Abstract
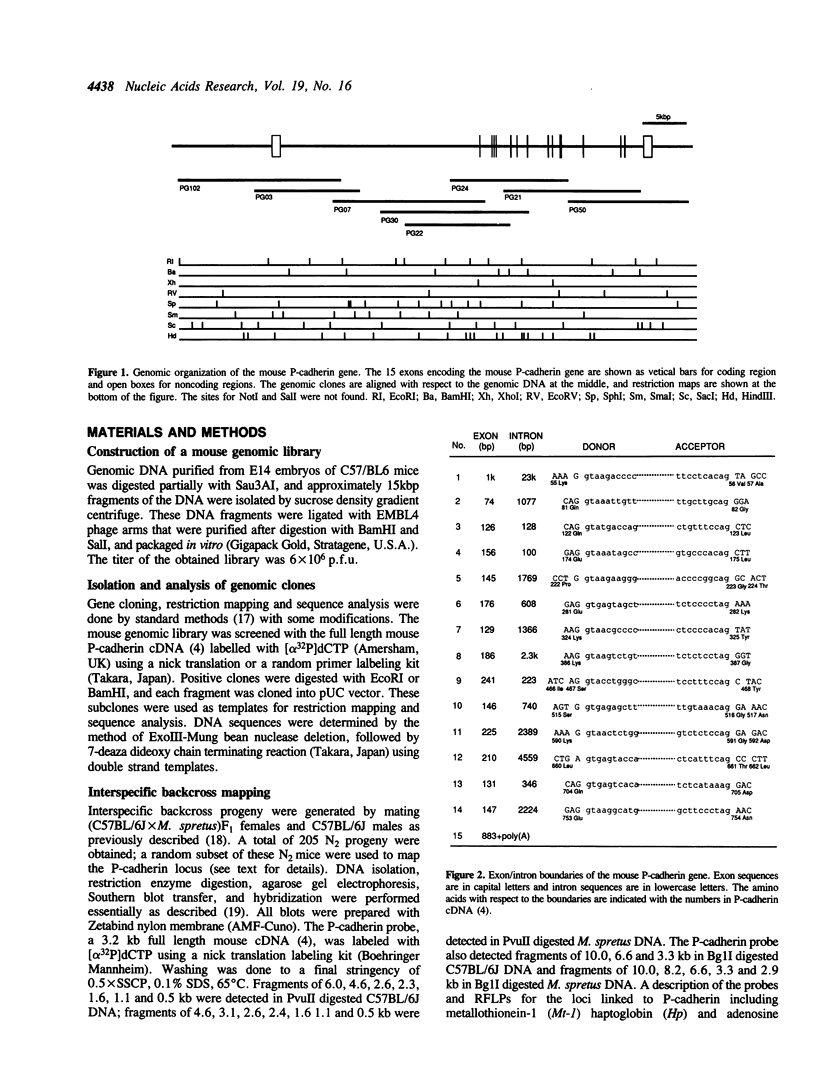
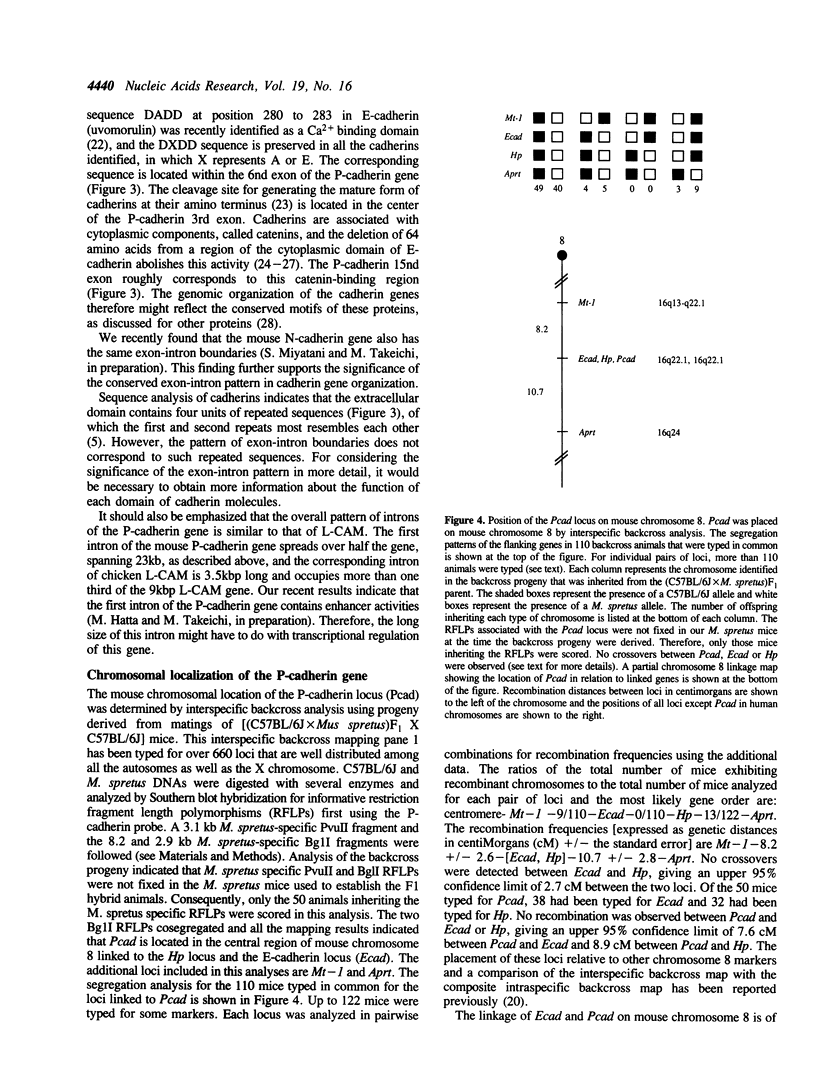
Cadherins are a family of Ca(2+)-dependent cell adhesion molecules, that includes P-cadherin, E-cadherin, N-cadherin and L-CAM. In this study, the genomic organization of the mouse P-cadherin gene was determined by analyzing overlapping DNA clones obtained from a mouse genomic library. The results showed that this gene spans over 45 kb and consists of 15 exons. A marked feature of this gene is that the first intron is 23 kbp long accounting for half its length. Comparisons of this structure with that of L-CAM, a chicken cadherin, revealed that the exon-intron boundaries are conserved between the two genes except that the P-cadherin first exon includes the correspoding first and second exons of the L-CAM gene. This gene was also similar to the other in that the second intron, which corresponds to the P-cadherin first intron, is exceptionally longer than other introns. These results suggest that the exon-intron pattern conserved in these genes is of significance for generation of domain structure of cadherin molecules or for their transcriptional regulation. We also determined the chromosomal localization of the P-cadherin gene by interspecific backcross analysis, and found that this gene is located in the central region of mouse chromosome 8 and linked with the E-cadherin locus. This is the first evidence for the linkage of different cadherin genes.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Buchberg A. M., Bedigian H. G., Taylor B. A., Brownell E., Ihle J. N., Nagata S., Jenkins N. A., Copeland N. G. Localization of Evi-2 to chromosome 11: linkage to other proto-oncogene and growth factor loci using interspecific backcross mice. Oncogene Res. 1988;2(2):149–165. [PubMed] [Google Scholar]
- Ceci J. D., Justice M. J., Lock L. F., Jenkins N. A., Copeland N. G. An interspecific backcross linkage map of mouse chromosome 8. Genomics. 1990 Jan;6(1):72–79. doi: 10.1016/0888-7543(90)90449-5. [DOI] [PubMed] [Google Scholar]
- Eistetter H. R., Adolph S., Ringwald M., Simon-Chazottes D., Schuh R., Guénet J. L., Kemler R. Chromosomal mapping of the structural gene coding for the mouse cell adhesion molecule uvomorulin. Proc Natl Acad Sci U S A. 1988 May;85(10):3489–3493. doi: 10.1073/pnas.85.10.3489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallin W. J., Sorkin B. C., Edelman G. M., Cunningham B. A. Sequence analysis of a cDNA clone encoding the liver cell adhesion molecule, L-CAM. Proc Natl Acad Sci U S A. 1987 May;84(9):2808–2812. doi: 10.1073/pnas.84.9.2808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginsberg D., DeSimone D., Geiger B. Expression of a novel cadherin (EP-cadherin) in unfertilized eggs and early Xenopus embryos. Development. 1991 Feb;111(2):315–325. doi: 10.1242/dev.111.2.315. [DOI] [PubMed] [Google Scholar]
- Hatta K., Nose A., Nagafuchi A., Takeichi M. Cloning and expression of cDNA encoding a neural calcium-dependent cell adhesion molecule: its identity in the cadherin gene family. J Cell Biol. 1988 Mar;106(3):873–881. doi: 10.1083/jcb.106.3.873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holton J. L., Kenny T. P., Legan P. K., Collins J. E., Keen J. N., Sharma R., Garrod D. R. Desmosomal glycoproteins 2 and 3 (desmocollins) show N-terminal similarity to calcium-dependent cell-cell adhesion molecules. J Cell Sci. 1990 Oct;97(Pt 2):239–246. doi: 10.1242/jcs.97.2.239. [DOI] [PubMed] [Google Scholar]
- Jenkins N. A., Copeland N. G., Taylor B. A., Lee B. K. Organization, distribution, and stability of endogenous ecotropic murine leukemia virus DNA sequences in chromosomes of Mus musculus. J Virol. 1982 Jul;43(1):26–36. doi: 10.1128/jvi.43.1.26-36.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch P. J., Walsh M. J., Schmelz M., Goldschmidt M. D., Zimbelmann R., Franke W. W. Identification of desmoglein, a constitutive desmosomal glycoprotein, as a member of the cadherin family of cell adhesion molecules. Eur J Cell Biol. 1990 Oct;53(1):1–12. [PubMed] [Google Scholar]
- Mansouri A., Spurr N., Goodfellow P. N., Kemler R. Characterization and chromosomal localization of the gene encoding the human cell adhesion molecule uvomorulin. Differentiation. 1988 Jun;38(1):67–71. doi: 10.1111/j.1432-0436.1988.tb00593.x. [DOI] [PubMed] [Google Scholar]
- Miyatani S., Shimamura K., Hatta M., Nagafuchi A., Nose A., Matsunaga M., Hatta K., Takeichi M. Neural cadherin: role in selective cell-cell adhesion. Science. 1989 Aug 11;245(4918):631–635. doi: 10.1126/science.2762814. [DOI] [PubMed] [Google Scholar]
- Nagafuchi A., Shirayoshi Y., Okazaki K., Yasuda K., Takeichi M. Transformation of cell adhesion properties by exogenously introduced E-cadherin cDNA. Nature. 1987 Sep 24;329(6137):341–343. doi: 10.1038/329341a0. [DOI] [PubMed] [Google Scholar]
- Nagafuchi A., Takeichi M. Cell binding function of E-cadherin is regulated by the cytoplasmic domain. EMBO J. 1988 Dec 1;7(12):3679–3684. doi: 10.1002/j.1460-2075.1988.tb03249.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagafuchi A., Takeichi M. Transmembrane control of cadherin-mediated cell adhesion: a 94 kDa protein functionally associated with a specific region of the cytoplasmic domain of E-cadherin. Cell Regul. 1989 Nov;1(1):37–44. doi: 10.1091/mbc.1.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nose A., Nagafuchi A., Takeichi M. Isolation of placental cadherin cDNA: identification of a novel gene family of cell-cell adhesion molecules. EMBO J. 1987 Dec 1;6(12):3655–3661. doi: 10.1002/j.1460-2075.1987.tb02698.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozawa M., Baribault H., Kemler R. The cytoplasmic domain of the cell adhesion molecule uvomorulin associates with three independent proteins structurally related in different species. EMBO J. 1989 Jun;8(6):1711–1717. doi: 10.1002/j.1460-2075.1989.tb03563.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozawa M., Engel J., Kemler R. Single amino acid substitutions in one Ca2+ binding site of uvomorulin abolish the adhesive function. Cell. 1990 Nov 30;63(5):1033–1038. doi: 10.1016/0092-8674(90)90506-a. [DOI] [PubMed] [Google Scholar]
- Ozawa M., Kemler R. Correct proteolytic cleavage is required for the cell adhesive function of uvomorulin. J Cell Biol. 1990 Oct;111(4):1645–1650. doi: 10.1083/jcb.111.4.1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozawa M., Ringwald M., Kemler R. Uvomorulin-catenin complex formation is regulated by a specific domain in the cytoplasmic region of the cell adhesion molecule. Proc Natl Acad Sci U S A. 1990 Jun;87(11):4246–4250. doi: 10.1073/pnas.87.11.4246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranscht B., Bronner-Fraser M. T-cadherin expression alternates with migrating neural crest cells in the trunk of the avian embryo. Development. 1991 Jan;111(1):15–22. doi: 10.1242/dev.111.1.15. [DOI] [PubMed] [Google Scholar]
- Ringwald M., Schuh R., Vestweber D., Eistetter H., Lottspeich F., Engel J., Dölz R., Jähnig F., Epplen J., Mayer S. The structure of cell adhesion molecule uvomorulin. Insights into the molecular mechanism of Ca2+-dependent cell adhesion. EMBO J. 1987 Dec 1;6(12):3647–3653. doi: 10.1002/j.1460-2075.1987.tb02697.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorkin B. C., Hemperly J. J., Edelman G. M., Cunningham B. A. Structure of the gene for the liver cell adhesion molecule, L-CAM. Proc Natl Acad Sci U S A. 1988 Oct;85(20):7617–7621. doi: 10.1073/pnas.85.20.7617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki S., Sano K., Tanihara H. Diversity of the cadherin family: evidence for eight new cadherins in nervous tissue. Cell Regul. 1991 Apr;2(4):261–270. doi: 10.1091/mbc.2.4.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeichi M. Cadherin cell adhesion receptors as a morphogenetic regulator. Science. 1991 Mar 22;251(5000):1451–1455. doi: 10.1126/science.2006419. [DOI] [PubMed] [Google Scholar]
- Traut T. W. Do exons code for structural or functional units in proteins? Proc Natl Acad Sci U S A. 1988 May;85(9):2944–2948. doi: 10.1073/pnas.85.9.2944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh F. S., Barton C. H., Putt W., Moore S. E., Kelsell D., Spurr N., Goodfellow P. N. N-cadherin gene maps to human chromosome 18 and is not linked to the E-cadherin gene. J Neurochem. 1990 Sep;55(3):805–812. doi: 10.1111/j.1471-4159.1990.tb04563.x. [DOI] [PubMed] [Google Scholar]
- Westphal E. M., Burmeister M., Wienker T. F., Lehrach H., Bender K., Scherer G. Tyrosine aminotransferase and chymotrypsinogen B are linked to haptoglobin on human chromosome 16q: comparison of genetic and physical distances. Genomics. 1987 Dec;1(4):313–319. doi: 10.1016/0888-7543(87)90030-9. [DOI] [PubMed] [Google Scholar]



