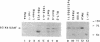Abstract
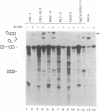
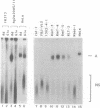
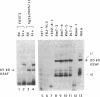
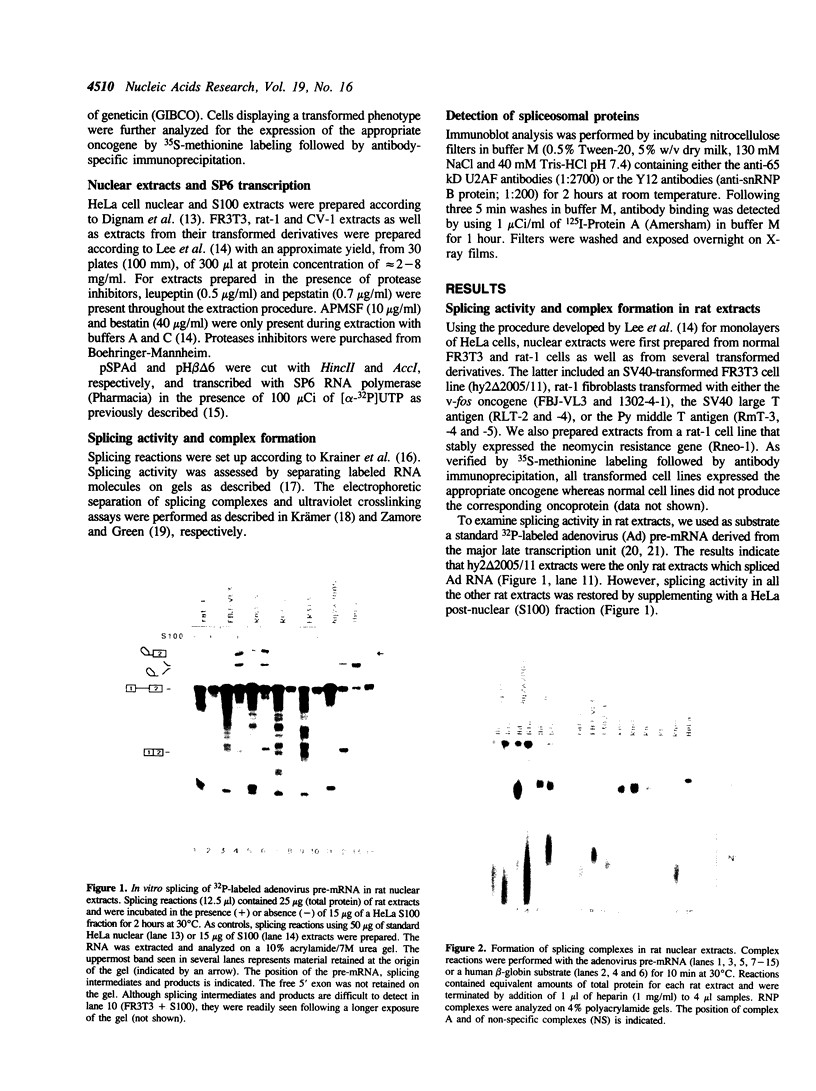
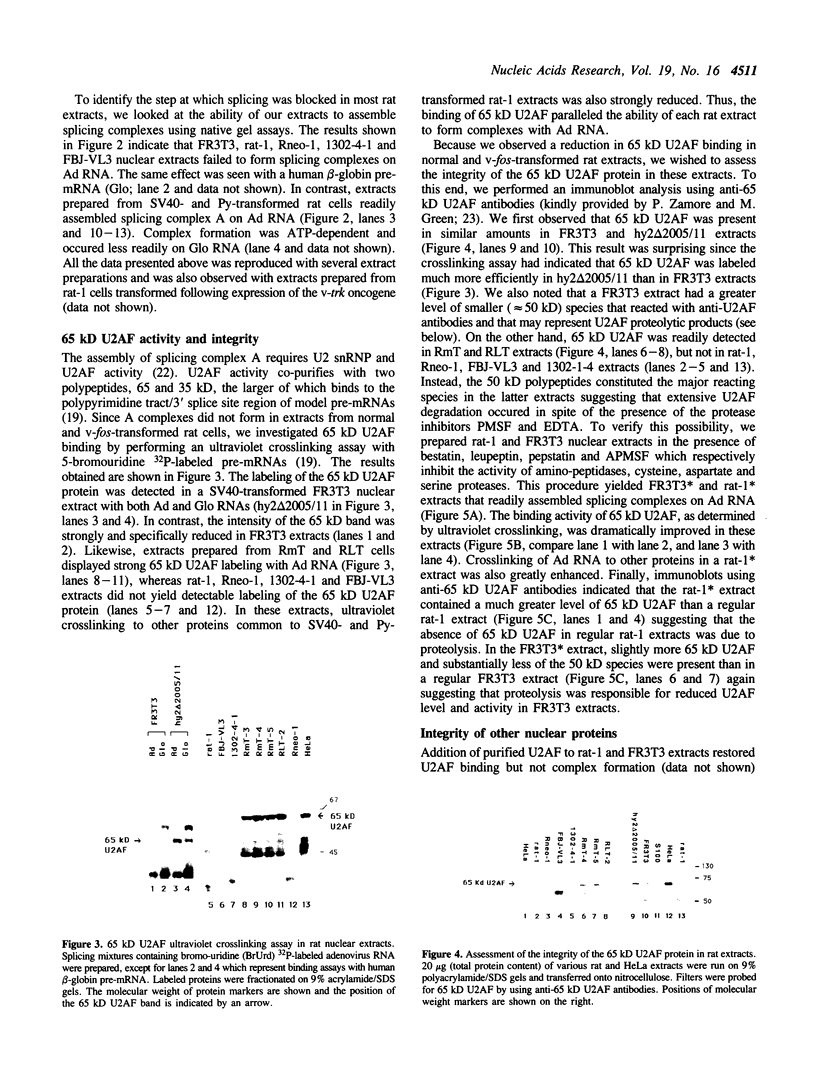
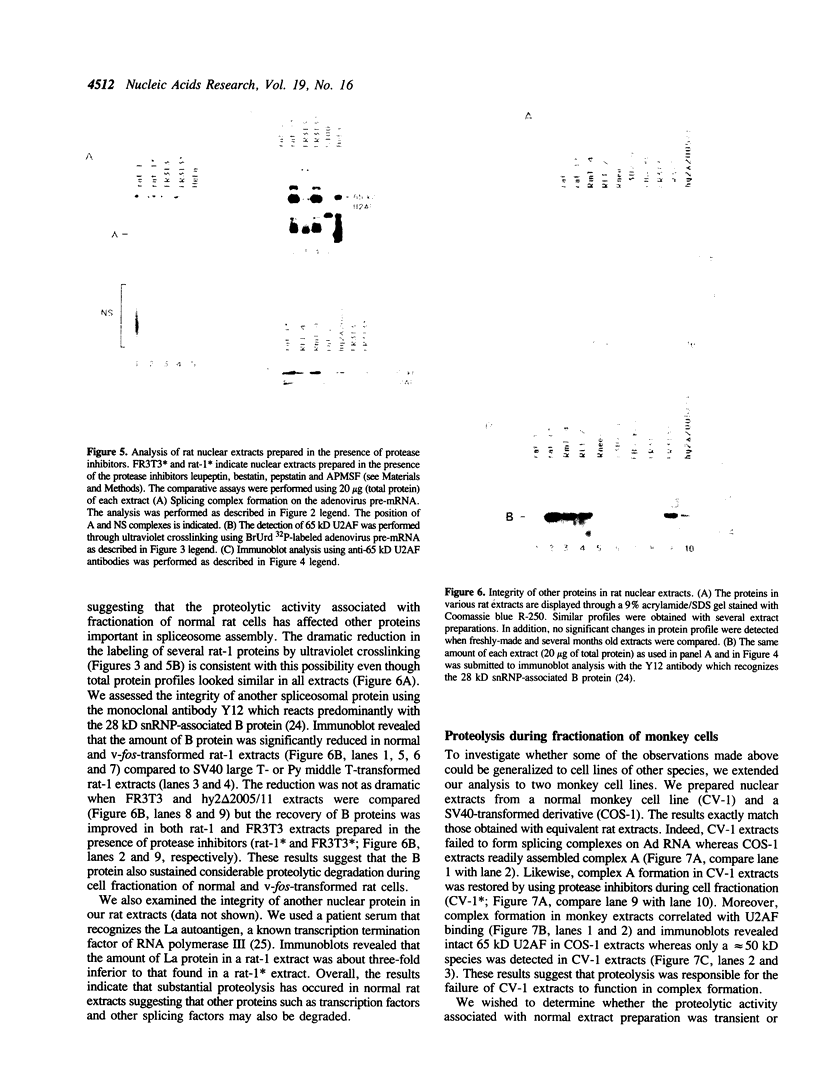
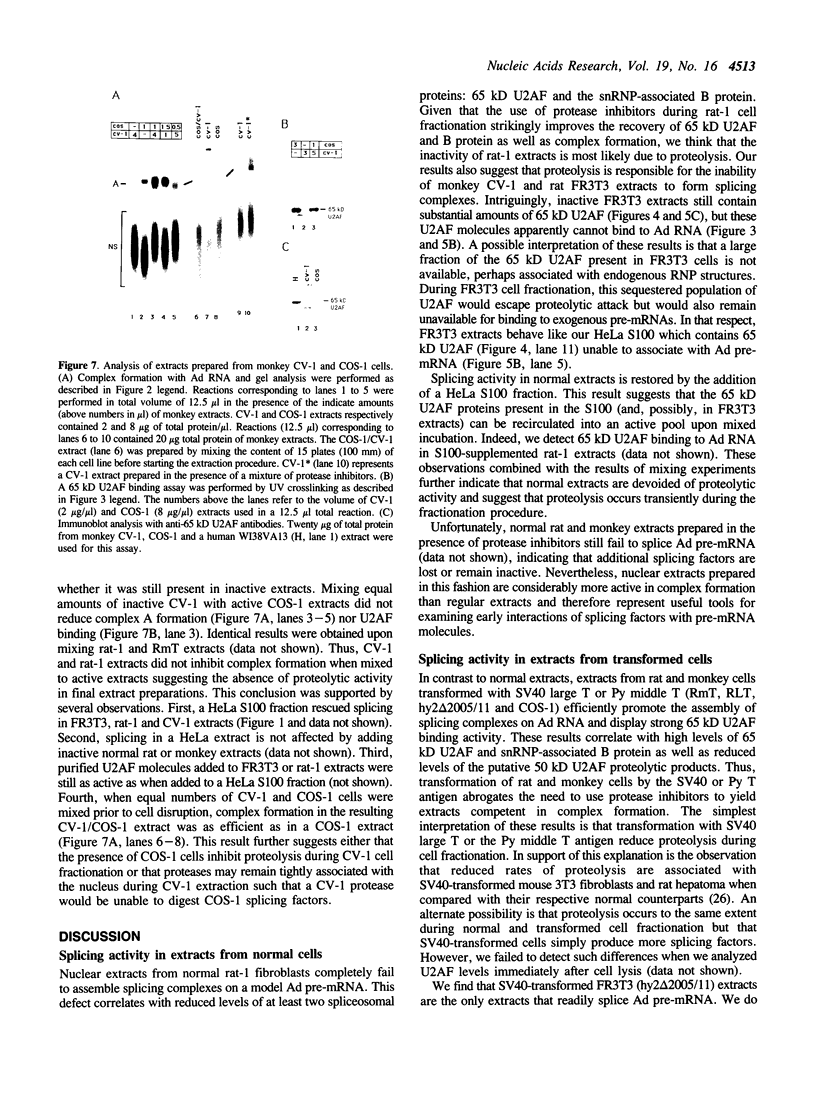
We have investigated the ability of various rat and monkey cell lines to yield nuclear extracts that would allow splicing of a model adenovirus pre-mRNA substrate. Extracts from normal FR3T3, rat-1 and CV-1 fibroblasts were unable to assemble splicing complexes and displayed a dramatic reduction in the binding activity of the splicing factor 65 kD U2AF. These results correlated with reduced levels of 65 kD U2AF and the snRNP-associated B protein. When a battery of protease inhibitors was used during cell fractionation, increased levels of 65 kD U2AF and B proteins were detected. Most importantly, U2AF binding and complex formation were dramatically improved in FR3T3, rat-1 and CV-1 extracts. Interestingly, transformation of rat and monkey cells with the SV40 large T antigen yielded extracts active in complex formation. Similar extracts were generated following transformation of rat-1 cells with the Py middle T antigen but not with the v-fos oncogene. Only SV40-transformed FR3T3 extracts displayed splicing activity. Our results indicate that proteolysis is a major obstacle encountered during the preparation of active extracts from normal rat and monkey cells and suggest that cells transformed with T antigens manifest reduced proteolysis during fractionation.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Black D. L., Chabot B., Steitz J. A. U2 as well as U1 small nuclear ribonucleoproteins are involved in premessenger RNA splicing. Cell. 1985 Oct;42(3):737–750. doi: 10.1016/0092-8674(85)90270-3. [DOI] [PubMed] [Google Scholar]
- Bouchard L., Vass-Marengo J., Bastin M. Expression of the malignant phenotype in rat fibroblasts transfected with the polyomavirus transforming genes. Virology. 1986 Nov;155(1):1–12. doi: 10.1016/0042-6822(86)90162-5. [DOI] [PubMed] [Google Scholar]
- Bovenberg R. A., Adema G. J., Jansz H. S., Baas P. D. Model for tissue specific Calcitonin/CGRP-I RNA processing from in vitro experiments. Nucleic Acids Res. 1988 Aug 25;16(16):7867–7883. doi: 10.1093/nar/16.16.7867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chabot B., Steitz J. A. Multiple interactions between the splicing substrate and small nuclear ribonucleoproteins in spliceosomes. Mol Cell Biol. 1987 Jan;7(1):281–293. doi: 10.1128/mcb.7.1.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curran T., Peters G., Van Beveren C., Teich N. M., Verma I. M. FBJ murine osteosarcoma virus: identification and molecular cloning of biologically active proviral DNA. J Virol. 1982 Nov;44(2):674–682. doi: 10.1128/jvi.44.2.674-682.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dignam J. D., Lebovitz R. M., Roeder R. G. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983 Mar 11;11(5):1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelinas C., Masse S., Bastin M. mlt Mutation in the polyomavirus genome impairing a function of the middle T protein. J Virol. 1984 Jul;51(1):242–246. doi: 10.1128/jvi.51.1.242-246.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldenberg C. J., Hauser S. D. Accurate and efficient in vitro splicing of purified precursor RNAs specified by early region 2 of the adenovirus 2 genome. Nucleic Acids Res. 1983 Mar 11;11(5):1337–1348. doi: 10.1093/nar/11.5.1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottlieb E., Steitz J. A. Function of the mammalian La protein: evidence for its action in transcription termination by RNA polymerase III. EMBO J. 1989 Mar;8(3):851–861. doi: 10.1002/j.1460-2075.1989.tb03446.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunn J. M., Clark M. G., Knowles S. E., Hopgood M. F., Ballard F. J. Reduced rates of proteolysis in transformed cells. Nature. 1977 Mar 3;266(5597):58–60. doi: 10.1038/266058a0. [DOI] [PubMed] [Google Scholar]
- Jolicoeur P., Rassart E., Massé G., Paquette Y. The specificity of the disease induced by defective murine retroviruses containing abl, fos, or Ha-ras is usually not determined by their LTR. Virology. 1991 Feb;180(2):831–836. doi: 10.1016/0042-6822(91)90102-h. [DOI] [PubMed] [Google Scholar]
- Kedes D. H., Steitz J. A. Accurate 5' splice-site selection in mouse kappa immunoglobulin light chain premessenger RNAs is not cell-type-specific. Proc Natl Acad Sci U S A. 1987 Nov;84(22):7928–7932. doi: 10.1073/pnas.84.22.7928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konarska M. M., Sharp P. A. Electrophoretic separation of complexes involved in the splicing of precursors to mRNAs. Cell. 1986 Sep 12;46(6):845–855. doi: 10.1016/0092-8674(86)90066-8. [DOI] [PubMed] [Google Scholar]
- Krainer A. R., Maniatis T., Ruskin B., Green M. R. Normal and mutant human beta-globin pre-mRNAs are faithfully and efficiently spliced in vitro. Cell. 1984 Apr;36(4):993–1005. doi: 10.1016/0092-8674(84)90049-7. [DOI] [PubMed] [Google Scholar]
- Krämer A. Presplicing complex formation requires two proteins and U2 snRNP. Genes Dev. 1988 Sep;2(9):1155–1167. doi: 10.1101/gad.2.9.1155. [DOI] [PubMed] [Google Scholar]
- Lee K. A., Bindereif A., Green M. R. A small-scale procedure for preparation of nuclear extracts that support efficient transcription and pre-mRNA splicing. Gene Anal Tech. 1988 Mar-Apr;5(2):22–31. doi: 10.1016/0735-0651(88)90023-4. [DOI] [PubMed] [Google Scholar]
- Mimori T., Hinterberger M., Pettersson I., Steitz J. A. Autoantibodies to the U2 small nuclear ribonucleoprotein in a patient with scleroderma-polymyositis overlap syndrome. J Biol Chem. 1984 Jan 10;259(1):560–565. [PubMed] [Google Scholar]
- Noble J. C., Pan Z. Q., Prives C., Manley J. L. Splicing of SV40 early pre-mRNA to large T and small t mRNAs utilizes different patterns of lariat branch sites. Cell. 1987 Jul 17;50(2):227–236. doi: 10.1016/0092-8674(87)90218-2. [DOI] [PubMed] [Google Scholar]
- Ruskin B., Zamore P. D., Green M. R. A factor, U2AF, is required for U2 snRNP binding and splicing complex assembly. Cell. 1988 Jan 29;52(2):207–219. doi: 10.1016/0092-8674(88)90509-0. [DOI] [PubMed] [Google Scholar]
- Solnick D. Alternative splicing caused by RNA secondary structure. Cell. 1985 Dec;43(3 Pt 2):667–676. doi: 10.1016/0092-8674(85)90239-9. [DOI] [PubMed] [Google Scholar]
- St-Onge L., Bouchard L., Laurent S., Bastin M. Intrachromosomal recombination mediated by papovavirus large T antigens. J Virol. 1990 Jun;64(6):2958–2966. doi: 10.1128/jvi.64.6.2958-2966.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber F., de Villiers J., Schaffner W. An SV40 "enhancer trap" incorporates exogenous enhancers or generates enhancers from its own sequences. Cell. 1984 Apr;36(4):983–992. doi: 10.1016/0092-8674(84)90048-5. [DOI] [PubMed] [Google Scholar]
- Zamore P. D., Green M. R. Biochemical characterization of U2 snRNP auxiliary factor: an essential pre-mRNA splicing factor with a novel intranuclear distribution. EMBO J. 1991 Jan;10(1):207–214. doi: 10.1002/j.1460-2075.1991.tb07937.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamore P. D., Green M. R. Identification, purification, and biochemical characterization of U2 small nuclear ribonucleoprotein auxiliary factor. Proc Natl Acad Sci U S A. 1989 Dec;86(23):9243–9247. doi: 10.1073/pnas.86.23.9243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarbl H., Latreille J., Jolicoeur P. Revertants of v-fos-transformed fibroblasts have mutations in cellular genes essential for transformation by other oncogenes. Cell. 1987 Nov 6;51(3):357–369. doi: 10.1016/0092-8674(87)90632-5. [DOI] [PubMed] [Google Scholar]