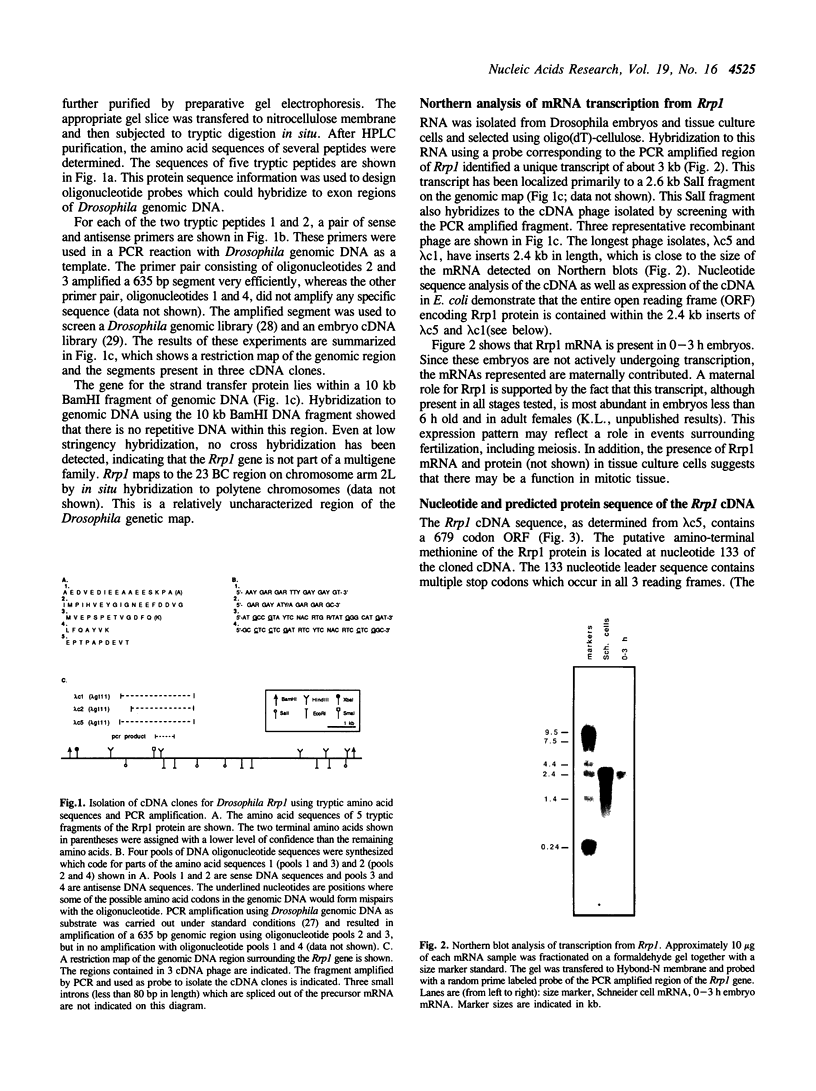
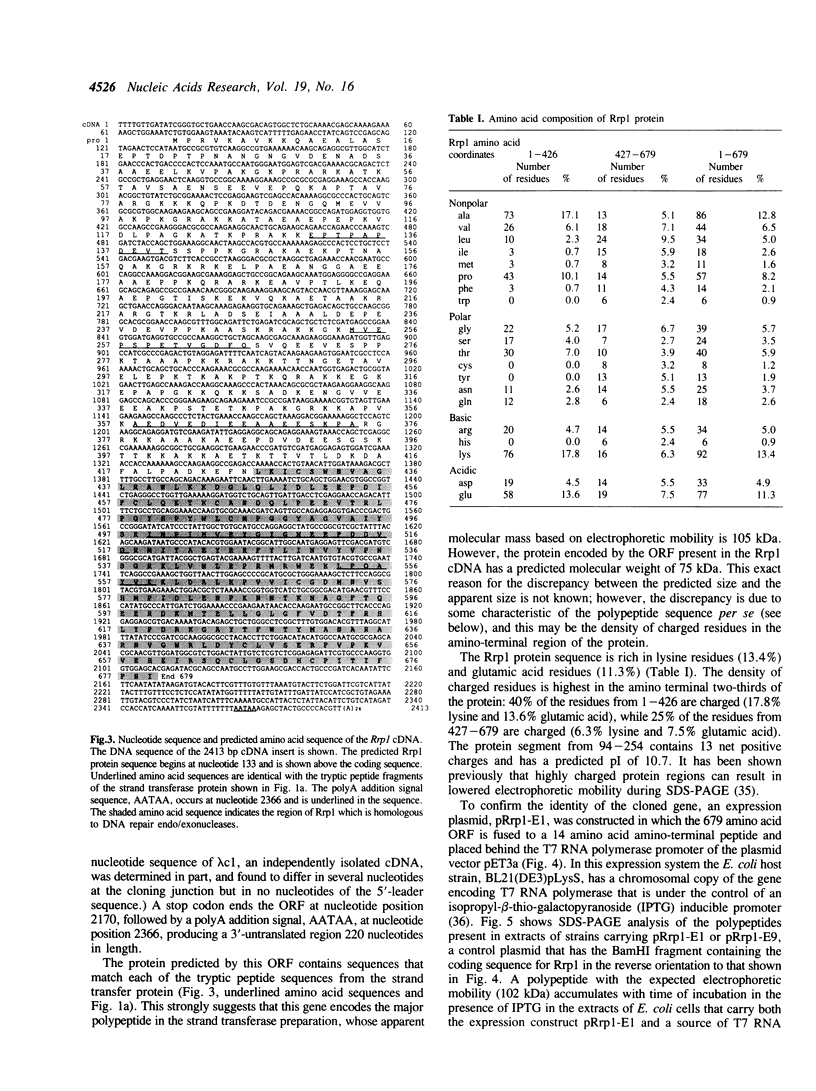
Abstract
We previously reported the purification of a protein from Drosophila embryo extracts that carries out the strand transfer step in homologous recombination (Lowenhaupt, K., Sander, M., Hauser, C. and A. Rich, 1989, J. Biol. Chem. 264, 20568). We report here the isolation of the gene encoding this protein. Partial amino acid sequence from a tryptic digest of gel purified strand transfer protein was used to design a pair of degenerate oligonucleotide primers which amplified a 635 bp region of Drosophila genomic DNA. Recombinant bacteriophage were isolated from genomic and embryo cDNA libraries by screening with the amplified DNA fragment. These bacteriophage clones identify a single copy gene that expresses a single mRNA transcript in early embryos and in embryo-derived tissue culture cells. The cDNA nucleotide sequence contains an open reading frame of 679 amino acids within which are found 5 tryptic peptides from the strand transfer protein. Expression of this cDNA in E. coli produces a polypeptide with the same electrophoretic mobility as the purified protein. The deduced protein sequence has two distinct regions. The first 427 residues are basic, rich in glutamic acid and lysine residues and unrelated to known proteins. The carboxy-terminal 252 residues are average in amino acid composition and are homologous to the DNA repair proteins, Escherichia coli exonuclease III and Streptococcus pneumoniae exonuclease A. This protein, which we name Rrp1 (Recombination Repair Protein 1), may facilitate recombinational repair of DNA damage.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aebersold R. H., Leavitt J., Saavedra R. A., Hood L. E., Kent S. B. Internal amino acid sequence analysis of proteins separated by one- or two-dimensional gel electrophoresis after in situ protease digestion on nitrocellulose. Proc Natl Acad Sci U S A. 1987 Oct;84(20):6970–6974. doi: 10.1073/pnas.84.20.6970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akaboshi E., Howard-Flanders P. Proteins induced by DNA-damaging agents in cultured Drosophila cells. Mutat Res. 1989 Sep;227(1):1–6. doi: 10.1016/0165-7992(89)90059-6. [DOI] [PubMed] [Google Scholar]
- Alani E., Padmore R., Kleckner N. Analysis of wild-type and rad50 mutants of yeast suggests an intimate relationship between meiotic chromosome synapsis and recombination. Cell. 1990 May 4;61(3):419–436. doi: 10.1016/0092-8674(90)90524-i. [DOI] [PubMed] [Google Scholar]
- Ayme A., Tissières A. Locus 67B of Drosophila melanogaster contains seven, not four, closely related heat shock genes. EMBO J. 1985 Nov;4(11):2949–2954. doi: 10.1002/j.1460-2075.1985.tb04028.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birnboim H. C. A rapid alkaline extraction method for the isolation of plasmid DNA. Methods Enzymol. 1983;100:243–255. doi: 10.1016/0076-6879(83)00059-2. [DOI] [PubMed] [Google Scholar]
- Blin N., Stafford D. W. A general method for isolation of high molecular weight DNA from eukaryotes. Nucleic Acids Res. 1976 Sep;3(9):2303–2308. doi: 10.1093/nar/3.9.2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark A. J. Recombination deficient mutants of E. coli and other bacteria. Annu Rev Genet. 1973;7:67–86. doi: 10.1146/annurev.ge.07.120173.000435. [DOI] [PubMed] [Google Scholar]
- Demple B., Halbrook J., Linn S. Escherichia coli xth mutants are hypersensitive to hydrogen peroxide. J Bacteriol. 1983 Feb;153(2):1079–1082. doi: 10.1128/jb.153.2.1079-1082.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devereux J., Haeberli P., Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen A., Camerini-Otero R. D. A recombinase from Drosophila melanogaster embryos. Proc Natl Acad Sci U S A. 1988 Oct;85(20):7481–7485. doi: 10.1073/pnas.85.20.7481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engebrecht J., Hirsch J., Roeder G. S. Meiotic gene conversion and crossing over: their relationship to each other and to chromosome synapsis and segregation. Cell. 1990 Sep 7;62(5):927–937. doi: 10.1016/0092-8674(90)90267-i. [DOI] [PubMed] [Google Scholar]
- Esposito M. S., Brown J. T. Conditional hyporecombination mutants of three REC genes of Saccharomyces cerevisiae. Curr Genet. 1990 Jan;17(1):7–12. doi: 10.1007/BF00313242. [DOI] [PubMed] [Google Scholar]
- Feinberg A. P., Vogelstein B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1983 Jul 1;132(1):6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- Fornace A. J., Jr, Alamo I., Jr, Hollander M. C. DNA damage-inducible transcripts in mammalian cells. Proc Natl Acad Sci U S A. 1988 Dec;85(23):8800–8804. doi: 10.1073/pnas.85.23.8800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganea D., Moore P., Chekuri L., Kucherlapati R. Characterization of an ATP-dependent DNA strand transferase from human cells. Mol Cell Biol. 1987 Sep;7(9):3124–3130. doi: 10.1128/mcb.7.9.3124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holliday R., Halliwell R. E., Evans M. W., Rowell V. Genetic characterization of rec-1, a mutant of Ustilago maydis defective in repair and recombination. Genet Res. 1976 Jun;27(3):413–453. doi: 10.1017/s0016672300016621. [DOI] [PubMed] [Google Scholar]
- Hotta Y., Tabata S., Bouchard R. A., Piñon R., Stern H. General recombination mechanisms in extracts of meiotic cells. Chromosoma. 1985;93(2):140–151. doi: 10.1007/BF00293161. [DOI] [PubMed] [Google Scholar]
- Hsieh P., Meyn M. S., Camerini-Otero R. D. Partial purification and characterization of a recombinase from human cells. Cell. 1986 Mar 28;44(6):885–894. doi: 10.1016/0092-8674(86)90011-5. [DOI] [PubMed] [Google Scholar]
- Kelley M. R., Venugopal S., Harless J., Deutsch W. A. Antibody to a human DNA repair protein allows for cloning of a Drosophila cDNA that encodes an apurinic endonuclease. Mol Cell Biol. 1989 Mar;9(3):965–973. doi: 10.1128/mcb.9.3.965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolodner R., Evans D. H., Morrison P. T. Purification and characterization of an activity from Saccharomyces cerevisiae that catalyzes homologous pairing and strand exchange. Proc Natl Acad Sci U S A. 1987 Aug;84(16):5560–5564. doi: 10.1073/pnas.84.16.5560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lowenhaupt K., Sander M., Hauser C., Rich A. Drosophila melanogaster strand transferase. A protein that forms heteroduplex DNA in the absence of both ATP and single-strand DNA binding protein. J Biol Chem. 1989 Dec 5;264(34):20568–20575. [PubMed] [Google Scholar]
- Maniatis T., Hardison R. C., Lacy E., Lauer J., O'Connell C., Quon D., Sim G. K., Efstratiadis A. The isolation of structural genes from libraries of eucaryotic DNA. Cell. 1978 Oct;15(2):687–701. doi: 10.1016/0092-8674(78)90036-3. [DOI] [PubMed] [Google Scholar]
- Moore S. P., Fishel R. Purification and characterization of a protein from human cells which promotes homologous pairing of DNA. J Biol Chem. 1990 Jul 5;265(19):11108–11117. [PubMed] [Google Scholar]
- Nolan J. M., Lee M. P., Wyckoff E., Hsieh T. S. Isolation and characterization of the gene encoding Drosophila DNA topoisomerase II. Proc Natl Acad Sci U S A. 1986 Jun;83(11):3664–3668. doi: 10.1073/pnas.83.11.3664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardue M. L., Lowenhaupt K., Rich A., Nordheim A. (dC-dA)n.(dG-dT)n sequences have evolutionarily conserved chromosomal locations in Drosophila with implications for roles in chromosome structure and function. EMBO J. 1987 Jun;6(6):1781–1789. doi: 10.1002/j.1460-2075.1987.tb02431.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prakash S., Prakash L., Burke W., Montelone B. A. Effects of the RAD52 Gene on Recombination in SACCHAROMYCES CEREVISIAE. Genetics. 1980 Jan;94(1):31–50. doi: 10.1093/genetics/94.1.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puyet A., Greenberg B., Lacks S. A. The exoA gene of Streptococcus pneumoniae and its product, a DNA exonuclease with apurinic endonuclease activity. J Bacteriol. 1989 May;171(5):2278–2286. doi: 10.1128/jb.171.5.2278-2286.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Query C. C., Bentley R. C., Keene J. D. A common RNA recognition motif identified within a defined U1 RNA binding domain of the 70K U1 snRNP protein. Cell. 1989 Apr 7;57(1):89–101. doi: 10.1016/0092-8674(89)90175-x. [DOI] [PubMed] [Google Scholar]
- Rogers S. G., Weiss B. Exonuclease III of Escherichia coli K-12, an AP endonuclease. Methods Enzymol. 1980;65(1):201–211. doi: 10.1016/s0076-6879(80)65028-9. [DOI] [PubMed] [Google Scholar]
- Sammartano L. J., Tuveson R. W. Escherichia coli xthA mutants are sensitive to inactivation by broad-spectrum near-UV (300- to 400-nm) radiation. J Bacteriol. 1983 Nov;156(2):904–906. doi: 10.1128/jb.156.2.904-906.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebastian J., Kraus B., Sancar G. B. Expression of the yeast PHR1 gene is induced by DNA-damaging agents. Mol Cell Biol. 1990 Sep;10(9):4630–4637. doi: 10.1128/mcb.10.9.4630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith G. R. Mechanism and control of homologous recombination in Escherichia coli. Annu Rev Genet. 1987;21:179–201. doi: 10.1146/annurev.ge.21.120187.001143. [DOI] [PubMed] [Google Scholar]
- Spiering A. L., Deutsch W. A. Drosophila apurinic/apyrimidinic DNA endonucleases. Characterization of mechanism of action and demonstration of a novel type of enzyme activity. J Biol Chem. 1986 Mar 5;261(7):3222–3228. [PubMed] [Google Scholar]
- Studier F. W., Rosenberg A. H., Dunn J. J., Dubendorff J. W. Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol. 1990;185:60–89. doi: 10.1016/0076-6879(90)85008-c. [DOI] [PubMed] [Google Scholar]
- Sugino A., Nitiss J., Resnick M. A. ATP-independent DNA strand transfer catalyzed by protein(s) from meiotic cells of the yeast Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1988 Jun;85(11):3683–3687. doi: 10.1073/pnas.85.11.3683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor A. F., Weiss B. Role of exonuclease III in the base excision repair of uracil-containing DNA. J Bacteriol. 1982 Jul;151(1):351–357. doi: 10.1128/jb.151.1.351-357.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker G. C. Inducible DNA repair systems. Annu Rev Biochem. 1985;54:425–457. doi: 10.1146/annurev.bi.54.070185.002233. [DOI] [PubMed] [Google Scholar]
- el-Hajj H. H., Zhang H., Weiss B. Lethality of a dut (deoxyuridine triphosphatase) mutation in Escherichia coli. J Bacteriol. 1988 Mar;170(3):1069–1075. doi: 10.1128/jb.170.3.1069-1075.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]