Abstract
Conditional temperature-sensitive (ts) mutations are valuable reagents for studying essential genes in the yeast Saccharomyces cerevisiae. We constructed 787 ts strains, covering 497 (~45%) of the 1,101 essential yeast genes, with ~30% of the genes represented by multiple alleles. All of the alleles are integrated into their native genomic locus in the S288C common reference strain and are linked to a kanMX selectable marker, allowing further genetic manipulation by synthetic genetic array (SGA)–based, high-throughput methods. We show two such manipulations: barcoding of 440 strains, which enables chemical-genetic suppression analysis, and the construction of arrays of strains carrying different fluorescent markers of subcellular structure, which enables quantitative analysis of phenotypes using high-content screening. Quantitative analysis of a GFP-tubulin marker identified roles for cohesin and condensin genes in spindle disassembly. This mutant collection should facilitate a wide range of systematic studies aimed at understanding the functions of essential genes.
The budding yeast S. cerevisiae is one of the most well-characterized model organisms for systematic analysis of fundamental eukaryotic processes. Approximately 19% of S. cerevisiae genes are considered essential, because haploid spores carrying a deletion allele of these genes fail to germinate and form colonies under standard laboratory conditions1. Comparative analysis revealed that gene dispensability is highly conserved (~83%) between S. cerevisiae and the distantly related yeast species, Schizosaccharomyces pombe, supporting the presence of strong selective pressure to retain essential gene activity across large evolutionary distances2. Indeed, essential genes tend to be more highly conserved from yeast to humans when compared to nonessential genes3. The set of essential yeast genes spans diverse biological processes and although the primary role of most essential yeast genes has been characterized, the full breadth of function associated with essential genes has not been examined, owing, at least in part, to the lack of adequate genetic reagents for their conditional and systematic perturbation. Here we present the construction and characterization of an extensive set of conditional ts alleles of S. cerevisiae essential genes. Other types of mutant alleles that perturb essential gene functions include cold-sensitive4, temperature-inducible degron5, tetracycline-regulatable promoter-replacement6 and decreased abundance by mRNA perturbation alleles7. However, ts alleles often provide the simplest and most finely tuned control of gene function, enabling permissive, semi-permissive and restrictive conditions to be established easily. At the permissive temperature, the phenotype of a ts mutant resembles that of the wild-type strain, whereas at the restrictive temperature, the activity of the essential gene is substantially reduced or abolished, resulting in a slow-growth or lethal phenotype. We collected hundreds of previously characterized ts alleles as yeast mutants or DNA clones from the yeast community and then amplified by PCR the ts alleles and integrated them at their native locus in the reference (S288c) genetic background, generating a set of 787 ts mutants spanning 497 different essential genes that are isogenic except for the ts allele we introduced. Our new set expands upon a nonoverlapping set of 250 ts strains constructed recently8, which combined cover ~65% of S. cerevisiae essential genes.
In this study, we characterize the ts mutant array as a resource for exploring the pleiotropic roles of highly conserved essential pathways in yeast using high-resolution growth profiling, chemical-genetic suppression and high content screening (HCS) experiments. The HCS experiments involved quantitative single-cell image analysis of fluorescent markers for specific pathways and structures in hundreds of different ts mutants. Our results highlight a role for two essential protein complexes, cohesin and condensin, in mitotic spindle disassembly.
RESULTS
Construction and confirmation of ts yeast strains
We collected hundreds of previously characterized yeast strains and DNA constructs carrying ts alleles and integrated the conditional alleles into the S288c reference background9. In total, we constructed a set of 787 ts strains representing 497 different essential genes or ~45% of the essential gene set (Supplementary Table 1), which covers a representative proportion of Gene Ontology (GO) molecular functions and biological processes for the complete essential gene set. Each ts allele was integrated into its native locus such that it was linked to a kanamycin-resistant cassette, kanMX, which was targeted to the 3′ UTR region of the gene and placed 0–300 bp downstream of the stop codon (Supplementary Table 2). We designed the integration such that the intervening sequences were not altered to avoid perturbing the expression of neighboring genes (Supplementary Fig. 1). Two PCR reactions were carried out to confirm that: (i) the kanMX cassette was linked to the expected target gene, and (ii) the kanMX cassette integrated at the expected genomic location (Supplementary Fig. 1). Correct integration was confirmed for all of 787 ts alleles using this PCR-based strategy (Supplementary Table 1).
The ts strain collection was further validated by plasmid complementation. Individual plasmids encoding wild-type genes corresponding to 786 of 787 alleles in the ts collection were taken from either the low-copy, molecular barcoded yeast open reading frame (MoBY-ORF) library10 or the high-copy yeast genomic tiling collection11 (neither library contained a plasmid encoding CDC39), transformed into the corresponding ts mutant strain and subsequently grown at permissive and restrictive temperatures. We observed plasmid-dependent rescue for 782 of 786 strains representing 494 essential genes in our collection, confirming that the expected allele was responsible for the growth phenotype (Supplementary Table 1). In summary, 782 of 787 ts strains representing 494 essential genes were confirmed by both PCR and plasmid complementation.
Profiling the temperature sensitivity of ts alleles
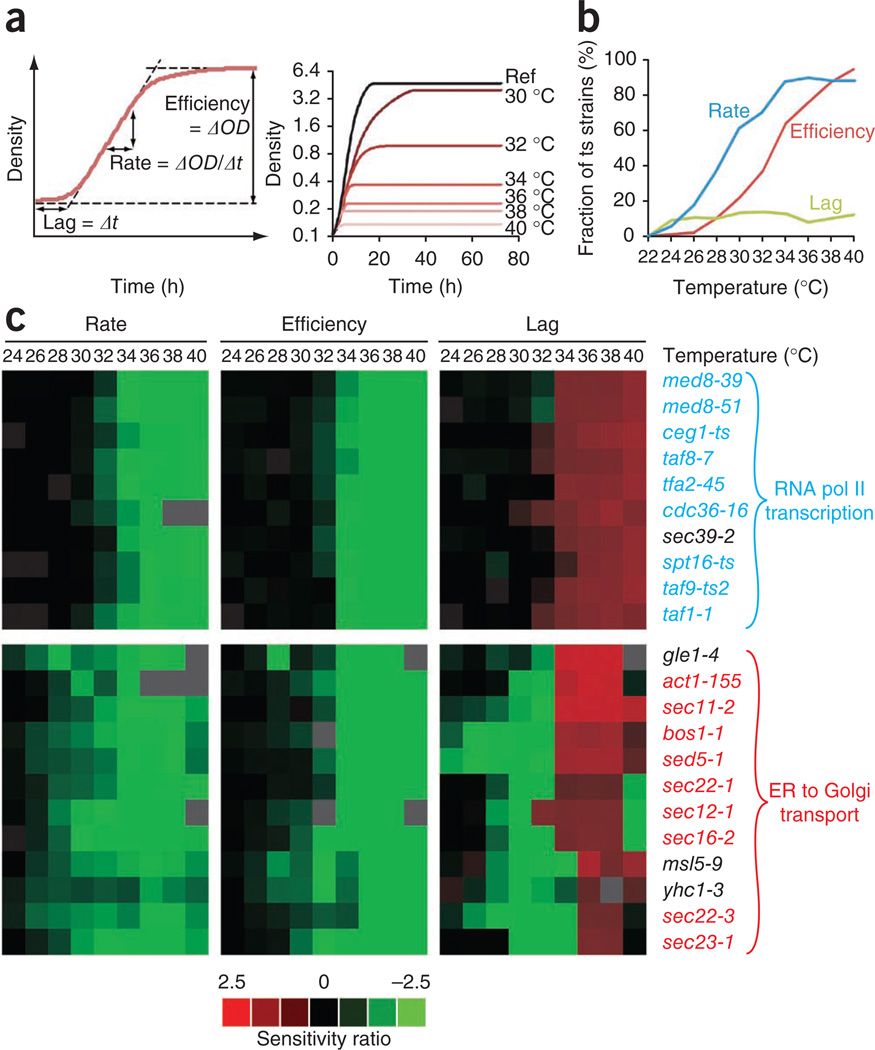
We next applied high-resolution growth profiling to characterize the temperature sensitivity of mutants in our strain collection. Lag time, growth rate and growth efficiency were quantified across a range of temperatures to generate a growth profile for each ts mutant (Fig. 1a and Supplementary Table 3). The reference strain BY4741 reached its maximal growth rate at 32–36 °C, whereas growth efficiency and growth lag continuously decreased from a maximum at 24 °C (Supplementary Fig. 2a). Individual ts-strain growth profiles were normalized against a reference wild-type profile to provide a measure of relative growth defects at each temperature. As expected, the frequency distribution of relative growth rates reflected the tendency toward exaggerated growth defects at higher temperatures (Supplementary Fig. 2b). In addition, after normalizing for defects at 22 °C, we found that the fraction of ts strains with clear temperature-dependent growth defects expanded with increasing temperature, from 24 °C to 34 °C (Fig. 1b).
Figure 1.
Profiling the temperature sensitivity of ts strains. (a) Left panel; general interpretation of high-resolution liquid growth profiles of strains harboring ts alleles. Growth rate (population doubling time) was determined by measuring the slope of the exponential phase of the growth curve. Lag was given by the intercept of the initial density and the slope, and growth efficiency was calculated as the total change in density for cells having reached stationary phase. Right panel; sample growth curves of the strains with the act1-105 ts allele, obtained by micro-cultivation over a temperature range. (b) The fraction of all ts strains that exhibit significant (P < 0.001) temperature sensitive defects in growth rate (blue), lag (green) and/or efficiency (red) phases compared to wild type (WT) at the indicated temperatures. (c) Uncentered hierarchical clustering of ts allele growth profiles over all three growth variables. Profile similarity was measured using a Pearson similarity metric and average linkage mapping. The ts-strain sensitivity ratio is expressed as log2(WT/ts strain) at temperature X − log2(WT/ts strains) at 22 °C. ER, endoplasmic reticulum.
Notably, comparison of average pair-wise Pearson correlation coefficients revealed that ts alleles corresponding to the same gene tended to show more similar temperature profiles than alleles of different essential genes (P = 10−23) (Supplementary Fig. 2c), suggesting that the ts-strain growth profiles may reflect essential gene function. Moreover, ts alleles of essential genes that participate in the same cellular process also tend to show more similar temperature profiles than essential gene alleles that do not share a similar function (comparison of average pair-wise Pearson correlation coefficients, excluding within-gene comparisons, P = 10−83), thus further supporting a relationship between ts-strain growth profiles and protein function. Consistent with this trend, hierarchical clustering of liquid growth profiles revealed that ts mutations in functionally related genes showed similar growth behavior in response to temperature (Fig. 1c and Supplementary Fig. 2d). Thus, the ts-strain growth profiles may represent distinct phenotypic fingerprints that reveal physiological defects associated with different essential cellular functions. However, we noted that whereas a general relationship between essential gene function and the ts-strain growth profile appears to exist, this trend may not hold true for all genes, especially those encoding multifunctional protein products involved in diverse cellular processes.
Chemical-genetic suppression analysis with a barcoded ts library
The ts mutant collection provides a reagent for performing systematic chemical-genetic suppression analysis. In this analysis, the ts mutants are grown in the presence of small-molecule compounds at both permissive and nonpermissive temperatures. Strains that grow at the nonpermissive temperature contain mutations that are suppressed by a particular compound. This contrasts with chemical-genetic sensitivity analysis that is often performed with the yeast deletion mutant collection, in which mutants are scored for growth sensitivity to a specific compound12,13. Notably, the use of suppression, rather than growth sensitivity, as a phenotype, enables screens for compounds that are not necessarily antifungals.
High-throughput chemical genetics of the deletion collection is possible because each strain contains two oligonucleotide molecular barcodes flanked by common primer sites. This enables mutants to be pooled, grown competitively in a single culture with a chemical and their relative fitnesses quantitatively estimated by measuring the abundance of each barcode in the culture using a microarray or sequencing14–16. To facilitate competitive growth assays using pools of ts mutant alleles, we applied the barcoder approach17 to introduce molecular barcodes into a subset of ~400 ts strains such that the HO allele is replaced by a drug-resistance marker flanked by two unique oligonucleotides.
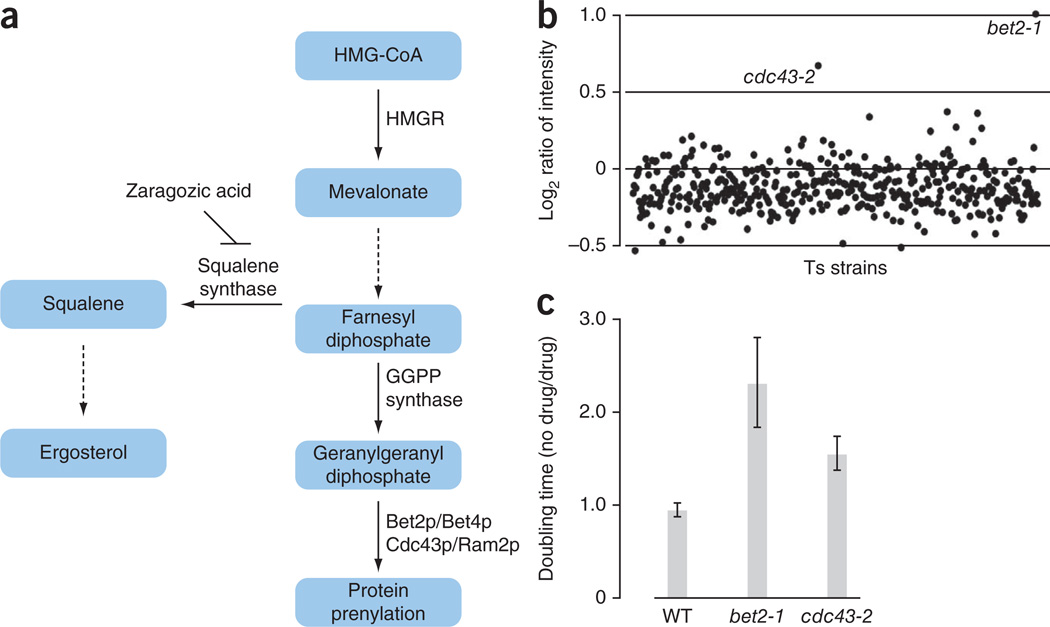
As a proof-of-principle analysis, we screened for mutations suppressed by zaragozic acid A (Squalestatin), an inhibitor of mammalian and fungal squalene synthase18,19. This is the first committed enzyme in sterol synthesis (Fig. 2a), and has received attention as a potential target for lipid-lowering agents20. We pooled the barcoded subset of ts strains, grew them at 36.5 °C in the presence or absence of zaragozic acid A and then quantified the molecular barcodes using barcode microarray analysis. Two strains, bet2-1 and cdc43-2, were overrepresented in the pooled population exposed to zaragozic acid A, indicating that this compound suppressed ts growth defects associated with these mutant alleles (Fig. 2b). Suppression of bet2-1 and cdc43-2 growth defects was also confirmed by liquid growth assays and spot dilution assays on solid media (Fig. 2c and data not shown). BET2 encodes the catalytic subunit of the type II geranylgeranyl transferase and CDC43 encodes the beta subunit of geranylgeranyltransferase type I involved in prenylation (Fig. 2a). Previous studies have shown that when squalene synthase is inhibited by zaragozic acid A, intracellular levels of squalene and post-squalene metabolites are decreased, whereas the levels of other metabolites synthesized from the squalene precursor are increased, such as farnesyl diphosphate (FPP)18 and farnesoic acid-derived metabolites19. Thus, zaragozic acid A may suppress the bet2-1 and cdc43-2 ts phenotype at restrictive temperature by increasing levels of cellular farnesyl diphosphate and geranylgeranyl diphosphate.
Figure 2.
Zaragozic acid rescues bet2-1 and cdc43-2 ts phenotype. (a) Isoprenoid pathway in S. cerevisiae. The solid lines indicate one step and the dashed lines indicate multiple synthetic steps in the ergosterol and protein prenylation pathways44–46. (b) A pool of 440 barcoded ts mutants was grown in rich (YPD) medium in the presence of 4 mM of zaragozic acid A (ZA) or DMSO at 36.5 °C. Genomic DNA was prepared from cells after five generations of growth. Molecular barcodes were amplified by PCR and hybridized to a microarray (GeneChip Genflex Tag 16K Array v2, Affymetrix). The x axis represents the 440 ts strains ordered alphabetically by systematic name. The y axis represents the log2 ratio of barcode hybridization intensity between the ZA treatment and the solvent (DMSO) treatment. Mutants with highest log2 ratios at the restrictive temperature were identified as suppressors. (c) Yeast cells were grown in YPD with or without ZA in a 96-well plate at 36.5 °C. Fitness was defined as the ratio of doubling times measured in DMSO compared to ZA treatment. Error bars represent s.d. for four independent experiments.
High-content screens to predict essential gene function
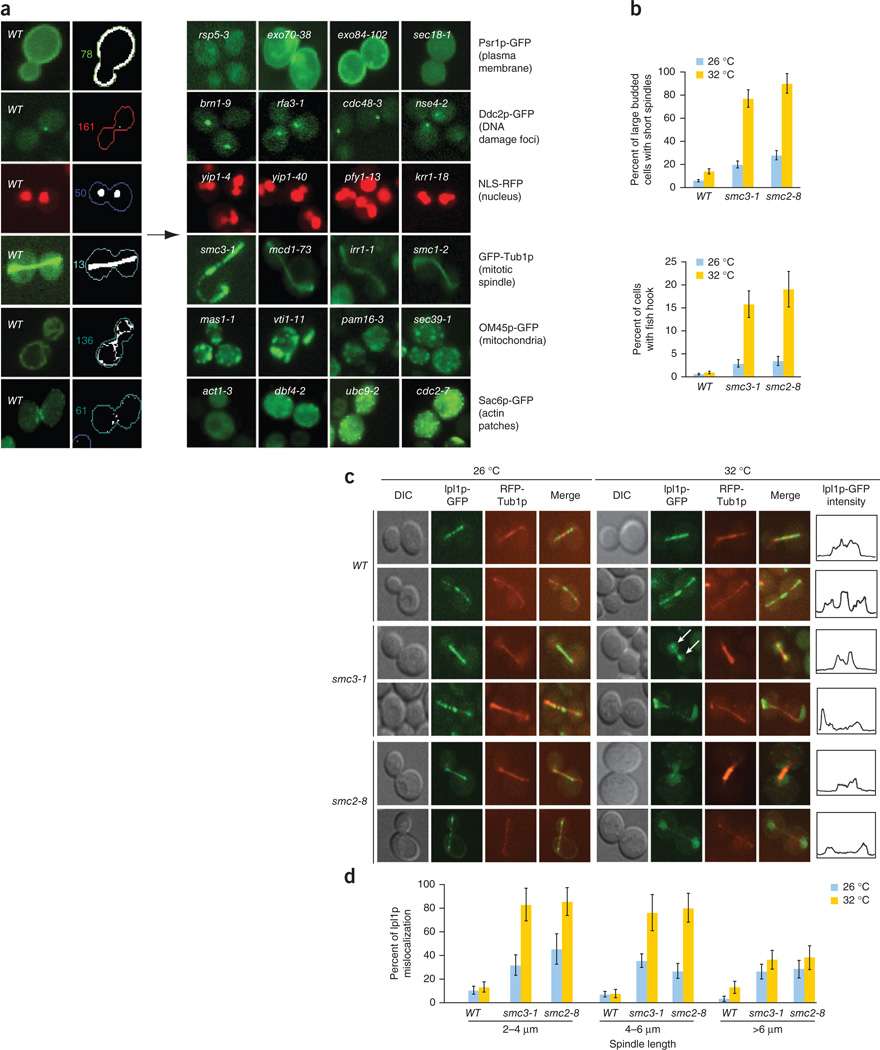
Having all of the ts alleles in an isogenic background allows the strain collection to be genetically modified using existing high-throughput strain manipulation methods, such as those developed to create double mutants for synthetic genetic array (SGA) analysis. Combining these manipulations with high-content screening enables functional discovery through the visualization and quantitative measurement of specific morphological features at a single-cell level21. We first constructed a panel of strains expressing fluorescent markers that represent six fundamental subcellular compartments or structures (Fig. 3a). Next, we crossed these reporters into an array of 762 ts strains (Supplementary Table 1) to create double mutants expressing a subcellular marker and a ts allele. Then, we imaged both the ts and the wild-type control strains at 26 °C and 32 °C. We chose 32 °C as a screening temperature because ~80% of the mutant alleles showed a growth defect at this temperature (Fig. 1b). We developed customized software21 that automates image analysis and measures several morphological features, generating a quantitative and unique cell morphology profile for each ts mutant strain. The cell morphology profile comprises features such as cell shape, budding index, organelle density and 85 reporter-specific parameters (Supplementary Table 4). Notably, we confirmed all computationally derived phenotypes by visual inspection of the corresponding images (Supplementary Table 5), generating a high confidence and systematic profile for each ts strain. The raw morphometric data measured at both permissive and restrictive temperatures are provided in Supplementary Table 6.
Figure 3.
High-content screening of the ts allele collection identifies abnormal spindle morphology associated with cohesin and condensin mutants. (a) Fluorescent reporters introduced into the ts collection. The left panels show the localization of six different GFP/RFP-tagged marker proteins expressed in wild-type cells as well as the corresponding computationally processed images. Representative mutants with defects in localization of particular markers are shown on the right. Markers included a plasma membrane marker (Psr1p-GFP), a reporter of DNA damage (Ddc2p-GFP), a nuclear marker (Mad1p-NLS-RFP), a mitotic spindle reporter (GFP-Tub1), a mitochondrial marker (OM45p-GFP) and an actin reporter (Sac6p-GFP). (b) A sub-population of cohesin and condensin mutants exhibit abnormal spindle morphology. Spindle length and morphology were quantified in large-budded wild-type cells and strains harboring a cohesin (smc3-1) or condensin (smc2-1) mutant allele grown asynchronously at permissive (26 °C) and restrictive temperatures (32 °C). The fraction of cells exhibiting either short (upper panel) or fishhook-shaped spindles (lower panel) were quantified in three independent populations; error bars represent s.d. across three independent experiments. (c) Ipl1p-GFP localization in cohesin (smc3-1) and condensin (smc2-8) mutants. Ipl1p-GFP localization to the mitotic spindle was monitored in wild-type, smc3-1 and smc2-8 mutants at permissive (26 °C) and restrictive temperatures (32 °C). Representative cells with short (top panels) and long (bottom panels) spindles are shown. White arrows indicate Ipl1p-GFP localization to the kinetochore in a cohesin mutant at the restrictive temperature. The fluorescent intensity line profile of Ipl1p-GFP for a representative cell is also shown. (d) Wild-type and mutant cells were divided into three separate groups based on spindle length (2–4 µm, 4–6 µm, >6 µm) as measured by assessment of an RFP-Tub1p marker. The percentage of cells in each spindle length group that exhibited mis-localized Ipl1p-GFP was measured at permissive and restrictive temperatures. Error bars represent the s.d. across three independent experiments.
In principle, a cell morphology profile should provide a rich phenotypic signature revealing functional relationships between genes. For example, mutants exhibiting increased DNA damage foci, as measured by Ddc2p-GFP localization, were enriched for essential genes involved in DNA replication (P = 2.1 × 10−10), prereplicative complex assembly (P = 1.4 × 10−5) and/or DNA synthesis during DNA repair (P = 2.0 × 10−5). The number of gene-specific profiles indicative of morphological defects increased at elevated temperature further validated our approach (Supplementary Fig. 3a). We therefore developed a method to predict functional relationships based on the similarity of cell morphology profiles (Supplementary Fig. 3b,c and Online Methods).
Our computational classification approach identified known functional relationships between previously characterized essential genes. For example, mutants with known roles in mitochondrial protein import, such as mas1-1 and pam16-3, exhibited defects in mitochondrial abundance whereas aberrant membrane targeting of the Psr1p-GFP reporter was observed in two exocyst mutants, exo70-38 and exo84-102 (Fig. 3a). Comparison of cell morphology profiles also uncovered gene-specific phenotypes, suggesting unanticipated functions for well-characterized essential genes. For example, components of cohesin, condensin and the chromosomal passenger complex (CPC), shared similar morphological profiles (Fig. 3a and Supplementary Fig. 3c). The products of IPL1 (Aurora B kinase), SLI15 (IN-CENP) and BIR1 (SURVIVIN) interact to form the chromosomal passenger complex (CPC) that relocalizes dynamically in dividing cells to perform key mitotic roles22–26. Specifically, we found that cohesin mutants, including smc3-1, mcd1-73, irr1-1 and smc1-2 (Fig. 3a), as well as condensin mutants, ycs4-1, smc2-8 and smc4-1 (Supplementary Table 5) were characterized by a hyperelongated spindle phenotype with a ‘fishhook’ shape, as visualized using the tubulin marker GFP-Tub1p, suggesting that, similar to the CPC complex, cohesin and condensin may also be involved in spindle disassembly.
A role for cohesin and condensin in spindle disassembly
Because the regulation of spindle disassembly is not completely understood, we decided to further examine the potential role of cohesin and condensin in this essential process. Cohesins bind the sister chromatids together during metaphase and are cleaved in a separase-dependent fashion during anaphase27,28. When cells are synchronized in the G1 phase with α-factor and then released into the cell cycle at a restrictive temperature, ts cohesin mutants activate the mitotic checkpoint and arrest as large-budded cells with short or partially elongated spindles29. However, in our screen, when asynchronous cohesin ts mutants were shifted to restrictive temperature, we found two populations of cells: (i) large budded cells with short or partially elongated spindles, as expected, and (ii) a subpopulation of large budded cells with hyperelongated fishhook spindles, which presumably represent cells exhibiting a post-checkpoint spindle disassembly defect (Fig. 3b).
The fishhook spindle phenotype we observed may result from changes in microtubule30 or CPC complex dynamics21,22. To examine this more carefully, we monitored spindle association and midzone enrichment of CPC components (Ipl1p-GFP, Sli15p-GFP and Bir1p-GFP) in a cohesin, smc3-1, and a condensin mutant, smc2-8. In wild-type cells, Ipl1p-GFP normally associates with short spindles ranging from ~1.5 to 2 µm in length, and this localization persists until the spindle disassembles, at which point Ipl1p localization is more prominent at the spindle midzone (Fig. 3c,d). Notably, in the cohesin mutants, Ipl1p-GFP, Sli15p-GFP and Bir1p-GFP associated with the kinetochore but failed to associate with the spindle until the spindle was >6 µm. In contrast, Ipl1p-GFP, Sli15p-GFP and Bir1p-GFP failed to localize to either the kinetochore or the spindle in condensin mutants (Fig. 3c,d and Supplementary Fig. 4a,d). Furthermore, the normal distribution of cohesin around the spindle, referred to as cohesin barrel, is lost in a mcm21Δ kinetochore mutant, suggesting that cohesin is required to stabilize the CPC on the spindle (Supplementary Fig. 5a)21,31.
Markers of late mitotic events enabled us to further explore the spindle defect in cohesin and condensin mutants. Specifically, we monitored localization of Ase1p, the spindle bundling protein that is reported to localize to the spindle midzone only at anaphase onset32 and Cdc14p, the mitotic exit trigger that regulates the CPC23. We found that Ase1p was strictly co-localized to the spindle midzone and the release of Cdc14p from the nucleolus was unaffected in the smc3-1 and smc2-8 mutants (Supplementary Fig. 5b,c). These observations are important because they suggest that cohesin and condensin mutants have initiated mitotic exit23,32–34 and further suggest that the fishhook spindle observed in these mutants is an anaphase-specific phenotype.
Reduced association of CPC components, Sli15p-GFP and Ipl1p-GFP, with elongated spindles was also observed in double mutants defective in cohesin or condensin components and lacking a functional spindle-assembly checkpoint (mad1Δ smc3-1 and mad1Δ smc2-8) (Supplementary Fig. 5d), indicating that CPC mislocalization observed in cohesin/condensin mutants is not due to checkpoint activation35,36. Finally, spindle midzone localization of a phosphorylation-deficient CPC mutant (Sli15p-6A-GFP), which binds to the spindle throughout mitosis23, was delayed in an smc3-1 mutant resulting in elongated fishhook spindles (Supplementary Fig. 5e). This finding is consistent with our checkpoint mutant observations because Sli15p dephosphorylation was previously shown to prevent mitotic checkpoint function during anaphase37. Taken together, our results suggest that the observed defect in spindle midzone accumulation and the fishhook phenotype associated with the cohesin and condensin complex is not dependent on activation of the Cdc14p pathway activation but rather is an anaphase-specific event.
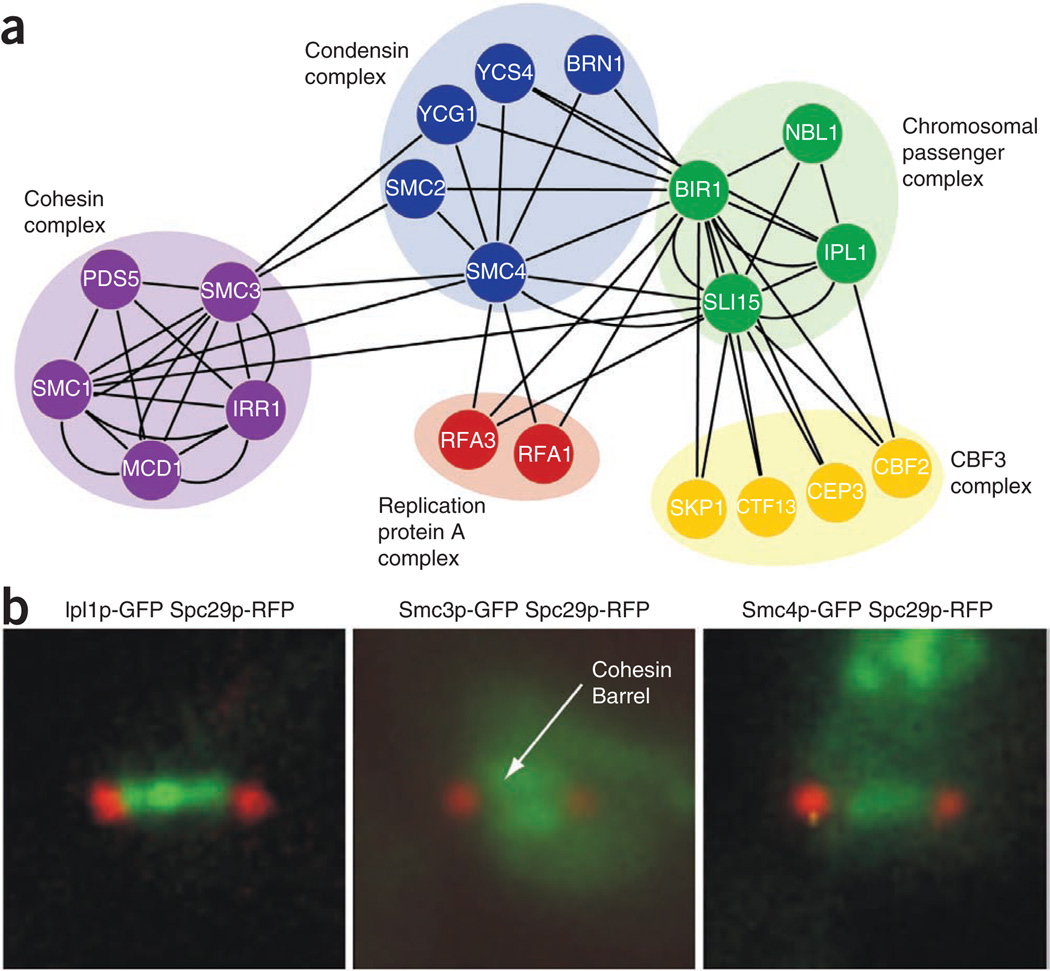
Cohesin is concentrated at the pericentromeric region of the yeast chromosomes that overlaps with the spindle midzone21,31 whereas condensin associates with the kinetochore, chromosomal arms and ribosomal DNA (rDNA)38–40. Because the CPC plays a major role in controlling spindle disassembly, we decided to test for physical interactions between CPC components, cohesin and condensin. Ipl1p-GFP, Sli15p-GFP, Bir1p-GFP, Smc3p-GFP and Smc4p-GFP were affinity purified from yeast extracts, and then liquid chromatography–tandem mass spectrometry (LC-MS/MS) analysis was carried out. These experiments recapitulated all previously reported interactions (Fig. 4a and Supplementary Table 7), and they uncovered reproducible interactions between the CPC component, Bir1p-GFP and members of the condensin complex, supporting a direct role for condensin components in stabilizing the CPC (Fig. 4a).
Figure 4.
Relationship between cohesin, condensin and chromosomal passenger complexes (CPC). (a) Physical interaction network illustrating protein-protein interactions involving cohesin, condensin and CPC complexes. Nodes represent individual proteins and edges represent physical interactions. (b) Cohesin, condensin and CPC complex localization. Ipl1p-GFP, Smc3p-GFP and Smc4p-GFP localization was assessed with respect to the spindle pole marker, Spc29p-RFP in a wild-type strain. Fluorescent micrograph images illustrate representative single cells.
To understand the spatial association of these components, we monitored the localization pattern of the CPC, cohesin and condensin complexes in wild-type cells. As reported previously31, cohesin forms a cylindrical barrel that surrounds Ipl1p (Fig. 4b and Supplementary Fig. 5f). During metaphase, Ipl1p-GFP localized to the spindle axis and was surrounded by the cohesin barrel, as identified with Smc3p-GFP (Supplementary Fig. 5f). As the cells enter anaphase, Ipl1p-GFP was largely restricted within the cohesin-enriched region (Supplementary Fig. 5f), suggesting that cohesin restricts the movement of the CPC along the spindle. The condensin component, Smc4p-GFP, displayed a localization pattern similar to that of Ipl1p-GFP along the spindle axis, distinct from the spindle pole body marker, Spc29p-RFP (Fig. 4b). Notably, whereas both cohesin and condensin were necessary for the spindle localization of Ipl1p, the localization of cohesin and condensin proteins were unaffected in an ipl1-2 mutant (Supplementary Fig. 5g), suggesting that cohesin and condensin complexes regulate the localization of the CPC and not vice versa. Thus, co-localization of CPC and condensin, within a cohesin barrel, is consistent with the physical interactions observed between the CPC and condensin and between condensin and cohesin.
DISCUSSION
Our ts mutant strain collection contains ts alleles of 497 (~45%) S. cerevisiae essential genes. Because each ts allele is linked to a kanMX marker, the alleles can be readily used in SGA-based automated genetic analysis, which enables their barcoding for competitive growth assays in pools17, genetic network mapping41, high-content screening21 and a host of other applications. Over ~30% of the essential genes in our collection have multiple alleles (Supplementary Table 1), which can be useful for dissecting different roles of multifunctional proteins, such as actin (Supplementary Fig. 6). Ultimately, we aim to expand the collection to include a range of ts alleles that cover all domains of essential genes. Indeed, previous efforts to dissect allele-specific interactions involving essential genes have provided significant insight into protein structure and function42,43.
High content microscopy-based assays of the ts array revealed several new functions for multiple genes (Fig. 3a, Supplementary Fig. 3c and Supplementary Table 5). For example, cellular profiling of our ts collection revealed a connection between the cohesin and condensin complexes and spindle disassembly. Spindle disassembly is a spatiotemporally regulated event such that disassembly of the mitotic spindle occurs only after separation of sister chromatids from the spindle midzone region. Our findings are consistent with a model whereby the CPC decorates the spindle in part through interactions with condensin and cohesin within the vicinity of the centromere31. Loss of cohesin/condensin function results in the mislocalization of the CPC, including the Ipl1p kinase that controls spindle disassembly. We speculate that the CPC accesses the spindle midzone only after the cohesin is cleaved, such that cohesin cleavage and clearing of chromosome midzone regions are prerequisites for the mitotic spindle disassembly.
Our collection of ts alleles builds upon a non-overlapping set of strains constructed recently8. The combined collections cover the majority of essential genes in the yeast cell and enable numerous systematic approaches for querying the pleiotropic roles of essential genes. For example, we highlighted the use of the collection in chemical-genetic screens for small-molecule suppressors of the ts phenotype, a general screen that may be of particular interest for yeast orthologs of human disease genes because in theory such suppressors may represent therapeutic leads.
Another powerful approach involving ts strains will involve the mapping of genetic interactions by systematic dosage suppression analysis using high-copy libraries, such as the molecular barcoded yeast ORF (MoBY-ORF) library10 (unpublished data) or the systematic library for comprehensive overexpression analysis11. Dosage suppression network interactions are highly abundant and functionally relevant and, even though some of these interactions overlap with protein-protein and negative genetic interactions, most of these suppression edges are unique and offer the potential for expanding our global view of the functional wiring diagram of the cell.
METHODS
Methods and any associated references are available in the online version of the paper at http://www.nature.com/naturebiotechnology/.
Supplementary Material
ACKNOWLEDGMENTS
We thank the members of the Boone, Andrews and Bloom laboratory for their input and discussions. We thank S. Biggins (Fred Hutchinson Cancer Research Center) for her insights on cohesin and condensin components. We thank M. Zackrisson (University of Gothenburg) for statistical support. The Sli15-6A-GFP construct was a gift from E. Schiebel (University of Heidelberg) and the Mad1-NLS construct was a gift from R. Wozniak (University of Alberta). We also thank Jennifer Reginold for manual inspection of HCS images. F.J.V. was supported by a postdoctoral fellowship from the Best Foundation. B.A. and C.B. were supported by Genome Canada through the Ontario Genomics Institute as per research agreement 2004-OGI-3-01 and the Canadian Institutes of Health Research (MOP-97939).
Footnotes
Note: Supplementary information is available on the Nature Biotechnology website.
AUTHOR CONTRIBUTIONS
Z.L., F.J.V., C.B. and B.A. conceived and designed the experiments. Z.L. and S.B. constructed the ts strains. J.W. and A.B. conducted and analyzed liquid growth profiling experiments. Z.L., F.J.V., F.S.V., J.L., R.M., K.J. and Z.Z. generated and analyzed the HCS data. M.C., A.B., B.V., J.B. and C.L.M. generated and analyzed the data. Z.Y.L. and A.C.G. performed mass spectrometry analyses. Z.L., Z.Y., M.D., J.A.F., C.B., G.G. and C.N. performed the chemical genetic experiments. Z.L., F.J.V., A.S., J.H. and K.B. performed the analysis on cohesin and condensin complexes. H.L., Z.Y., S.B., A.D., G.G., C.N., C.B., M.C., F.S.V., A.D., A.G., Z.Z. and K.B. provided technical support/reagents/materials/analysis tools. Z.L., F.J.V., M.C., B.A. and C.B. wrote the paper.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
References
- 1.Giaever G, et al. Functional profiling of the Saccharomyces cerevisiae genome. Nature. 2002;418:387–391. doi: 10.1038/nature00935. [DOI] [PubMed] [Google Scholar]
- 2.Kim DU, et al. Analysis of a genome-wide set of gene deletions in the fission yeast Schizosaccharomyces pombe. Nat. Biotechnol. 2010;28:617–623. doi: 10.1038/nbt.1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hughes TR. Yeast and drug discovery. Funct. Integr. Genomics. 2002;2:199–211. doi: 10.1007/s10142-002-0059-1. [DOI] [PubMed] [Google Scholar]
- 4.Moir D, Stewart SE, Osmond BC, Botstein D. Cold-sensitive cell-division-cycle mutants of yeast: isolation, properties, and pseudoreversion studies. Genetics. 1982;100:547–563. doi: 10.1093/genetics/100.4.547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kanemaki M, Sanchez-Diaz A, Gambus A, Labib K. Functional proteomic identification of DNA replication proteins by induced proteolysis in vivo. Nature. 2003;423:720–724. doi: 10.1038/nature01692. [DOI] [PubMed] [Google Scholar]
- 6.Mnaimneh S, et al. Exploration of essential gene functions via titratable promoter alleles. Cell. 2004;118:31–44. doi: 10.1016/j.cell.2004.06.013. [DOI] [PubMed] [Google Scholar]
- 7.Schuldiner M, et al. Exploration of the function and organization of the yeast early secretory pathway through an epistatic miniarray profile. Cell. 2005;123:507–519. doi: 10.1016/j.cell.2005.08.031. [DOI] [PubMed] [Google Scholar]
- 8.Ben-Aroya S, et al. Toward a comprehensive temperature-sensitive mutant repository of the essential genes of Saccharomyces cerevisiae. Mol. Cell. 2008;30:248–258. doi: 10.1016/j.molcel.2008.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brachmann CB, et al. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast. 1998;14:115–132. doi: 10.1002/(SICI)1097-0061(19980130)14:2<115::AID-YEA204>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 10.Ho CH, et al. A molecular barcoded yeast ORF library enables mode-of-action analysis of bioactive compounds. Nat. Biotechnol. 2009;27:369–377. doi: 10.1038/nbt.1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jones GM, et al. A systematic library for comprehensive overexpression screens in Saccharomyces cerevisiae. Nat. Methods. 2008;5:239–241. doi: 10.1038/nmeth.1181. [DOI] [PubMed] [Google Scholar]
- 12.Giaever G, et al. Chemogenomic profiling: identifying the functional interactions of small molecules in yeast. Proc. Natl. Acad. Sci. USA. 2004;101:793–798. doi: 10.1073/pnas.0307490100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hillenmeyer ME, et al. The chemical genomic portrait of yeast: uncovering a phenotype for all genes. Science. 2008;320:362–365. doi: 10.1126/science.1150021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shoemaker DD, Lashkari DA, Morris D, Mittmann M, Davis RW. Quantitative phenotypic analysis of yeast deletion mutants using a highly parallel molecular bar-coding strategy. Nat. Genet. 1996;14:450–456. doi: 10.1038/ng1296-450. [DOI] [PubMed] [Google Scholar]
- 15.Smith AM, et al. Quantitative phenotyping via deep barcode sequencing. Genome Res. 2009;19:1836–1842. doi: 10.1101/gr.093955.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pierce SE, Davis RW, Nislow C, Giaever G. Genome-wide analysis of barcoded Saccharomyces cerevisiae gene-deletion mutants in pooled cultures. Nat. Protoc. 2007;2:2958–2974. doi: 10.1038/nprot.2007.427. [DOI] [PubMed] [Google Scholar]
- 17.Yan Z, et al. Yeast Barcoders: a chemogenomic application of a universal donorstrain collection carrying bar-code identifiers. Nat. Methods. 2008;5:719–725. doi: 10.1038/nmeth.1231. [DOI] [PubMed] [Google Scholar]
- 18.Baxter A, et al. Squalestatin 1, a potent inhibitor of squalene synthase, which lowers serum cholesterol in vivo. J. Biol. Chem. 1992;267:11705–11708. [PubMed] [Google Scholar]
- 19.Bergstrom JD, et al. Zaragozic acids: a family of fungal metabolites that are picomolar competitive inhibitors of squalene synthase. Proc. Natl. Acad. Sci. USA. 1993;90:80–84. doi: 10.1073/pnas.90.1.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Charlton-Menys V, Durrington PN. Human cholesterol metabolism and therapeutic molecules. Exp. Physiol. 2008;93:27–42. doi: 10.1113/expphysiol.2007.035147. [DOI] [PubMed] [Google Scholar]
- 21.Vizeacoumar FJ, et al. Integrating high-throughput genetic interaction mapping and high-content screening to explore yeast spindle morphogenesis. J. Cell Biol. 2010;188:69–81. doi: 10.1083/jcb.200909013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Buvelot S, Tatsutani SY, Vermaak D, Biggins S. The budding yeast Ipl1/Aurora protein kinase regulates mitotic spindle disassembly. J. Cell Biol. 2003;160:329–339. doi: 10.1083/jcb.200209018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pereira G, Schiebel E. Separase regulates INCENP-Aurora B anaphase spindle function through Cdc14. Science. 2003;302:2120–2124. doi: 10.1126/science.1091936. [DOI] [PubMed] [Google Scholar]
- 24.Ruchaud S, Carmena M, Earnshaw WC. The chromosomal passenger complex: one for all and all for one. Cell. 2007;131:230–231. doi: 10.1016/j.cell.2007.10.002. [DOI] [PubMed] [Google Scholar]
- 25.Sullivan M, Lehane C, Uhlmann F. Orchestrating anaphase and mitotic exit: separase cleavage and localization of Slk19. Nat. Cell Biol. 2001;3:771–777. doi: 10.1038/ncb0901-771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zeng X, et al. Slk19p is a centromere protein that functions to stabilize mitotic spindles. J. Cell Biol. 1999;146:415–425. doi: 10.1083/jcb.146.2.415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang CE, Milutinovich M, Koshland D. Rings, bracelet or snaps: fashionable alternatives for Smc complexes. Phil. Trans. R. Soc. Lond. B. 2005;360:537–542. doi: 10.1098/rstb.2004.1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nasmyth K, Haering CH. The structure and function of SMC and kleisin complexes. Annu. Rev. Biochem. 2005;74:595–648. doi: 10.1146/annurev.biochem.74.082803.133219. [DOI] [PubMed] [Google Scholar]
- 29.Guacci V, Koshland D, Strunnikov A. A direct link between sister chromatid cohesion and chromosome condensation revealed through the analysis of MCD1 in S. cerevisiae. Cell. 1997;91:47–57. doi: 10.1016/s0092-8674(01)80008-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Straight AF, Sedat JW, Murray AW. Time-lapse microscopy reveals unique roles for kinesins during anaphase in budding yeast. J. Cell Biol. 1998;143:687–694. doi: 10.1083/jcb.143.3.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yeh E, et al. Pericentric chromatin is organized into an intramolecular loop in mitosis. Curr. Biol. 2008;18:81–90. doi: 10.1016/j.cub.2007.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Khmelinskii A, Lawrence C, Roostalu J, Schiebel E. Cdc14-regulated midzone assembly controls anaphase B. J. Cell Biol. 2007;177:981–993. doi: 10.1083/jcb.200702145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fridman V, Gerson-Gurwitz A, Movshovich N, Kupiec M, Gheber L. Midzone organization restricts interpolar microtubule plus-end dynamics during spindle elongation. EMBO Rep. 2009;10:387–393. doi: 10.1038/embor.2009.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schuyler SC, Liu JY, Pellman D. The molecular function of Ase1p: evidence for a MAP-dependent midzone-specific spindle matrix. Microtubule-associated proteins. J. Cell Biol. 2003;160:517–528. doi: 10.1083/jcb.200210021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pinsky BA, Biggins S. The spindle checkpoint: tension versus attachment. Trends Cell Biol. 2005;15:486–493. doi: 10.1016/j.tcb.2005.07.005. [DOI] [PubMed] [Google Scholar]
- 36.Stern BM, Murray AW. Lack of tension at kinetochores activates the spindle checkpoint in budding yeast. Curr. Biol. 2001;11:1462–1467. doi: 10.1016/s0960-9822(01)00451-1. [DOI] [PubMed] [Google Scholar]
- 37.Mirchenko L, Uhlmann F. Sli15(INCENP) Dephosphorylation prevents mitotic checkpoint reengagement due to loss of tension at anaphase onset. Curr. Biol. 2010;20:1396–1401. doi: 10.1016/j.cub.2010.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bachellier-Bassi S, Gadal O, Bourout G, Nehrbass U. Cell cycle-dependent kinetochore localization of condensin complex in Saccharomyces cerevisiae. J. Struct. Biol. 2008;162:248–259. doi: 10.1016/j.jsb.2008.01.002. [DOI] [PubMed] [Google Scholar]
- 39.Freeman L, Aragon-Alcaide L, Strunnikov A. The condensin complex governs chromosome condensation and mitotic transmission of rDNA. J. Cell Biol. 2000;149:811–824. doi: 10.1083/jcb.149.4.811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lavoie BD, Hogan E, Koshland D. In vivo requirements for rDNA chromosome condensation reveal two cell-cycle-regulated pathways for mitotic chromosome folding. Genes Dev. 2004;18:76–87. doi: 10.1101/gad.1150404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Costanzo M, et al. The genetic landscape of a cell. Science. 2010;327:425–431. doi: 10.1126/science.1180823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Amberg DC, Basart E, Botstein D. Defining protein interactions with yeast actin in vivo. Nat. Struct. Biol. 1995;2:28–35. doi: 10.1038/nsb0195-28. [DOI] [PubMed] [Google Scholar]
- 43.Dreze M, et al. ‘Edgetic‘ perturbation of a C. elegans BCL2 ortholog. Nat. Methods. 2009;6:843–849. doi: 10.1038/nmeth.1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dimster-Denk D, et al. Comprehensive evaluation of isoprenoid biosynthesis regulation in Saccharomyces cerevisiae utilizing the Genome Reporter Matrix. J. Lipid Res. 1999;40:850–860. [PubMed] [Google Scholar]
- 45.Kuranda K, Francois J, Palamarczyk G. The isoprenoid pathway and transcriptional response to its inhibitors in the yeast Saccharomyces cerevisiae. FEM. Yeast Res. 2010;10:14–27. doi: 10.1111/j.1567-1364.2009.00560.x. [DOI] [PubMed] [Google Scholar]
- 46.Song JL, Lyons CN, Holleman S, Oliver BG, White TC. Antifungal activity of fluconazole in combination with lovastatin and their effects on gene expression in the ergosterol and prenylation pathways in Candida albicans. Med. Mycol. 2003;41:417–425. doi: 10.1080/1369378031000137233. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.