Abstract
A major recent discovery was the identification of an expansion of a non-coding GGGGCC hexanucleotide repeat in the C9ORF72 gene in patients with frontotemporal dementia and amyotrophic lateral sclerosis. Mutations in two other genes are known to account for familial frontotemporal dementia: microtubule-associated protein tau and progranulin. Although imaging features have been previously reported in subjects with mutations in tau and progranulin, no imaging features have been published in C9ORF72. Furthermore, it remains unknown whether there are differences in atrophy patterns across these mutations, and whether regional differences could help differentiate C9ORF72 from the other two mutations at the single-subject level. We aimed to determine the regional pattern of brain atrophy associated with the C9ORF72 gene mutation, and to determine which regions best differentiate C9ORF72 from subjects with mutations in tau and progranulin, and from sporadic frontotemporal dementia. A total of 76 subjects, including 56 with a clinical diagnosis of behavioural variant frontotemporal dementia and a mutation in one of these genes (19 with C9ORF72 mutations, 25 with tau mutations and 12 with progranulin mutations) and 20 sporadic subjects with behavioural variant frontotemporal dementia (including 50% with amyotrophic lateral sclerosis), with magnetic resonance imaging were included in this study. Voxel-based morphometry was used to assess and compare patterns of grey matter atrophy. Atlas-based parcellation was performed utilizing the automated anatomical labelling atlas and Statistical Parametric Mapping software to compute volumes of 37 regions of interest. Hemispheric asymmetry was calculated. Penalized multinomial logistic regression was utilized to create a prediction model to discriminate among groups using regional volumes and asymmetry score. Principal component analysis assessed for variance within groups. C9ORF72 was associated with symmetric atrophy predominantly involving dorsolateral, medial and orbitofrontal lobes, with additional loss in anterior temporal lobes, parietal lobes, occipital lobes and cerebellum. In contrast, striking anteromedial temporal atrophy was associated with tau mutations and temporoparietal atrophy was associated with progranulin mutations. The sporadic group was associated with frontal and anterior temporal atrophy. A conservative penalized multinomial logistic regression model identified 14 variables that could accurately classify subjects, including frontal, temporal, parietal, occipital and cerebellum volume. The principal component analysis revealed similar degrees of heterogeneity within all disease groups. Patterns of atrophy therefore differed across subjects with C9ORF72, tau and progranulin mutations and sporadic frontotemporal dementia. Our analysis suggested that imaging has the potential to be useful to help differentiate C9ORF72 from these other groups at the single-subject level.
Keywords: frontotemporal dementia, magnetic resonance imaging, C9ORF72, tau, progranulin
Introduction
Autosomal-dominant familial frontotemporal dementia (FTD) accounts for a large proportion of FTD cases. Mutations in the microtubule associated protein tau (MAPT) (Hutton et al., 1998) and progranulin (GRN) (Baker et al., 2006; Cruts et al., 2006) genes account for ∼10–20% of familial cases and are both located on chromosome 17q21. However, a major recent discovery has identified a third gene mutation (Dejesus-Hernandez et al., 2011; Renton et al., 2011) that appears to be the most common genetic abnormality both in familial FTD and amyotrophic lateral sclerosis (ALS), accounting for 11.7% of familial FTD and 23.5% of familial ALS (Dejesus-Hernandez et al., 2011). This new mutation is an expansion of a non-coding GGGGCC hexanucleotide repeat in the C9ORF72 gene and is genetically linked to chromosome 9p (Dejesus-Hernandez et al., 2011; Renton et al., 2011). Pathologically, both the GRN and C9ORF72 gene mutations are associated with deposition of the transactive response DNA binding protein of 43 kDa (TDP-43), whereas mutations in MAPT are associated with deposition of the hyperphosphorylated protein tau.
Previous studies have shown that the most common clinical phenotype associated with both MAPT and GRN mutations is behavioural variant FTD, although a more varied clinical spectrum has been associated with GRN mutations, including diagnoses such as primary progressive aphasia and corticobasal syndrome (Masellis et al., 2006; Mesulam et al., 2007; Beck et al., 2008; Pickering-Brown et al., 2008; Kelley et al., 2009). The C9ORF72 mutation is also commonly associated with behavioural variant FTD, although also with ALS (Dejesus-Hernandez et al., 2011; Renton et al., 2011). Neuroanatomical differences have also been identified between the MAPT and GRN mutations using MRI (Whitwell et al., 2009a, b; Rohrer et al., 2010b). Mutations in MAPT have been associated with atrophy predominantly in the anteromedial temporal lobes (Whitwell et al., 2005, 2009a, b; Ghetti et al., 2008; Spina et al., 2008; Rohrer et al., 2010b), whereas mutations in GRN have been associated with more widespread and asymmetric patterns of atrophy that involve the frontal, temporal and parietal lobes (Whitwell et al., 2007, 2009b; Beck et al., 2008; Rohrer et al., 2010b). These findings suggest that imaging could be useful to help predict the specific mutation, and hence the underlying pathology. Importantly, characterizing the patterns of atrophy associated with these mutations also provides important insight into the biology of the disease and helps to further define neuroanatomical correlations with clinical and pathological features in FTD. No studies have yet assessed patterns of atrophy in the C9ORF72 mutation, and hence it is unknown whether the neuroanatomical signature of this mutation differs from MAPT and GRN mutations.
The aim of our study was therefore to determine the pattern of brain atrophy associated with C9ORF72, and to determine which regions best differentiate C9ORF72 from MAPT and GRN, as well as from sporadic FTD including ALS. Furthermore, although MAPT and GRN mutations have been associated with differing clinical and neuroanatomical features, a number of studies have highlighted a high degree of variability within subjects with each of these mutations, as well as within subjects, within the same family (van Swieten et al., 1999; Janssen et al., 2002; Boeve et al., 2005; Kelley et al., 2009). Therefore, we also aimed to assess the degree of neuroanatomical variability within each of the genetic mutations and sporadic FTD, in order to determine whether variability in the new C9ORF72 gene mutation is comparable with the other groups.
Materials and methods
Subjects
We identified all subjects seen at Mayo Clinic, Rochester, MN, USA who had screened positive for mutations in C9ORF72 with a volumetric MRI. A total of 19 subjects, representing 16 families, were identified.
We then identified three disease comparison groups. First, we identified all subjects who had screened positive for mutations in GRN or MAPT with volumetric MRI. Since all C9ORF72 subjects had a clinical diagnosis of behavioural variant FTD we only included GRN or MAPT subjects that also fulfilled clinical criteria for behavioural variant FTD (Neary et al., 1998). Eighteen subjects were identified with GRN mutations. Six subjects were excluded due to other clinical diagnoses [four with primary progressive aphasia (Mesulam, 1982), one with corticobasal syndrome (Boeve et al., 2003) and one with Alzheimer's dementia (McKhann et al., 1984)], resulting in a total of 12 subjects, representing six families, with behavioural variant FTD that were included in the study. Five different GRN mutations were present in our cohort: c.154delA (p.Thr52HisfsX2), c.910_911insTG (p.Trp304LeufsX58), c.1395_1396insC (p.Cys466LeufsX46), c.1145delC (p.Thr382SerfsX30) and c.138+1G>A (IVS1+1G>A). Twenty-six subjects were identified with MAPT mutations. One subject with a clinical diagnosis of primary progressive aphasia (Mesulam, 1982) was excluded, resulting in 25 subjects with behavioural variant FTD, representing 12 families that were included in the study. Nine different MAPT mutations were present in our cohort: P301L (c.1907C>T;p.Pro301Leu), IVS10+16 (c.1920+16C>T;IVS10+16C>T), IVS10+3 (c.1920+3G>A;IVS10+3G>A), N279K (c.1842T>G;p.Asn279Lys), V337M (c.2014G>A; Val337Met), S305N (c.1919G>A;p.Ser305Asn) (Boeve et al., 2005), G389R (c.2170G>A;p.Gly389Arg), R406W (c.2221C>T;p.Arg406Trp) and IVS9-10 (c.1827-10G>T;IVS9-10G>T) (Malkani et al., 2006). In all cases, the first MRI that was performed after the subject had a dementia diagnosis was used.
Secondly, the C9ORF72 group was age- and gender-matched to a group of 20 subjects with behavioural variant FTD (Neary et al., 1998) that included 50% that had clinicopathological evidence of ALS (Josephs, 2008). This was important as some C9ORF72 subjects also had clinicopathological evidence of ALS. These subjects did not have family history of either an FTD syndrome or ALS in a first- or second-degree relative, and were screened negative for genetic mutations, and hence were considered to have sporadic behavioural variant FTD.
The majority of subjects had been prospectively studied in our Alzheimer's Disease Research Centre (ADRC). Clinical diagnoses were made according to consensus criteria for behavioural variant FTD (Neary et al., 1998), primary progressive aphasia (Mesulam, 1982), corticobasal syndrome (Boeve et al., 2003) and Alzheimer's dementia (McKhann et al., 1984). Diagnoses were retrospectively fitted for 10/76 cases that met Lund–Manchester criteria for FTD (The Lund and Manchester Groups, 1994). Imaging results have previously been published from seven of the GRN subjects and 22 of the MAPT subjects (Whitwell et al., 2007). The total cohort of 76 FTD cases was frequency matched by age, gender and MRI field strength to a cohort of 40 cognitively normal control subjects. Subject demographics for all groups are shown in Table 1. Informed consent was obtained from all subjects and/or their proxies for participation in the studies, which were approved by the Mayo Institutional Review Board.
Table 1.
Subject demographics
| Demographics | Controls | MAPT | GRN | C9ORF72 | Sporadic FTD | P-values across disease groups |
|---|---|---|---|---|---|---|
| n | 40 | 25 | 12 | 19 | 20 | NA |
| Gender, female, n (%) | 20 (50) | 12 (48) | 6 (50) | 10 (53) | 10 (50) | 0.99 |
| Education (years) | 15 (10–20) | 14.0 (9–19) | 14.0 (12–20) | 14.0 (12–20) | 15.0 (11–18) | 0.76 |
| Age at onset (years) | NA | 43.4 (21–63) | 56.0 (47–83) | 51.0 (33–67) | 51.5 (38–62) | <0.0001 |
| Age at death (years)a | NA | 55.3 (42–70) | 64.1 (57–87) | 56.5 (35–75) | 58.3 (43–70) | 0.08 |
| Disease duration (years) | NA | 8.0 (4–21) | 7.0 (5–10) | 5.5 (2–12) | 4.4 (1–15) | 0.17 |
| Age at scan (years) | 58.0 (34–84) | 51.6 (25–65) | 62.6 (50–84) | 54.8 (35–70) | 54.5 (41–67) | 0.01b |
| Onset to scan (years) | NA | 5.0 (0.2–24) | 2.7 (1.3–7.1) | 2.7 (1.2–15.0) | 2.0 (0–6) | 0.08 |
Data are shown as median (range). NA = not applicable.
a A total of 10 subjects with the C9ORF72 mutation, 14 subjects with the MAPT mutation, 8 subjects with the GRN mutation, and 14 sporadic FTD have died.
b Significant differences were observed when including controls, at P < 0.05.
Genetic analysis
Exons 0–12 and the 3′-untranslated region of the GRN gene were amplified by polymerase chain reaction using our previously published primers and protocol (Baker et al., 2006; Gass et al., 2006). Analysis of MAPT exons 1, 7 and 9–13 was also performed using primers and conditions that were previously described (Hutton et al., 1998). For GRN and MAPT, the polymerase chain reaction amplicons were purified using the Multiscreen system (Millipore) and then sequenced in both directions using Big Dye® chemistry following manufacturer's protocol (Applied Biosystems). Sequence products were purified using the Montage system (Millipore) prior to being run on an ABI 3730 DNA Analyzer. Sequence data were analysed using either SeqScape or Sequencher software. To determine the presence of an expanded GGGGCC hexanucleotide repeat in C9ORF72, we used the repeat primed polymerase chain reaction method previously published (Dejesus-Hernandez et al., 2011).
Pathological analysis
Neuropathological examinations were performed according to the Consortium to Establish a Registry for Alzheimer's Disease (CERAD) recommendations (Mirra et al., 1991). Pathological assessment and diagnosis was conducted by one of two expert neuropathologists (D.W.D. or J.E.P.). All cases were reclassified based on recent consensus recommendations for neuropathological subtypes of frontotemporal lobar degeneration (FTLD) (Mackenzie et al., 2009). Cases were classified according to abnormal protein into FTLD with tau immunoreactive inclusions (FTLD-tau), FTLD with TDP-43 (FTLD-TDP) and FTLD with fused in sarcoma (FTLD-FUS). FTLD-TDP was subclassified into types 1–3 according to MacKenzie et al. (2006).
Magnetic resonance imaging analysis
Magnetic resonance imaging protocol
All subjects underwent a standardized protocol head MRI that included a T1-weighted 3D volumetric sequence. In the majority of cases this consisted of a spoiled gradient echo sequence performed at 1.5T, although 35% of C9ORF72, 32% of MAPT, 33% of GRN, 20% of sporadic FTD and 37% controls had a magnetization-prepared rapid acquisition gradient echo sequence performed at 3T. All images underwent pre-processing that included corrections for gradient non-linearity (Jovicich et al., 2006) and intensity inhomogeneity using both the N3 bias correction (Sled et al., 1998) followed by the SPM5-based bias correction.
Voxel-based morphometry
Patterns of grey matter atrophy were assessed using the automated and unbiased technique of voxel-based morphometry (Ashburner and Friston, 2000). An optimized method of voxel-based morphometry was applied using both customized templates and prior probability maps (Senjem et al., 2005), implemented using SPM5 (http://www.fil.ion.ucl.ac.uk/spm). Briefly, all images were normalized to a customized template and segmented using unified segmentation (Ashburner and Friston, 2005), followed by the hidden Markov random-field clean-up step (Zhang et al., 2001). All images were modulated and smoothed with an 8-mm full-width at half-maximum smoothing kernel. A full factorial statistical model was used to assess patterns of grey matter loss in each disease group compared with controls. The C9ORF72 group was also compared directly with MAPT, GRN and sporadic FTD. Age at scan, gender, total intracranial volume and field strength were included as covariates in the statistical model. Results were assessed after correction for multiple comparisons using the family-wise error correction at P < 0.05. Comparisons across disease groups were investigated uncorrected for multiple comparisons at P < 0.001.
Atlas-based parcellation
Atlas-based parcellation was employed using SPM5 and an in-house modified version of the automated anatomic labelling atlas (Tzourio-Mazoyer et al., 2002), as previously described (Whitwell et al., 2009c), in order to generate grey matter volumes for 37 regions of interest that covered cortical and subcortical grey matter: left and right dorsolateral frontal, medial frontal, orbitofrontal, sensorimotor, inferior temporal (fusiform + inferior temporal gyrus), lateral temporal (middle + superior temporal gyri), medial temporal (amygdala + hippocampus), temporal pole, lateral parietal, precuneus, occipital lobe, anterior cingulate, middle cingulate, posterior cingulate, insula, thalamus, striatum (caudate + putamen), cerebellum and total vermis.
Whole-brain volume and total intracranial volume estimates were obtained by propagating region of interest masks from a template. All regional grey matter volumes were divided by whole brain volume to correct for differences in global atrophy between subjects. This step was performed because we were interested in assessing the relative involvement of each region, without confounds of global severity. In addition, an asymmetry score was calculated for the frontal, temporal and parietal lobes for each subject as follows: (left volume − right volume)*2/(left volume + right volume).
Statistics
All statistical analyses were performed using R version 2.13.2 (http://www.R-project.org) and the glmnet package version 1.7.1. Comparisons of demographic and clinical variables of interest among groups were performed using Kruskal–Wallis tests for continuous variables and chi-squared or Fisher's exact tests for categorical variables.
Four-category multinomial logistic regression was used to discriminate among the four disease groups (Agresti, 2002) using whole brain volume-corrected volumes of the 37 regions of interest and frontal lobe asymmetry score as predictors. Due to the large number of predictors relative to the sample size, we used the elastic net penalization approach to develop a parsimonious multivariable model (Friedman et al., 2010).
In multivariable modelling, the degree of overfitting depends on the number of variables considered rather than the number of variables in the final model (Harrell, 2001). Roughly speaking, in traditional model-selection techniques, and in particular with step-wise variable selection, when a large number of variables are considered, the most significant variables are included ‘as is’ even though they are very likely overfitting to the sample and thus overestimates of the magnitude of the association. This is due to the regression to the mean phenomena; in a different sample, such variables would likely still show an association but probably to a lesser degree. In repeated sample, the coefficients would tend to regress to their ‘rightful place’. Penalization methods ‘shrink’ regression coefficients towards zero as a way to reduce the effect of extreme coefficients and the resulting bias that is present in overfitting. In a sense, penalized methods are used to help the coefficients find their rightful place by penalizing those that are overly large. In the following paragraph, we describe how the penalty was applied and how the specific value of the penalty was chosen.
The elastic net penalization approach we used is a combination of ridge regression penalization, where the sum of the squares of the coefficients are controlled, and the lasso penalty, where the sum of the absolute values of the coefficients are controlled. The elastic net requires the analyst to select two tuning parameters. The first, α, was set to 0.9, so that the lasso penalty was predominant but there remained a degree of ridge regression, a balance that tends to perform well when there are a large number of possibly correlated predictors (Friedman et al., 2010). The second parameter, λ, determines the degree of penalization and can range from zero corresponding to an unpenalized full model up to infinity corresponding to a model in which all non-intercept coefficients are shrunk to zero. We selected λ to optimize goodness of fit using the multinomial deviance based on leave-one-out cross-validation. Informally, the method omitted a subject from the sample, fit the model for 100 different values λ and calculated the goodness-of-fit error for each value of λ. After doing this 76 times, we obtained 76 estimates of goodness-of-fit error at each value of λ and from these the mean and SD errors are calculated. The value of λ corresponding to where the mean error was smallest defined the ‘optimal’ model in terms of cross-validation. That is, the model that is likely to be most generalizable. To be conservative, our final model was chosen based on the ‘1SE rule’ in which we selected the largest λ value, or equivalently the most parsimonious model, such that the goodness-of-fit error was still within 1 standard error (1SE) of the optimum.
In order to assess the dimensionality of the imaging data in each disease group, principal component analysis was performed within the C9ORF72, MAPT, GRN and sporadic FTD groups separately using the whole brain volume-corrected volumes of the 37 regions of interest, and the frontal lobe asymmetry score.
Results
Clinical, demographic and pathological findings
Significant differences were observed in age at onset and MRI across disease groups, with the youngest median age observed in MAPT, and oldest age observed in GRN (Table 1). No differences were observed across disease groups in gender, education, disease duration, age at death or time from onset to MRI. The degree of cognitive, functional and behavioural impairment, as well as the degree of parkinsonism, also did not differ across groups (Table 2). The C9ORF72 mutation was associated with behavioural variant FTD. Two subjects had co-existing ALS (Table 2). None of the MAPT or GRN subjects had ALS. Although pathology was not available for all subjects, the pathological diagnoses were strikingly different (Table 2). The C9ORF72 mutation was associated with FTLD-TDP types 1 (Mackenzie et al., 2006) (harmonized type A; Mackenzie et al., 2011) and 3 (Mackenzie et al., 2006) (harmonized type B; Mackenzie et al., 2011) and a pathological diagnosis of ALS. This contrasted with GRN that was associated with only FTLD-TDP type 1 (Mackenzie et al., 2006), and MAPT that was associated with FTLD-tau. By design, the sporadic FTD group consisted of subjects with behavioural variant FTD of which 50% had clinicopathological evidence of ALS (FTLD-TDP type; Mackenzie et al., 2006).
Table 2.
Clinical, pathological and imaging features
| Features | Controls | MAPT | GRN | C9ORF72 | Sporadic FTD | P-values across disease groups |
|---|---|---|---|---|---|---|
| n | 40 | 25 | 12 | 19 | 20 | NA |
| MMSE at scan | 30 (25–30) | 25.0 (1–30) | 19.0 (4–27) | 25.0 (11–29) | 25.0 (2–30) | 0.16a |
| CDR-SB at scan | 0 (0–0) | 5.5 (0.5–18) | 7.0 (4.5–18) | 5.0 (1–18) | 6.5 (2–12) | 0.28a |
| UPDRS at scan | 0.0 (0–2) | 2.5 (0–48) | 4.0 (0–25) | 0.0 (0–15) | 1.0 (0–9) | 0.43a |
| NPI-Q at scan | 0.0 (0–3) | 8.0 (1–20) | 7.0 (2–18) | 8.0 (2–24) | 11 (1–22) | 0.78a |
| Presence/absence of clinical features at scan | ||||||
| Amyotrophic lateral sclerosis, n (%) | 0 (0) | 0 (0) | 0 (0) | 2 (11) | 7 (35) | NA |
| Cerebellar ataxia, n (%) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | NA |
| Limb apraxia, n (%) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | NA |
| Autonomic dysfunction, n (%) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | NA |
| Pathological diagnoses | ||||||
| FTLD-Tau | NA | 6 | 0 | 0 | 3 | <0.0001 |
| FTLD-FUS | NA | 0 | 0 | 0 | 1 | |
| FTLD-TDP type 1 | NA | 0 | 8 | 5 (2 with ALS) | 0 | |
| FTLD-TDP type 3 | NA | 0 | 0 | 3 (2 with ALS) | 8 (6 with ALS) | |
| Asymmetry score | ||||||
| Frontal lobe | 0.05 | 0.06 | 0.21 | 0.06 | 0.08 | 0.0035a |
| Temporal lobe | 0.06 | 0.11 | 0.20 | 0.08 | 0.09 | 0.0083a |
| Parietal lobe | 0.04 | 0.07 | 0.13 | 0.05 | 0.07 | 0.0003a |
Data are shown as median (range).
CDR-SB = Clinical Dementia Rating Scale Sum of Boxes; MMSE = Mini-Mental Status Examination; NPI-Q = brief questionnaire version of the Neuropsychiatric Inventory; UPDRS = Unified Parkinson's Disease Rating Scale.
a Significant differences were observed when including controls, at P < 0.05.
Patterns of atrophy
Patterns of regional atrophy compared with controls as revealed by voxel-based morphometry can be observed in Fig. 1A, and the regional volumes for all disease groups are shown in Figs 2 and 3. The C9ORF72 group showed a widespread pattern of grey matter loss compared with controls. The most striking loss was observed in frontal lobes, involving orbitofrontal, medial and dorsolateral regions, followed by temporal lobes. Loss was also observed in parietal and occipital lobes and in cerebellum. Patterns of loss observed in MAPT and GRN were typical of those we have previously published (Whitwell et al., 2009b), with striking temporal lobe volume loss observed in MAPT and temporoparietal loss observed in GRN. The sporadic FTD group showed predominant frontal lobe atrophy, with only mild temporal involvement.
Figure 1.
Results of the voxel-based morphometry analysis of grey matter volume. (A) Patterns of grey matter loss in MAPT, GRN, C9ORF72 and sporadic FTD groups compared with controls, at P < 0.05 (corrected for multiple comparisons using family-wise error). (B) Differences in grey matter volume between the C9ORF72 group and the other disease groups, at P < 0.001 (uncorrected for multiple comparisons). Results are shown in 3D renderings of the brain.
Figure 2.
Box plots of all variables selected in the penalized multinomial logistic regression model as best for differentiating disease groups. All regional grey matter volumes were divided by whole brain volume to correct for differences in global atrophy between subjects. This step was performed because we were interested in assessing the relative involvement of each region, without confounds of global severity. L = left; R = right; ROI = region of interest.
Figure 3.
Box plots of variables that were not selected in the penalized multinomial logistic regression model as useful for differentiating disease groups, and hence not shown in Fig. 2. All regional grey matter volumes were divided by whole brain volume to correct for differences in global atrophy between subjects. This step was performed because we were interested in assessing the relative involvement of each region, without confounds of global severity. L = left; R = right; ROI = region of interest.
On direct comparison across groups (Fig. 1B), C9ORF72 showed greater loss in parietal and occipital lobes, particularly medially, as well as lateral inferior frontal lobe and cerebellum, compared with MAPT and sporadic FTD. Conversely, MAPT showed greater grey matter loss throughout the anterior temporal lobes, bilaterally, compared with C9ORF72. The C9ORF72 group did not show any regions of greater loss than GRN, although GRN showed greater involvement of left inferior temporal lobe and bilateral parietal lobe than C9ORF72. The sporadic FTD group only showed a small region of greater loss than C9ORF72 in the medial frontal lobe.
Asymmetry
The degree of frontal, temporal and parietal asymmetry differed across disease groups (Table 2), with greatest asymmetry observed in the GRN group across all three regions. The C9ORF72 group was not associated with asymmetry in any region.
Classification accuracy
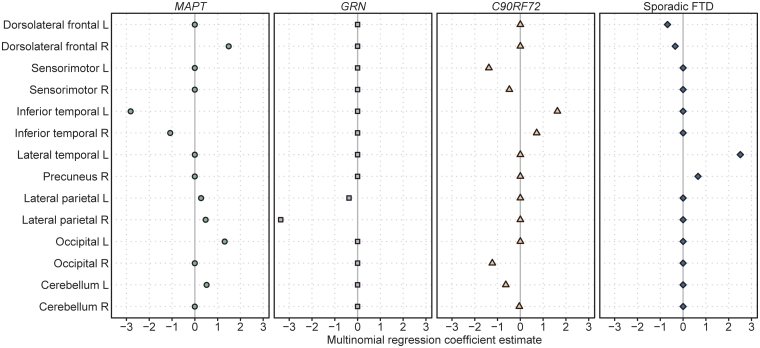
The optimal penalized multinomial logistic regression model was able to accurately classify 71/76 (93%) subjects using 26 variables (Table 3). In a more conservative model that required only 14 variables and hence potentially more clinically meaningful, the regression model was still able to correctly classify 56/76 (74%) subjects (Table 3). Box plots of these 14 variables are shown in Fig. 2 and the regression coefficients of the 14 variables are summarized graphically in Fig. 4. The predictive value of each variable is demonstrated in Fig. 5. Small volumes of left sensorimotor cortices, right occipital lobe and left cerebellum, and relatively large volumes of left inferior temporal lobe were independently predictive of C9ORF72; small volumes of inferior temporal lobe and large volumes of right dorsolateral frontal lobe, parietal lobe and left occipital lobe were independently predictive of MAPT; small volumes of right lateral parietal lobe were independently predictive of GRN; and small volumes of dorsolateral frontal lobes and large volumes of left lateral temporal lobe were independently predictive of sporadic FTD.
Table 3.
Classification tables for both the optimum and conservative penalized multinomial logistic regression models
| Predicted patient groups | MAPT (n = 25) | GRN (n = 12) | C9ORF72 (n = 19) | Sporadic FTD (n = 20) |
|---|---|---|---|---|
| Optimal model with 26 variables (overall classification 93%) | ||||
| MAPT | 24 | 0 | 0 | 1 |
| GRN | 0 | 12 | 0 | 0 |
| C9ORF72 | 0 | 0 | 17 | 1 |
| Sporadic FTD | 1 | 0 | 2 | 18 |
| Conservative model with 14 variables (overall classification 74%) | ||||
| MAPT | 23 | 3 | 0 | 3 |
| GRN | 0 | 7 | 1 | 0 |
| C9ORF72 | 1 | 1 | 13 | 4 |
| Sporadic FTD | 1 | 1 | 5 | 13 |
Figure 4.
Coefficients for the 14 variables that were retained in the final penalized multinomial logistic regression for the MAPT, GRN, C9ORF72 and sporadic FTD groups. A negative coefficient means that lower values (i.e. more atrophy) on that variable increases the estimated probability of being in that group, holding all other variables constant. L = left; R = right.
Figure 5.
Plots showing the estimated probability of MAPT, GRN, C9ORF72 and sporadic FTD according to volume for each of the 14 variables that were retained in the final model. In each panel, the adjustment covariates are set to their mean values. L = left; R = right; ROI = region of interest.
Variability within groups
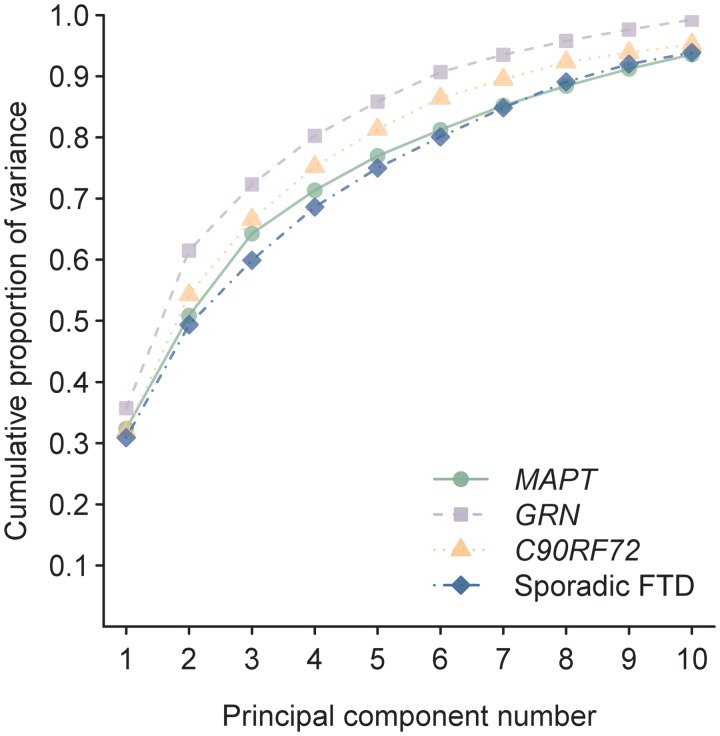
The results of the within-group principal component analysis are shown in Fig. 6. Approximately six principal components explained 80% of the variability in the imaging data in all four groups. In addition, the percentage of variability explained by each principal component was similar across groups.
Figure 6.
Plot of the cumulative proportion of variability explained by each principal component for the C9ORF72, MAPT, GRN and sporadic FTD groups.
Discussion
This study demonstrates that the newly discovered mutation in the C9ORF72 gene is associated with predominant frontal lobe atrophy, with involvement of the anterior temporal, parietal lobes and cerebellum. This specific pattern differed from that observed in MAPT and GRN mutations, and sporadic behavioural variant FTD. These patterns of regional atrophy accurately classified the majority of C9ORF72, MAPT, GRN and sporadic FTD subjects at the single-subject level.
Widespread and severe patterns of atrophy were observed in C9ORF72, although the most striking atrophy was observed in frontal lobes, followed by anterior temporal lobes. Frontal atrophy was not focal and involved medial, dorsolateral and orbitofrontal regions, demonstrating the lack of regional specificity within the frontal lobe for this mutation. In addition, we identified parietal lobe atrophy. Once again, parietal atrophy was not focal and involved medial and lateral regions. Involvement of both lateral and medial parietal lobes is unusual in FTD, and hence may be particularly associated with this mutation. Even more unusual was the identification of atrophy of the cerebellum, which has not been emphasized in any clinical or pathological FTD variants. This finding is intriguing since a family with the C9ORF72 gene mutation was found to exhibit numerous neuronal cytoplasmic inclusions and short neurites in the granule cell layer of the cerebellar cortex (Boxer et al., 2011); a finding that has now been confirmed in two recent studies assessing relatively large cohorts of subjects with the C9ORF72 gene mutation that also identified cerebellar inclusions (Al-Sarraj et al., 2011; Murray et al., 2011). Therefore, cerebellar pathology, both macroscopic and microscopic, could be a characteristic feature of C9ORF72. Cerebellar clinical signs, such as ataxia, were not recorded in our C9ORF72 subjects, although cerebellar ataxia has been observed previously in a family with chromosome 9 mutations (Pearson et al., 2011). It is possible that cerebellar signs are absent in our study but it is also possible that cerebellar signs are present, yet not recorded. Regardless, the imaging findings of cerebellar atrophy in this study, supports the notion that future prospective studies should include standardized neurological evaluations of the cerebellar system. It would also appear surprising that we did not observe significant atrophy of the primary motor cortex in C9ORF72; however, it must be emphasized that only two of our C9ORF72 subjects had clinical evidence of ALS. Furthermore, previous imaging studies have similarly not identified atrophy or hypometabolism in the primary motor cortex of subjects with FTD with ALS (Jeong et al., 2005; Whitwell et al., 2006; Boxer et al., 2011). It is possible that diffusion tensor imaging assessment of specific white matter tracts may be more sensitive to deficits in primary motor regions, or to loss of projection fibres originating from the primary motor cortex in FTD subjects with ALS (Verstraete et al., 2011).
The specific pathological diagnoses associated with C9ORF72 were FTLD-TDP types 1 and 3. Both of these pathologies are associated with frontotemporal atrophy, with a frontal predominance in type 3 and parietal involvement in type 1 (Rohrer et al., 2010a; Whitwell et al., 2010). Hence, the patterns we observe in C9ORF72 may be driven by a combination of these pathologies. Neither of these two pathologies, however, has been associated with cerebellar atrophy, which is not necessarily surprising since previous studies would not have stratified FTLD-TDP types by specific mutation type. More specifically, previous studies assessing FTLD-TDP type 1 would have grouped cases with C9ORF72, with GRN mutation cases, and with cases with neither mutation.
The patterns observed in C9ORF72 contrasted with MAPT mutations, in which the most striking atrophy occurred in anteromedial temporal lobes and with GRN mutations where the most striking atrophy occurred in temporal and parietal lobes. These findings confirm our previous studies of MAPT and GRN using many of the same subjects (Whitwell et al., 2009a, b) and previous studies using independent cohorts that have associated MAPT mutations with anteromedial temporal atrophy (Arvanitakis et al., 2007; Spina et al., 2008; Miyoshi et al., 2010; Rohrer et al., 2010b; Seelaar et al., 2011) and GRN mutations with parietal atrophy (Spina et al., 2007; Cruchaga et al., 2009; Rohrer et al., 2010b; Seelaar et al., 2011), despite clinical variability across studies. Both MAPT and GRN showed significantly greater temporal lobe involvement than C9ORF72, with the temporal involvement focused on anteromedial regions in MAPT and inferior temporal lobe in GRN. This result helps to further clarify our previous finding that subjects with FTLD-TDP type 1 with GRN mutations have greater lateral temporal atrophy than those without GRN mutations (Whitwell et al., 2010); we now know that many of the GRN negative FTLD-TDP type 1 subjects in our cohort have C9ORF72 mutations. Although all the MAPT subjects had a clinical diagnosis of behavioural variant FTD, semantic deficits are common (Rizzini et al., 2000; Snowden et al., 2006; Pickering-Brown et al., 2008), concurring with the anteromedial involvement. The C9ORF72 group did, however, show significant atrophy in posterior regions of the brain, which is distinct from MAPT and sporadic FTD, but similar to GRN. Both GRN and C9ORF72 gene mutations are associated with FTLD-TDP type 1 pathology as mentioned earlier, and this pathology is particularly associated with atrophy in posterior regions of the brain (Whitwell et al., 2010). Atrophy of the cerebellum was not observed in the MAPT group; with C9ORF72 showing significantly greater cerebellar atrophy than the MAPT subjects. Cerebellar atrophy was however observed, although to a lesser degree, in the GRN group, again suggesting a possible link to FTLD-TDP type 1 pathology. Another important feature of the C9ORF72 mutation was that it was associated with relatively symmetric patterns of frontal, temporal and parietal atrophy at the group level. In contrast, GRN mutations were associated with asymmetric patterns of atrophy, as previously noted (Beck et al., 2008; Ghetti et al., 2008; Le Ber et al., 2008; Kelley et al., 2009). The C9ORF72 gene mutation showed a different pattern of atrophy from sporadic FTD in spite of the fact that both groups consisted of subjects with behavioural variant FTD with and without ALS. Both groups showed striking frontal lobe atrophy, which is not surprising given the clinical diagnoses (Chang et al., 2005; Jeong et al., 2005; Whitwell et al., 2006), although greater involvement of posterior cortices and cerebellum was observed in C9ORF72. These findings argue that genetics has a strong influence over patterns of neuroanatomical damage in FTD and suggest that each disease group has a different path of pathological progression through the brain.
We have demonstrated that these neuroanatomical differences across groups could classify the subjects in our cohort at the individual level. The penalized multinomial logistic regression model showed that atrophy of sensorimotor cortex, precuneus, occipital lobe and cerebellum, with sparing of the inferior temporal lobe, was particularly useful to help predict the presence of the C9ORF72 gene mutation in the context of MAPT, GRN and sporadic FTD. Although the sensorimotor cortex and occipital lobes were not the most heavily affected regions in C9ORF72, they were relatively more affected than in the other groups, and hence useful for differentiation.
These findings further validate the differences observed at the group level. Furthermore, they suggest that patterns of atrophy have the potential to be useful to help differentiate these groups and help predict the presence of the C9ORF72 gene mutation in subjects with behavioural variant FTD, regardless of the presence of family history. This is particularly useful since the pathologies underlying behavioural variant FTD are notoriously heterogeneous and difficult to predict based on clinical features alone (Josephs et al., 2011). The findings could help, for example, to tailor an approach to genetic testing to avoid performing costly and unnecessary multiple genetic tests. Taken into context, if a patient presents to the clinic with behavioural variant FTD the easiest mutation to identify, or rule out, using MRI appears to be mutations in MAPT since it is the only group in which atrophy predominantly and focally affects the anteromedial temporal lobes. If a mutation in MAPT can be ruled out, our data suggest that it would then be helpful to assess the degree of involvement of the lateral temporal and parietal lobes. If these regions are affected, then one would suspect either a GRN or C9ORF72 mutation, since atrophy in sporadic FTD does not tend to heavily involve these regions. The differentiation of C9ORF72 and GRN appears to be more challenging due to the degree of overlap observed in these groups, although there is a suggestion that greater involvement of the sensorimotor cortices and occipital lobe would suggest C9ORF72, while striking atrophy of the parietal lobe would be more suggestive of GRN.
All four disease groups showed some involvement of the frontal lobes so it may be difficult to differentiate groups based only on this region; however, the absence of frontal atrophy would point towards a MAPT mutation and striking dorsolateral frontal atrophy would more suggest sporadic FTD. An important point to stress is that the regional volumes used in our model were divided by the size of the brain to allow us to assess the relative involvement of each region, without confounds of global severity. It is therefore important when assessing these patients to not just look at the degree of atrophy but to instead assess the relative involvement of each region in comparison with other regions of the brain. Therefore, a GRN subject that is early in the disease course may have the same amount of parietal involvement as a C9ORF72 subject later in the disease course, but importantly GRN would still have relatively greater involvement of the parietal lobe compared with other regions. One caveat to take into account, however, is that differentiation is likely to be much more difficult in patients with advanced disease since regions that were previously unaffected may start to ‘catch up’ to the regions that showed a greater degree of atrophy early in the disease, resulting in a less focal pattern of loss. Imaging is therefore most likely to be helpful early in the disease course.
It is important to point out that while our analysis focused only on imaging, the addition of clinical features will even further improve prediction. The presence of a strong family history argues against sporadic FTD; the presence of ALS is very suggestive of the C9ORF72 mutation (Dejesus-Hernandez et al., 2011; Renton et al., 2011), rather than either MAPT or GRN; young age would be particularly suggestive of a MAPT mutation, and older age of GRN mutations (Beck et al., 2008; Pickering-Brown et al., 2008; Whitwell et al., 2009b). Ultimately, optimum prediction is likely to require both imaging and clinical information.
While there could be concern that the results of our model are specific to the subjects in our study and not generalizable to the population, we specifically chose penalization methods, instead of more traditional regression methodology, with leave-one-out cross-validation, since it is a good way to fit a model that generalizes well to the population rather than to a particular set of patients. We therefore do not think that the classification rates are over estimates based on the patients in the sample but are instead reflective of the population. We acknowledge that due to sampling variability the classification rates are estimates only, but because of our methods and choice of a conservative model, we do not think they are biased.
We also demonstrated that the dimensionality or richness of the imaging data did not differ across these groups, and hence the imaging data in each group was similarly varied and similarly complex. The C9ORF72 group therefore does not appear to be more heterogeneous than the other groups. Anatomical heterogeneity has been previously reported in GRN and MAPT cohorts and families, although these cohorts often included subjects with varying clinical diagnoses (van Swieten et al., 1999; Janssen et al., 2002; Boeve et al., 2005; Kelley et al., 2009). Our findings show that the degree of heterogeneity associated with these mutations in behavioural variant FTD is very similar to that observed in a typical sporadic FTD cohort.
This study describes for the first time the patterns of atrophy associated with the newly discovered C9ORF72 gene mutation. These findings provide important understanding of the biology of these mutations, have the potential to be useful clinically and will likely pave the way for future studies that further investigate neuroanatomical correlations with clinical and pathological features.
Funding
NIH (grants R21 AG38736, R01 DC010367, R01 AG037491, R01 AG11378, P50 AG16574, U01 AG024904, R01-AG023195, U01 AG06786, R01 HL70825, U24 AG026395, U01 AG03949, RO1 NS065782-01, R56 AG26251-03, P50-AG25711, P50-NS40256, P01-AG17216, R01-AG15866, R01-NS65782, R01-AG26251, 1RC2NS070276, NS057567, P50 NS072187-01S2); the Dana Foundation; the Pacific Alzheimer Research Foundation (Canada); the Amyotrophic Lateral Sclerosis Association; Mayo Clinic Florida (MCF) Research Committee CR program (MCF #90052030); Dystonia Medical Research Foundation; a gift from Carl Edward Bolch, Jr and Susan Bass Bolch (MCF #90052031/PAU #90052); the Alexander Family Alzheimer's Disease Research Professorship of the Mayo Foundation; and the Robert H. and Clarice Smith and Abigail Van Buren Alzheimer's Disease Research Program of the Mayo Foundation.
Acknowledgements
We would like to acknowledge Dr Jay Mandrekar, PhD, and Seth Slettedahl, BS, for contributions to the statistical analysis.
Glossary
Abbreviations
- ALS
amyotrophic lateral sclerosis
- FTD
frontotemporal dementia
- FTLD
frontotemporal lobar degeneration
- GRN
progranulin
- MAPT
microtubule-associated protein tau
- TDP-43
transactive response DNA binding protein of 43 kDa
References
- Agresti A. New York: Wiley-Interscience; 2002. Categorical data analysis. [Google Scholar]
- Al-Sarraj S, King A, Troakes C, Smith B, Maekawa S, Bodi I, et al. p62 positive, TDP-43 negative, neuronal cytoplasmic and intranuclear inclusions in the cerebellum and hippocampus define the pathology of C9orf72-linked FTLD and MND/ALS. Acta Neuropathol. 2011;122:691–702. doi: 10.1007/s00401-011-0911-2. [DOI] [PubMed] [Google Scholar]
- Arvanitakis Z, Witte RJ, Dickson DW, Tsuboi Y, Uitti RJ, Slowinski J, et al. Clinical-pathologic study of biomarkers in FTDP-17 (PPND family with N279K tau mutation) Parkinsonism Relat Disord. 2007;13:230–9. doi: 10.1016/j.parkreldis.2006.10.007. [DOI] [PubMed] [Google Scholar]
- Ashburner J, Friston KJ. Voxel-based morphometry–the methods. Neuroimage. 2000;11:805–21. doi: 10.1006/nimg.2000.0582. [DOI] [PubMed] [Google Scholar]
- Ashburner J, Friston KJ. Unified segmentation. Neuroimage. 2005;26:839–51. doi: 10.1016/j.neuroimage.2005.02.018. [DOI] [PubMed] [Google Scholar]
- Baker M, Mackenzie IR, Pickering-Brown SM, Gass J, Rademakers R, Lindholm C, et al. Mutations in progranulin cause tau-negative frontotemporal dementia linked to chromosome 17. Nature. 2006;442:916–9. doi: 10.1038/nature05016. [DOI] [PubMed] [Google Scholar]
- Beck J, Rohrer JD, Campbell T, Isaacs A, Morrison KE, Goodall EF, et al. A distinct clinical, neuropsychological and radiological phenotype is associated with progranulin gene mutations in a large UK series. Brain. 2008;131:706–20. doi: 10.1093/brain/awm320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeve BF, Lang AE, Litvan I. Corticobasal degeneration and its relationship to progressive supranuclear palsy and frontotemporal dementia. Ann Neurol. 2003;54(Suppl. 5):S15–9. doi: 10.1002/ana.10570. [DOI] [PubMed] [Google Scholar]
- Boeve BF, Tremont-Lukats IW, Waclawik AJ, Murrell JR, Hermann B, Jack CR, Jr, et al. Longitudinal characterization of two siblings with frontotemporal dementia and parkinsonism linked to chromosome 17 associated with the S305N tau mutation. Brain. 2005;128:752–72. doi: 10.1093/brain/awh356. [DOI] [PubMed] [Google Scholar]
- Boxer AL, Mackenzie IR, Boeve BF, Baker M, Seeley WW, Crook R, et al. Clinical, neuroimaging and neuropathological features of a new chromosome 9p-linked FTD-ALS family. J Neurol Neurosurg Psychiatry. 2011;82:196–203. doi: 10.1136/jnnp.2009.204081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang JL, Lomen-Hoerth C, Murphy J, Henry RG, Kramer JH, Miller BL, et al. A voxel-based morphometry study of patterns of brain atrophy in ALS and ALS/FTLD. Neurology. 2005;65:75–80. doi: 10.1212/01.wnl.0000167602.38643.29. [DOI] [PubMed] [Google Scholar]
- Cruchaga C, Fernandez-Seara MA, Seijo-Martinez M, Samaranch L, Lorenzo E, Hinrichs A, et al. Cortical atrophy and language network reorganization associated with a novel progranulin mutation. Cereb Cortex. 2009;19:1751–60. doi: 10.1093/cercor/bhn202. [DOI] [PubMed] [Google Scholar]
- Cruts M, Gijselinck I, van der Zee J, Engelborghs S, Wils H, Pirici D, et al. Null mutations in progranulin cause ubiquitin-positive frontotemporal dementia linked to chromosome 17q21. Nature. 2006;442:920–4. doi: 10.1038/nature05017. [DOI] [PubMed] [Google Scholar]
- Dejesus-Hernandez M, Mackenzie IR, Boeve BF, Boxer AL, Baker M, Rutherford NJ, et al. Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron. 2011;72:245–56. doi: 10.1016/j.neuron.2011.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman J, Hastie T, Tibshirani R. Regularization paths for generalized linear models via coordinate descent. J Stat Software. 2010;33:1–22. [PMC free article] [PubMed] [Google Scholar]
- Gass J, Cannon A, Mackenzie IR, Boeve B, Baker M, Adamson J, et al. Mutations in progranulin are a major cause of ubiquitin-positive frontotemporal lobar degeneration. Hum Mol Genet. 2006;15:2988–3001. doi: 10.1093/hmg/ddl241. [DOI] [PubMed] [Google Scholar]
- Ghetti B, Spina S, Murrell JR, Huey ED, Pietrini P, Sweeney B, et al. In vivo and postmortem clinicoanatomical correlations in frontotemporal dementia and parkinsonism linked to chromosome 17. Neurodegener Dis. 2008;5:215–7. doi: 10.1159/000113706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrell FE. Regression modeling strategies: with applications to linear models, logistic regression, and survival analysis. New York: Springer. 2001 [Google Scholar]
- Hutton M, Lendon CL, Rizzu P, Baker M, Froelich S, Houlden H, et al. Association of missense and 5′-splice-site mutations in tau with the inherited dementia FTDP-17. Nature. 1998;393:702–5. doi: 10.1038/31508. [DOI] [PubMed] [Google Scholar]
- Janssen JC, Warrington EK, Morris HR, Lantos P, Brown J, Revesz T, et al. Clinical features of frontotemporal dementia due to the intronic tau 10(+16) mutation. Neurology. 2002;58:1161–8. doi: 10.1212/wnl.58.8.1161. [DOI] [PubMed] [Google Scholar]
- Jeong Y, Park KC, Cho SS, Kim EJ, Kang SJ, Kim SE, et al. Pattern of glucose hypometabolism in frontotemporal dementia with motor neuron disease. Neurology. 2005;64:734–6. doi: 10.1212/01.WNL.0000152047.58767.9D. [DOI] [PubMed] [Google Scholar]
- Josephs KA. Frontotemporal dementia and related disorders: deciphering the enigma. Ann Neurol. 2008;64:4–14. doi: 10.1002/ana.21426. [DOI] [PubMed] [Google Scholar]
- Josephs KA, Hodges JR, Snowden J, Mackenzie IR, Neumann M, Mann D, et al. Neuropathological background of phenotypical variability in frontotemporal dementia. Acta Neuropathol. 2011;122:137–53. doi: 10.1007/s00401-011-0839-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jovicich J, Czanner S, Greve D, Haley E, van der Kouwe A, Gollub R, et al. Reliability in multi-site structural MRI studies: effects of gradient non-linearity correction on phantom and human data. Neuroimage. 2006;30:436–43. doi: 10.1016/j.neuroimage.2005.09.046. [DOI] [PubMed] [Google Scholar]
- Kelley BJ, Haidar W, Boeve BF, Baker M, Graff-Radford NR, Krefft T, et al. Prominent phenotypic variability associated with mutations in Progranulin. Neurobiol Aging. 2009;30:739–51. doi: 10.1016/j.neurobiolaging.2007.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Ber I, Camuzat A, Hannequin D, Pasquier F, Guedj E, Rovelet-Lecrux A, et al. Phenotype variability in progranulin mutation carriers: a clinical, neuropsychological, imaging and genetic study. Brain. 2008;131:732–46. doi: 10.1093/brain/awn012. [DOI] [PubMed] [Google Scholar]
- Mackenzie IR, Baborie A, Pickering-Brown S, Du Plessis D, Jaros E, Perry RH, et al. Heterogeneity of ubiquitin pathology in frontotemporal lobar degeneration: classification and relation to clinical phenotype. Acta Neuropathol. 2006;112:539–49. doi: 10.1007/s00401-006-0138-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackenzie IR, Neumann M, Baborie A, Sampathu DM, Du Plessis D, Jaros E, et al. A harmonized classification system for FTLD-TDP pathology. Acta Neuropathol. 2011;122:111–3. doi: 10.1007/s00401-011-0845-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackenzie IR, Neumann M, Bigio EH, Cairns NJ, Alafuzoff I, Kril J, et al. Nomenclature for neuropathologic subtypes of frontotemporal lobar degeneration: consensus recommendations. Acta Neuropathol. 2009;117:15–8. doi: 10.1007/s00401-008-0460-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malkani R, D'Souza I, Gwinn-Hardy K, Schellenberg GD, Hardy J, Momeni P. A MAPT mutation in a regulatory element upstream of exon 10 causes frontotemporal dementia. Neurobiol Dis. 2006;22:401–3. doi: 10.1016/j.nbd.2005.12.001. [DOI] [PubMed] [Google Scholar]
- Masellis M, Momeni P, Meschino W, Heffner R, Jr, Elder J, Sato C, et al. Novel splicing mutation in the progranulin gene causing familial corticobasal syndrome. Brain. 2006;129:3115–23. doi: 10.1093/brain/awl276. [DOI] [PubMed] [Google Scholar]
- McKhann G, Drachman D, Folstein M, Katzman R, Price D, Stadlan EM. Clinical diagnosis of Alzheimer's disease: report of the NINCDS-ADRDA Work Group under the auspices of Department of Health and Human Services Task Force on Alzheimer's Disease. Neurology. 1984;34:939–44. doi: 10.1212/wnl.34.7.939. [DOI] [PubMed] [Google Scholar]
- Mesulam M, Johnson N, Krefft TA, Gass JM, Cannon AD, Adamson JL, et al. Progranulin mutations in primary progressive aphasia: the PPA1 and PPA3 families. Arch Neurol. 2007;64:43–7. doi: 10.1001/archneur.64.1.43. [DOI] [PubMed] [Google Scholar]
- Mesulam MM. Slowly progressive aphasia without generalized dementia. Ann Neurol. 1982;11:592–8. doi: 10.1002/ana.410110607. [DOI] [PubMed] [Google Scholar]
- Mirra SS, Heyman A, McKeel D, Sumi SM, Crain BJ, Brownlee LM, et al. The Consortium to Establish a Registry for Alzheimer's Disease (CERAD). Part II. Standardization of the neuropathologic assessment of Alzheimer's disease. Neurology. 1991;41:479–86. doi: 10.1212/wnl.41.4.479. [DOI] [PubMed] [Google Scholar]
- Miyoshi M, Shinotoh H, Wszolek ZK, Strongosky AJ, Shimada H, Arakawa R, et al. In vivo detection of neuropathologic changes in presymptomatic MAPT mutation carriers: a PET and MRI study. Parkinsonism Relat Disord. 2010;16:404–8. doi: 10.1016/j.parkreldis.2010.04.004. [DOI] [PubMed] [Google Scholar]
- Murray ME, Dejesus-Hernandez M, Rutherford N, Baker M, Duara R, Graff-Radford N, et al. Clinical and neuropathologic heterogeneity of c9FTD/ALS associated with hexanucleotide repeat expansion in C9ORF72. Acta Neuropathol. 2011;122:673–90. doi: 10.1007/s00401-011-0907-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neary D, Snowden JS, Gustafson L, Passant U, Stuss D, Black S, et al. Frontotemporal lobar degeneration: a consensus on clinical diagnostic criteria. Neurology. 1998;51:1546–54. doi: 10.1212/wnl.51.6.1546. [DOI] [PubMed] [Google Scholar]
- Pearson JP, Williams NM, Majounie E, Waite A, Stott J, Newsway V, et al. Familial frontotemporal dementia with amyotrophic lateral sclerosis and a shared haplotype on chromosome 9p. J Neurol. 2011;258:647–55. doi: 10.1007/s00415-010-5815-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickering-Brown SM, Rollinson S, Du Plessis D, Morrison KE, Varma A, Richardson AM, et al. Frequency and clinical characteristics of progranulin mutation carriers in the Manchester frontotemporal lobar degeneration cohort: comparison with patients with MAPT and no known mutations. Brain. 2008;131:721–31. doi: 10.1093/brain/awm331. [DOI] [PubMed] [Google Scholar]
- Renton AE, Majounie E, Waite A, Simon-Sanchez J, Rollinson S, Gibbs JR, et al. A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21-Linked ALS-FTD. Neuron. 2011;72:257–68. doi: 10.1016/j.neuron.2011.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizzini C, Goedert M, Hodges JR, Smith MJ, Jakes R, Hills R, et al. Tau gene mutation K257T causes a tauopathy similar to Pick's disease. J Neuropathol Exp Neurol. 2000;59:990–1001. doi: 10.1093/jnen/59.11.990. [DOI] [PubMed] [Google Scholar]
- Rohrer JD, Geser F, Zhou J, Gennatas ED, Sidhu M, Trojanowski JQ, et al. TDP-43 subtypes are associated with distinct atrophy patterns in frontotemporal dementia. Neurology. 2010a;75:2204–11. doi: 10.1212/WNL.0b013e318202038c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohrer JD, Ridgway GR, Modat M, Ourselin S, Mead S, Fox NC, et al. Distinct profiles of brain atrophy in frontotemporal lobar degeneration caused by progranulin and tau mutations. Neuroimage. 2010b;53:1070–6. doi: 10.1016/j.neuroimage.2009.12.088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seelaar H, Papma JM, Garraux G, de Koning I, Reijs AE, Valkema R, et al. Brain perfusion patterns in familial frontotemporal lobar degeneration. Neurology. 2011;77:384–92. doi: 10.1212/WNL.0b013e3182270456. [DOI] [PubMed] [Google Scholar]
- Senjem ML, Gunter JL, Shiung MM, Petersen RC, Jack CR., Jr Comparison of different methodological implementations of voxel-based morphometry in neurodegenerative disease. Neuroimage. 2005;26:600–8. doi: 10.1016/j.neuroimage.2005.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sled JG, Zijdenbos AP, Evans AC. A nonparametric method for automatic correction of intensity nonuniformity in MRI data. IEEE Trans Med Imag. 1998;17:87–97. doi: 10.1109/42.668698. [DOI] [PubMed] [Google Scholar]
- Snowden JS, Pickering-Brown SM, Mackenzie IR, Richardson AM, Varma A, Neary D, et al. Progranulin gene mutations associated with frontotemporal dementia and progressive non-fluent aphasia. Brain. 2006;129:3091–102. doi: 10.1093/brain/awl267. [DOI] [PubMed] [Google Scholar]
- Spina S, Farlow MR, Unverzagt FW, Kareken DA, Murrell JR, Fraser G, et al. The tauopathy associated with mutation +3 in intron 10 of Tau: characterization of the MSTD family. Brain. 2008;131:72–89. doi: 10.1093/brain/awm280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spina S, Murrell JR, Huey ED, Wassermann EM, Pietrini P, Grafman J, et al. Corticobasal syndrome associated with the A9D progranulin mutation. J Neuropathol Exp Neurol. 2007;66:892–900. doi: 10.1097/nen.0b013e3181567873. [DOI] [PubMed] [Google Scholar]
- The Lund and Manchester Groups. Clinical and neuropathological criteria for frontotemporal dementia. J Neurol Neurosurg Psychiatry. 1994;57:416–8. doi: 10.1136/jnnp.57.4.416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzourio-Mazoyer N, Landeau B, Papathanassiou D, Crivello F, Etard O, Delcroix N, et al. Automated anatomical labeling of activations in SPM using a macroscopic anatomical parcellation of the MNI MRI single-subject brain. Neuroimage. 2002;15:273–89. doi: 10.1006/nimg.2001.0978. [DOI] [PubMed] [Google Scholar]
- van Swieten JC, Stevens M, Rosso SM, Rizzu P, Joosse M, de Koning I, et al. Phenotypic variation in hereditary frontotemporal dementia with tau mutations. Ann Neurol. 1999;46:617–26. doi: 10.1002/1531-8249(199910)46:4<617::aid-ana10>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]
- Verstraete E, Veldink JH, Mandl RC, van den Berg LH, van den Heuvel MP. Impaired structural motor connectome in amyotrophic lateral sclerosis. PLoS One. 2011;6:e24239. doi: 10.1371/journal.pone.0024239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitwell JL, Jack CR, Jr, Baker M, Rademakers R, Adamson J, Boeve BF, et al. Voxel-based morphometry in frontotemporal lobar degeneration with ubiquitin-positive inclusions with and without progranulin mutations. Arch Neurol. 2007;64:371–6. doi: 10.1001/archneur.64.3.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitwell JL, Jack CR, Jr, Boeve BF, Senjem ML, Baker M, Ivnik RJ, et al. Atrophy patterns in IVS10+16, IVS10+3, N279K, S305N, P301L, and V337M MAPT mutations. Neurology. 2009a;73:1058–65. doi: 10.1212/WNL.0b013e3181b9c8b9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitwell JL, Jack CR, Jr, Boeve BF, Senjem ML, Baker M, Rademakers R, et al. Voxel-based morphometry patterns of atrophy in FTLD with mutations in MAPT or PGRN. Neurology. 2009b;72:813. doi: 10.1212/01.wnl.0000343851.46573.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitwell JL, Jack CR, Jr, Parisi JE, Senjem ML, Knopman DS, Boeve BF, et al. Does TDP-43 type confer a distinct pattern of atrophy in frontotemporal lobar degeneration? Neurology. 2010;75:2212–20. doi: 10.1212/WNL.0b013e31820203c2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitwell JL, Jack CR, Jr, Senjem ML, Josephs KA. Patterns of atrophy in pathologically confirmed FTLD with and without motor neuron degeneration. Neurology. 2006;66:102–4. doi: 10.1212/01.wnl.0000191395.69438.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitwell JL, Josephs KA, Rossor MN, Stevens JM, Revesz T, Holton JL, et al. Magnetic resonance imaging signatures of tissue pathology in frontotemporal dementia. Arch Neurol. 2005;62:1402–8. doi: 10.1001/archneur.62.9.1402. [DOI] [PubMed] [Google Scholar]
- Whitwell JL, Przybelski SA, Weigand SD, Ivnik RJ, Vemuri P, Gunter JL, et al. Distinct anatomical subtypes of the behavioural variant of frontotemporal dementia: a cluster analysis study. Brain. 2009c;132:2932–46. doi: 10.1093/brain/awp232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Brady M, Smith S. Segmentation of brain MR images through a hidden Markov random field model and the expectation-maximization algorithm. IEEE Trans Med Imag. 2001;20:45–57. doi: 10.1109/42.906424. [DOI] [PubMed] [Google Scholar]