a, or DYRK) autophosphorylate at their activation loops (sites that do not conform to conventional substrate sequences) only when stabilized in unique conformations as a result of intramolecular interactions or docking interactions with protein chaperones (Lochhead, 2009). Second, these results indicate that Ser742 phosphorylation plays little to no role in the control of PKD1 activity, because WT-PKD1 and the PKD1-S910A mutant (which is not phosphorylated at Ser742) display similar high levels of activity toward target substrates (Rybin et al., 2009). The notion that PKD1 activity is regulated by activation loop phosphorylation at Ser738—and not at Ser742—is at odds with previous conclusions derived from mutagenesis studies, where a single S738A or S742A substitution decreases—and a double S738A/S742A substitution abrogates—PKD1 catalytic activity; the previous studies were interpreted as evidence that phosphorylation reactions at Ser738 and Ser742 play similar roles to regulate PKD1 activity (Iglesias et al., 1998). However, activation loop Ser→Ala substitutions can have dual effects to prevent phosphorylation and to remove hydroxyl groups that may engage in structurally important electrostatic interactions (Steichen et al., 2010). Our results with the PKD1-S910A mutant—a catalytically active enzyme with a nonphosphorylated position 742 serine—argue that the position 742 serine plays a structural role to stabilize the active site of the enzyme—and that phosphorylation at this site has little to no effect on PKD1 activity. Finally, it is interesting to note that the observation that PKD1 activity is regulated primarily via a phosphorylation at Ser738 (and not Ser742) resonates with results obtained for the Caenorhabditis elegans PKD enzyme D kinase family-2, where sites in the homologous 925SFRRS929 activation loop sequence (corresponding to 738SFRRS742 in PKD1) play distinct roles to regulate activity (through phosphorylation at Ser925) or the duration of the signaling response (through phosphorylation at Ser929) (Feng et al., 2007).
Although activation loop phosphorylation is critical for PKD1 activation by agonists that signal via PKC, PKD1 also is activated via a PKC-independent mechanism that is not associated with (and does not require) activation loop phosphorylation in bone morphogenetic protein 2-treated MC3T3-E1 osteoblast-like cells (Lemonnier et al., 2004), reactive oxygen species-activated endothelial cells (Zhang et al., 2005), and UVB-treated keratinocytes (Arun et al., 2011). Although the molecular underpinnings for PKC-independent modes of PKD1 activation remain uncertain, mutagenesis studies provide some hints regarding mechanism. In particular, the observation that S738A/S742A substitutions abrogate WT-PKD1 catalytic activity, but PKD1 truncation mutants lacking either the isolated PH domain or the entire regulatory domain tolerate S738A/S742A substitutions without a significant loss of catalytic activity, have been interpreted as evidence that activation loop phosphorylation activates the enzyme by relieving intramolecular autoinhibitory constraints involving the regulatory domain that limit catalytic activity (Waldron and Rozengurt, 2003). According to this formulation, other events (such as other post-translation modifications or protein-protein interactions) that disrupt autoinhibitory constraints might also increase PKD1 activity via a mechanism that does not involve (or require) activation loop phosphorylation. In this regard, dextran sulfate activates PKD1 without increasing activation loop phosphorylation—and dextran sulfate is a potent agonist for both WT and S738A/S742A-substituted PKD1 enzymes (Gschwendt et al., 1997; Rybin et al., 2009). Gschwendt et al. (1997) speculated that dextran sulfate activates PKD1 by disrupting an intramolecular interaction between a highly acidic region in the C1-PH interdomain and basic regions elsewhere in the enzyme. This formulation provides a framework to consider whether some agonist-dependent increases in PKD1 activity that develop over protracted intervals and are not associated with increased activation loop phosphorylation might be attributable to the de novo synthesis of a PKD1 binding partner that disrupts intramolecular autoinhibitory constraints in the enzyme (Lemonnier et al., 2004).
Other Phosphorylation Sites That Regulate PKD1 Activity
The PKD1 regulatory domain contains other phosphorylation sites that are known or predicted to regulate signaling by PKD1 (Fig. 1). For example, the C1A-C1B interdomain contains a cluster of autophosphorylation sites (at Ser205, Ser208, Ser219, and Ser223) that reside in 14-3-3 consensus binding motifs. Autophosphorylation at these sites leads to the formation of PKD1-14-3-3τ complexes, recruitment of apoptosis signal-regulated kinase 1 to the PKD1-PH domain, activation of the apoptosis signal-regulated kinase 1-JNK pathway, c-Jun phosphorylation, and induction of apoptosis in H2O2-treated endothelial cells (Fig. 2D) (Hausser et al., 1999; Zhang et al., 2005). This seems to be a kinase-independent mechanism for PKD1 activation of the JNK signaling pathway, because C1A-C1B interdomain autophosphorylation is not linked to gross changes in PKD1 activity; the docking interaction between 14-3-3τ and PKD1 actually decreases PKD1 catalytic activity. Moreover, catalytically active PKD1 phosphorylates c-Jun at N-terminal regulatory sites (that are distinct from the sites phosphorylated by JNK) and actually inhibits JNK-dependent c-Jun phosphorylation (Hurd et al., 2002; Waldron et al., 2007). The PKD1 C1A-C1B interdomain also contains another phosphorylation site at position 249; studies to date suggest that Ser249 is a target for trans-phosphorylation by PKC and that Ser249 phosphorylation may contribute to optimal PKD1 activation by PKC (but it is not required for PKD1 activation by lipid cofactors) (Vertommen et al., 2000).
Two additional phosphorylation sites have recently been identified adjacent to the autoinhibitory PH domain at positions 421 and 412 (Fig. 1). Ser421 is a target for an autophosphorylation reaction or trans-phosphorylation by protein kinase A (Smith et al., 2011). Ser412 is phosphorylated via a PKC-dependent mechanism (and not an autophosphorylation reaction) in neonatal cardiomyocytes treated with PMA or hypertrophic agonists such phenylephrine or endothelin-1 (Phan et al., 2011). Phosphorylation at these sites could in theory influence intramolecular interactions involving the PH domain that limit catalytic activity (or influence the PH domain-mediated mechanism that controls nuclear export of PKD1) (Rey et al., 2001). However, mutagenesis studies to date do not link Ser421 phosphorylation to changes in PKD1 localization or catalytic activity; the consequences of a S412A substitution have not been examined (Phan et al., 2011; Smith et al., 2011). Finally, p38MAPK-dependent phosphorylation of PKD1 at Ser397 and Ser401 in the C1-PH interdomain is identified in pancreatic β-cells, where it is implicated as a mechanism that controls insulin secretion by inhibiting PKD1 activity (Sumara et al., 2009).
Recent improvements in methods for large scale phosphoproteomics analyses have led to the identification of a large number of additional phosphorylation sites in PKD1 (Villén et al., 2007; Cantin et al., 2008; Zanivan et al., 2008; Brill et al., 2009; Old et al., 2009; Oppermann et al., 2009; Chen et al., 2010; Huttlin et al., 2010). It is interesting to note that these phosphorylation sites map primarily to the C1A-C1B and C1B-PH interdomains. These phosphorylation “hot spots”—in unstructured regions of the enzyme that share little homology with corresponding regions of PKD2 and PKD3—might contribute to PKD1 isoform- and organelle-specific functions.
Conclusions and Future Directions
PKD1 has recently emerged as a signaling enzyme with multifunctional roles in both physiologic and pathologic cellular responses. Recent studies identify an elaborate network of phosphorylation reactions at the activation loop, C terminus, and other regions of the enzyme that contribute to the control of PKD1's cellular actions. These multisite phosphorylations also provide a mechanism to integrate input from diverse inciting stimuli; stimulus-specific differences in the ensemble phosphorylation pattern (or pools of PKD1 with different phosphorylation profiles) could underlie stimulus-specific PKD1 signaling repertoires and cellular responses. Although this review focused primarily on the phosphorylation reactions that influence PKD1 signaling efficiency or specificity, docking interactions with small molecules, protein scaffolds, or protein substrates also may contribute to the allosteric control of PKD1 activity (Sharlow et al., 2008). With this in mind, it is worth noting that high-throughput screens currently in use to identify PKD1 inhibitors typically screen for compounds that inhibit peptide substrate phosphorylation by the resting (unphosphorylated) form of PKD1. This approach may miss clinically useful PKD1 inhibitors for two reasons. First, it assumes that PKD1 inhibitor sensitivity is an inherent property of the enzyme that is not altered (or fine-tuned) by events that accompany enzyme activation. This approach does not allow for possible phosphorylation-dependent changes in PKD1 inhibitor sensitivity (i.e., the notion that differentially phosphorylated forms of PKD1 might effectively constitute distinct drug targets). Second, although peptide substrates typically bind to a single site within the catalytic pocket, physiologically relevant protein substrates typically bind to protein kinases at both the active site and at distal docking motifs outside the catalytic cleft. These docking interactions may serve two functions. First, by tethering or orienting protein substrates on the enzyme, they may facilitate phosphorylation of sites that do not conform to optimal consensus phosphorylation motifs (perhaps explaining the known effects of PKD1 to phosphorylate sites in c-Jun, β-catenin, c-TnI, and type IIα phosphatidylinositol 4-phosphate kinase that do not conform to LxRxxpS/T motifs) (Hinchliffe and Irvine, 2006; Qin et al., 2006; Waldron et al., 2007; Du et al., 2009). Second, a docking interaction with a protein substrate or scaffold may influence inhibitor sensitivity; there is recent evidence that PKC is rendered insensitive to inhibitors that compete with ATP when anchored to AKAP79 (Hoshi et al., 2010). Although similar docking interactions that alter the pharmacologic profile of PKD1 have not yet been identified, PKD1 is a conformationally flexible enzyme that could be regulated in this manner. The phosphorylation reactions and docking mechanisms that influence the pharmacologic profile or signaling specificity of PKD1 present both challenges and opportunities for the development of novel PKD1-targeted pharmaceuticals for the treatment of cardiac hypertrophy/failure and certain intractable cancers.
This work was supported by the National Institutes of Health National Heart, Lung, and Blood Institute [Grant HL77860].
Article, publication date, and citation information can be found at http://molpharm.aspetjournals.org.
- PKD
- protein kinase D
- PH
- pleckstrin homology
- PKC
- protein kinase C
- nPKC
- novel PKC isoform (δ, ε, η, θ)
- AR
- adrenergic receptor
- CREB
- cAMP response element-binding protein
- cTnI
- cardiac troponin I
- NFκB
- nuclear factor κB
- WT
- wild type
- PMA
- phorbol 12-myristate 13-acetate
- SB203580
- 4-(4-fluorophenyl)-2-(4-methylsulfinylphenyl)-5-(4-pyridyl)1H-imidazole
- DYRK
- dual-specificity tyrosine-phosphorylation regulated kinase
- GSK-3
- glycogen synthase kinase-3
- Gö6976
- 12-(2-cyanoethyl)-6,7,12,13-tetrahydro-13-methyl-5-oxo-5H-indolo(2,3-a)pyrrolo(3,4-c)-carbazole
- HADC
- histone deacetylase
- BPKDi
- bipyridyl PKD inhibitor
- PSSA
- phosphorylation site-specific antibody
- PDZ
- postsynaptic density 95/discs-large/zona occludens
- JNK
- c-jun-N-terminal kinase.
Authorship Contributions
Wrote or contributed to the writing of the manuscript: Steinberg.
References
- Arun SN, Kaddour-Djebbar I, Shapiro BA, Bollag WB. (2011) Ultraviolet B irradiation and activation of protein kinase D in primary mouse epidermal keratinocytes. Oncogene 30:1586–1596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avkiran M, Rowland AJ, Cuello F, Haworth RS. (2008) Protein kinase D in the cardiovascular system: emerging roles in health and disease. Circ Res 102:157–163 [DOI] [PubMed] [Google Scholar]
- Bardswell SC, Cuello F, Rowland AJ, Sadayappan S, Robbins J, Gautel M, Walker JW, Kentish JC, Avkiran M. (2010) Distinct sarcomeric substrates are responsible for protein kinase D-mediated regulation of cardiac myofilament Ca2+ sensitivity and cross-bridge cycling. J Biol Chem 285:5674–5682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barišić S, Nagel AC, Franz-Wachtel M, Macek B, Preiss A, Link G, Maier D, Hausser A. (2011) Phosphorylation of Ser 402 impedes phosphatase activity of slingshot 1. EMBO Rep 12:527–533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biliran H, Jan Y, Chen R, Pasquale EB, Ruoslahti E. (2008) Protein kinase D is a positive regulator of Bit1 apoptotic function. J Biol Chem 283:28029–28037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bossuyt J, Chang CW, Helmstadter K, Kunkel MT, Newton AC, Campbell KS, Martin JL, Bossuyt S, Robia SL, Bers DM. (2011) Spatiotemporally distinct protein kinase D activation in adult cardiomyocytes in response to phenylephrine and endothelin. J Biol Chem 286:33390–33400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bossuyt J, Helmstadter K, Wu X, Clements-Jewery H, Haworth RS, Avkiran M, Martin JL, Pogwizd SM, Bers DM. (2008) Ca2+/calmodulin-dependent protein kinase IIδ and protein kinase D overexpression reinforce the histone deacetylase 5 redistribution in heart failure. Circ Res 102:695–702 [DOI] [PubMed] [Google Scholar]
- Brändlin I, Hübner S, Eiseler T, Martinez-Moya M, Horschinek A, Hausser A, Link G, Rupp S, Storz P, Pfizenmaier K, et al. (2002) Protein kinase C (PKC)η-mediated PKCμ activation modulates ERK and JNK signal pathways. J Biol Chem 277:6490–6496 [DOI] [PubMed] [Google Scholar]
- Brill LM, Xiong W, Lee KB, Ficarro SB, Crain A, Xu Y, Terskikh A, Snyder EY, Ding S. (2009) Phosphoproteomic analysis of human embryonic stem cells. Cell Stem Cell 5:204–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantin GT, Yi W, Lu B, Park SK, Xu T, Lee JD, Yates JR., 3rd (2008) Combining protein-based IMAC, peptide-based IMAC, and MudPIT for efficient phosphoproteomic analysis. J Proteome Res 7:1346–1351 [DOI] [PubMed] [Google Scholar]
- Celil AB, Campbell PG. (2005) BMP-2 and insulin-like growth factor-I mediate Osterix (Osx) expression in human mesenchymal stem cells via the MAPK and protein kinase D signaling pathways. J Biol Chem 280:31353–31359 [DOI] [PubMed] [Google Scholar]
- Chen J, Deng F, Li J, Wang QJ. (2008) Selective binding of phorbol esters and diacylglycerol by individual C1 domains of the PKD family. Biochem J 411:333–342 [DOI] [PubMed] [Google Scholar]
- Chen L, Giorgianni F, Beranova-Giorgianni S. (2010) Characterization of the phosphoproteome in LNCaP prostate cancer cells by in-gel isoelectric focusing and tandem mass spectrometry. J Proteome Res 9:174–178 [DOI] [PubMed] [Google Scholar]
- Döppler H, Storz P. (2007) A novel tyrosine phosphorylation site in protein kinase D contributes to oxidative stress-mediated activation. J Biol Chem 282:31873–31881 [DOI] [PubMed] [Google Scholar]
- Döppler H, Storz P, Li J, Comb MJ, Toker A. (2005) A phosphorylation state-specific antibody recognizes Hsp27, a novel substrate of protein kinase D. J Biol Chem 280:15013–15019 [DOI] [PubMed] [Google Scholar]
- Du C, Jaggi M, Zhang C, Balaji KC. (2009) Protein kinase D1-mediated phosphorylation and subcellular localization of β-catenin. Cancer Res 69:1117–1124 [DOI] [PubMed] [Google Scholar]
- Eiseler T, Döppler H, Yan IK, Kitatani K, Mizuno K, Storz P. (2009) Protein kinase D1 regulates cofilin-mediated F-actin reorganization and cell motility through slingshot. Nat Cell Biol 11:545–556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eiseler T, Hausser A, De Kimpe L, Van Lint J, Pfizenmaier K. (2010) Protein kinase D controls actin polymerization and cell motility through phosphorylation of cortactin. J Biol Chem 285:18672–18683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endo K, Oki E, Biedermann V, Kojima H, Yoshida K, Johannes FJ, Kufe D, Datta R. (2000) Proteolytic cleavage and activation of protein kinase Cμ by caspase-3 in the apoptotic response of cells to 1-β-D-arabinofuranosylcytosine and other genotoxic agents. J Biol Chem 275:18476–18481 [DOI] [PubMed] [Google Scholar]
- Feng H, Ren M, Chen L, Rubin CS. (2007) Properties, regulation, and in vivo functions of a novel protein kinase D: Caenorhabditis elegans DKF-2 links diacylglycerol second messenger to the regulation of stress responses and life span. J Biol Chem 282:31273–31288 [DOI] [PubMed] [Google Scholar]
- Ge B, Gram H, Di Padova F, Huang B, New L, Ulevitch RJ, Luo Y, Han J. (2002) MAPKK-independent activation of p38α mediated by TAB1-dependent autophosphorylation of p38α. Science 295:1291–1294 [DOI] [PubMed] [Google Scholar]
- Gschwendt M, Johannes FJ, Kittstein W, Marks F. (1997) Regulation of protein kinase Cμ by basic peptides and heparin. Putative role of an acidic domain in the activation of the kinase. J Biol Chem 272:20742–20746 [DOI] [PubMed] [Google Scholar]
- Guha S, Tanasanvimon S, Sinnett-Smith J, Rozengurt E. (2010) Role of protein kinase D signaling in pancreatic cancer. Biochem Pharmacol 80:1946–1954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J, Gertsberg Z, Ozgen N, Sabri A, Steinberg SF. (2011) Protein kinase D isoforms are activated in an agonist-specific manner in cardiomyocytes. J Biol Chem 286:6500–6509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison BC, Kim MS, van Rooij E, Plato CF, Papst PJ, Vega RB, McAnally JA, Richardson JA, Bassel-Duby R, Olson EN, et al. (2006) Regulation of cardiac stress signaling by protein kinase D1. Mol Cell Biol 26:3875–3888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hausser A, Storz P, Link G, Stoll H, Liu YC, Altman A, Pfizenmaier K, Johannes FJ. (1999) Protein kinase Cμ is negatively regulated by 14-3-3 signal transduction proteins. J Biol Chem 274:9258–9264 [DOI] [PubMed] [Google Scholar]
- Häussermann S, Kittstein W, Rincke G, Johannes FJ, Marks F, Gschwendt M. (1999) Proteolytic cleavage of protein kinase Cμ upon induction of apoptosis in U937 cells. FEBS Lett 462:442–446 [DOI] [PubMed] [Google Scholar]
- Haworth RS, Goss MW, Rozengurt E, Avkiran M. (2000) Expression and activity of protein kinase D/protein kinase Cμ in myocardium: evidence for α1-adrenergic receptor- and protein kinase C-mediated regulation. J Mol Cell Cardiol 32:1013–1023 [DOI] [PubMed] [Google Scholar]
- Hinchliffe KA, Irvine RF. (2006) Regulation of type II PIP kinase by PKD phosphorylation. Cell Signal 18:1906–1913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoshi N, Langeberg LK, Gould CM, Newton AC, Scott JD. (2010) Interaction with AKAP79 modifies the cellular pharmacology of PKC. Mol Cell 37:541–550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurd C, Waldron RT, Rozengurt E. (2002) Protein kinase D complexes with c-Jun N-terminal kinase via activation loop phosphorylation and phosphorylates the c-Jun N-terminus. Oncogene 21:2154–2160 [DOI] [PubMed] [Google Scholar]
- Huttlin EL, Jedrychowski MP, Elias JE, Goswami T, Rad R, Beausoleil SA, Villén J, Haas W, Sowa ME, Gygi SP. (2010) A tissue-specific atlas of mouse protein phosphorylation and expression. Cell 143:1174–1189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iglesias T, Rozengurt E. (1998) Protein kinase D activation by mutations within its pleckstrin homology domain. J Biol Chem 273:410–416 [DOI] [PubMed] [Google Scholar]
- Iglesias T, Waldron RT, Rozengurt E. (1998) Identification of in vivo phosphorylation sites required for protein kinase D activation. J Biol Chem 273:27662–27667 [DOI] [PubMed] [Google Scholar]
- Jacamo R, Sinnett-Smith J, Rey O, Waldron RT, Rozengurt E. (2008) Sequential protein kinase C (PKC)-dependent and PKC-independent protein kinase D catalytic activation via Gq-coupled receptors. Differential regulation of activation loop Ser(744) and Ser(748) phosphorylation. J Biol Chem 283:12877–12887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim MS, Wang F, Puthanveetil P, Kewalramani G, Innis S, Marzban L, Steinberg SF, Webber TD, Kieffer TJ, Abrahani A, et al. (2009) Cleavage of protein kinase D after acute hypoinsulinemia prevents excessive lipoprotein lipase-mediated cardiac triglyceride accumulation. Diabetes 58:2464–2475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel MT, Garcia EL, Kajimoto T, Hall RA, Newton AC. (2009) The protein scaffold NHERF-1 controls the amplitude and duration of localized protein kinase D activity. J Biol Chem 284:24653–24661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaValle CR, George KM, Sharlow ER, Lazo JS, Wipf P, Wang QJ. (2010) Protein kinase D as a potential new target for cancer therapy. Biochim Biophys Acta 1806:183–192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JY, Chiu YH, Asara J, Cantley LC. (2011) Inhibition of PI3K binding to activators by serine phosphorylation of PI3K regulatory subunit p85α Src homology-2 domains. Proc Natl Acad Sci USA 108:14157–14162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemonnier J, Ghayor C, Guicheux J, Caverzasio J. (2004) Protein kinase C-independent activation of protein kinase D is involved in BMP-2-induced activation of stress mitogen-activated protein kinases JNK and p38 and osteoblastic cell differentiation. J Biol Chem 279:259–264 [DOI] [PubMed] [Google Scholar]
- Lochhead PA. (2009) Protein kinase activation loop autophosphorylation in cis: overcoming a Catch-22 situation. Sci Signal 2:pe4. [DOI] [PubMed] [Google Scholar]
- Lochhead PA, Kinstrie R, Sibbet G, Rawjee T, Morrice N, Cleghon V. (2006) A chaperone-dependent GSK3β transitional intermediate mediates activation-loop autophosphorylation. Mol Cell 24:627–633 [DOI] [PubMed] [Google Scholar]
- Matthews SA, Rozengurt E, Cantrell D. (1999) Characterization of serine 916 as an in vivo autophosphorylation site for protein kinase D/protein kinase Cμ. J Biol Chem 274:26543–26549 [DOI] [PubMed] [Google Scholar]
- Meredith EL, Beattie K, Burgis R, Capparelli M, Chapo J, Dipietro L, Gamber G, Enyedy I, Hood DB, Hosagrahara V, et al. (2010) Identification of potent and selective amidobipyridyl inhibitors of protein kinase D. J Med Chem 53:5422–5438 [DOI] [PubMed] [Google Scholar]
- Nishikawa K, Toker A, Johannes FJ, Songyang Z, Cantley LC. (1997) Determination of the specific substrate sequence motifs of protein kinase C isozymes. J Biol Chem 272:952–960 [DOI] [PubMed] [Google Scholar]
- Old WM, Shabb JB, Houel S, Wang H, Couts KL, Yen CY, Litman ES, Croy CH, Meyer-Arendt K, Miranda JG, et al. (2009) Functional proteomics identifies targets of phosphorylation by B-Raf signaling in melanoma. Mol Cell 34:115–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oppermann FS, Gnad F, Olsen JV, Hornberger R, Greff Z, Kéri G, Mann M, Daub H. (2009) Large-scale proteomics analysis of the human kinome. Mol Cell Proteomics 8:1751–1764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozgen N, Obreztchikova M, Guo J, Elouardighi H, Dorn GW, 2nd, Wilson BA, Steinberg SF. (2008) Protein kinase D links Gq-coupled receptors to cAMP response element-binding protein (CREB)-Ser133 phosphorylation in the heart. J Biol Chem 283:17009–17019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterburs P, Heering J, Link G, Pfizenmaier K, Olayioye MA, Hausser A. (2009) Protein kinase D regulates cell migration by direct phosphorylation of the cofilin phosphatase slingshot 1 like. Cancer Res 69:5634–5638 [DOI] [PubMed] [Google Scholar]
- Phan D, Stratton MS, Huynh QK, McKinsey TA. (2011) A novel protein kinase C target site in protein kinase D is phosphorylated in response to signals for cardiac hypertrophy. Biochem Biophys Res Commun 411:335–341 [DOI] [PubMed] [Google Scholar]
- Qin L, Zeng H, Zhao D. (2006) Requirement of protein kinase D tyrosine phosphorylation for VEGF-A165-induced angiogenesis through its interaction and regulation of phospholipase Cγ phosphorylation. J Biol Chem 281:32550–32558 [DOI] [PubMed] [Google Scholar]
- Rey O, Sinnett-Smith J, Zhukova E, Rozengurt E. (2001) Regulated nucleocytoplasmic transport of protein kinase D in response to G protein-coupled receptor activation. J Biol Chem 276:49228–49235 [DOI] [PubMed] [Google Scholar]
- Rozengurt E, Rey O, Waldron RT. (2005) Protein kinase D signaling. J Biol Chem 280:13205–13208 [DOI] [PubMed] [Google Scholar]
- Rybin VO, Guo J, Steinberg SF. (2009) Protein kinase D1 Autophosphorylation via distinct mechanisms at Ser744/Ser748 and Ser916. J Biol Chem 284:2332–2343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez-Ruiloba L, Cabrera-Poch N, Rodríguez-Martínez M, López-Menéndez C, Jean-Mairet RM, Higuero AM, Iglesias T. (2006) Protein kinase D intracellular localization and activity control kinase D-interacting substrate of 220-kDa traffic through a postsynaptic density-95/discs large/zonula occludens-1-binding motif. J Biol Chem 281:18888–18900 [DOI] [PubMed] [Google Scholar]
- Sharlow ER, Giridhar KV, LaValle CR, Chen J, Leimgruber S, Barrett R, Bravo-Altamirano K, Wipf P, Lazo JS, Wang QJ. (2008) Potent and selective disruption of protein kinase D functionality by a benzoxoloazepinolone. J Biol Chem 283:33516–33526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi F, Telesco SE, Liu Y, Radhakrishnan R, Lemmon MA. (2010) ErbB3/HER3 intracellular domain is competent to bind ATP and catalyze autophosphorylation. Proc Natl Acad Sci USA 107:7692–7697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinnett-Smith J, Jacamo R, Kui R, Wang YM, Young SH, Rey O, Waldron RT, Rozengurt E. (2009) Protein kinase D mediates mitogenic signaling by Gq-coupled receptors through protein kinase C-independent regulation of activation loop Ser744 and Ser748 phosphorylation. J Biol Chem 284:13434–13445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith FD, Samelson BK, Scott JD. (2011) Discovery of cellular substrates for protein kinase A using a peptide array screening protocol. Biochem J 438:103–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spratley SJ, Bastea LI, Döppler H, Mizuno K, Storz P. (2011) Protein kinase D regulates cofilin activity through p21-activated kinase 4. J Biol Chem 286:34254–34261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steichen JM, Iyer GH, Li S, Saldanha SA, Deal MS, Woods VL, Jr, Taylor SS. (2010) Global consequences of activation loop phosphorylation on protein kinase A. J Biol Chem 285:3825–3832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiner TS, Ivison SM, Yao Y, Kifayet A. (2010) Protein kinase D1 and D2 are involved in chemokine release induced by toll-like receptors 2, 4, and 5. Cell Immunol 264:135–142 [DOI] [PubMed] [Google Scholar]
- Storz P, Döppler H, Johannes FJ, Toker A. (2003) Tyrosine phosphorylation of protein kinase D in the pleckstrin homology domain leads to activation. J Biol Chem 278:17969–17976 [DOI] [PubMed] [Google Scholar]
- Storz P, Döppler H, Toker A. (2004) Activation loop phosphorylation controls protein kinase D-dependent activation of nuclear factor κB. Mol Pharmacol 66:870–879 [DOI] [PubMed] [Google Scholar]
- Storz P, Döppler H, Toker A. (2005) Protein kinase D mediates mitochondrion-to-nucleus signaling and detoxification from mitochondrial reactive oxygen species. Mol Cell Biol 25:8520–8530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storz P, Toker A. (2003) Protein kinase D mediates a stress-induced NF-κB activation and survival pathway. EMBO J 22:109–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumara G, Formentini I, Collins S, Sumara I, Windak R, Bodenmiller B, Ramracheya R, Caille D, Jiang H, Platt KA, et al. (2009) Regulation of PKD by the MAPK p38δ in insulin secretion and glucose homeostasis. Cell 136:235–248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vántus T, Vertommen D, Saelens X, Rykx A, De Kimpe L, Vancauwenbergh S, Mikhalap S, Waelkens E, Kéri G, Seufferlein T, et al. (2004) Doxorubicin-induced activation of protein kinase D1 through caspase-mediated proteolytic cleavage: identification of two cleavage sites by microsequencing. Cell Signal 16:703–709 [DOI] [PubMed] [Google Scholar]
- Vertommen D, Rider M, Ni Y, Waelkens E, Merlevede W, Vandenheede JR, Van Lint J. (2000) Regulation of protein kinase D by multisite phosphorylation. Identification of phosphorylation sites by mass spectrometry and characterization by site-directed mutagenesis. J Biol Chem 275:19567–19576 [DOI] [PubMed] [Google Scholar]
- Villén J, Beausoleil SA, Gerber SA, Gygi SP. (2007) Large-scale phosphorylation analysis of mouse liver. Proc Natl Acad Sci USA 104:1488–1493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waldron RT, Iglesias T, Rozengurt E. (1999) The pleckstrin homology domain of protein kinase D interacts preferentially with the eta isoform of protein kinase C. J Biol Chem 274:9224–9230 [DOI] [PubMed] [Google Scholar]
- Waldron RT, Rozengurt E. (2003) Protein kinase C phosphorylates protein kinase D activation loop Ser744 and Ser748 and releases autoinhibition by the pleckstrin homology domain. J Biol Chem 278:154–163 [DOI] [PubMed] [Google Scholar]
- Waldron RT, Whitelegge JP, Faull KF, Rozengurt E. (2007) Identification of a novel phosphorylation site in c-Jun directly targeted in vitro by protein kinase D. Biochem Biophys Res Commun 356:361–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanivan S, Gnad F, Wickström SA, Geiger T, Macek B, Cox J, Fässler R, Mann M. (2008) Solid tumor proteome and phosphoproteome analysis by high resolution mass spectrometry. J Proteome Res 7:5314–5326 [DOI] [PubMed] [Google Scholar]
- Zhang W, Zheng S, Storz P, Min W. (2005) Protein kinase D specifically mediates apoptosis signal-regulating kinase 1-JNK signaling induced by H2O2 but not tumor necrosis factor. J Biol Chem 280:19036–19044 [DOI] [PubMed] [Google Scholar]
- Ziegler S, Eiseler T, Scholz RP, Beck A, Link G, Hausser A. (2011) A novel protein kinase D phosphorylation site in the tumor suppressor Rab interactor 1 is critical for coordination of cell migration. Mol Biol Cell 22:570–580 [DOI] [PMC free article] [PubMed] [Google Scholar]