The first mouse carrying a deletion mutation in its apolipoprotein E (apoE) gene was born in our mouse colony in the summer of 1991. Our ongoing experiments to that point had indicated that this would happen, but the realization that the mouse in front of my eyes had inherited a mutation that we had designed and made in the laboratory was awesome. It did not occur to me immediately that this mouse would develop hypercholesterolemia and atherosclerosis and dictate the direction of my research for the next 20 years. I here briefly describe the background and my personal experience that led to the development of apoE-deficient mice as a model for human atherosclerosis.
Homologous Recombination
As a research scientist in the Department of Genetics at the University of Wisconsin, my main interest during the early 1980’s was directed towards understanding how different types of DNA recombination contribute to the architecture and diversity of individual genomes within human populations and to DNA differences between humans and non-human primates. My studies clearly showed that new alleles and new genes are constantly being formed in the human genome by recombinational events particularly between existing members of multigene families. There also appeared to be “hot spots” for recombination. By tracing a history of the human haptoglobin-gene cluster from the present day structures in human populations and in primates, I found that a single recombination event between related genes often produces much more drastic changes in the genome than those that result from simple point mutations.1
Whether this frequent yet natural occurrence of “homologous recombination” in the mammalian genome could be used to alter the genome of mammalian cells in culture in a pre-planned fashion became a question of interest to several laboratories.2,3 An affirmative answer was first obtained in 1985 when Oliver Smithies and his colleagues at the University of Wisconsin demonstrated that a predicted recombinant between the endogenous human β-globin gene locus and the incoming targeting DNA fragment could be found in cultured cells.2 This experiment was strongly motivated by the thought that correction of an inherited genetic mistake in a precise manner through homologous recombination would be a means of achieving gene therapy. However, the frequency of the desired recombinational event was one in a million treated cells, which was far too small to be practical for therapeutic use. Instead, Smithies and Capecchi3, who was another leader in the field of gene targeting at the University of Utah, both turned towards applying the method to alter genes in mice.
Embryonic stem cells and transgenic mice
Manipulation of the expression of specific gene products in mice was first accomplished in the early 1980s by pronuclear injection with exogenous DNA to produce transgenic mice in which the exogenous DNA was incorporated randomly into the mouse genome.4 Although transgenes were often unpredictable for the levels of expression and were frequently associated with the occurrence of large genomic rearrangements at or near the site of integration, mice generated in this way provided biological insights into the in vivo functions of many genes and in some cases provided useful models of human conditions. At about the same time, Evans and Kaufman5 demonstrated that cells of the inner cell mass of early mouse embryos (blastocysts) could be grown in culture without losing their capacity to differentiate into multiple cell types. Remarkably, when these cells, later called embryonic stem (ES) cells, were reintroduced into blastocysts and allowed to complete their development in a foster mother, they contributed to many tissues of resulting embryos, including germ cells.6 ES cells can therefore be considered to be “mice in tissue culture”. A path to making mutant mice was now open. The ES cell genome can be modified, cells with a correct modification identified and expanded in culture, and the modified cells injected into blastocysts to generate a living mouse having the intended mutation. Gene targeting in ES cells consequently allows us to generate a mouse carrying any specific alteration in any given gene. Disrupting (knocking out) a gene is the best approach to elucidate the function of the gene. Altering the protein made by a gene or changing the expression is the best approach to understand the consequences in vivo of specific genetic mutations associated with human disease.
Apolipoprotein and Atherosclerosis
The association between plasma cholesterol and cardiovascular disease has long been recognized and extensively studied. Historical accounts of the many important discoveries in this area are covered in other articles in this series. The early 1980s were particularly active in advancing the molecular biology of lipoproteins and lipoprotein metabolism. Multiple apolipoproteins, cDNAs and genomic loci coding for apolipoproteins and lipid metabolism-related enzymes were cloned, sequenced and their regulatory mechanisms began to be elucidated.7,8
My own involvement in the field of lipid metabolism began in 1984 in New Glarus, a town in Wisconsin called “little Switzerland”, where Oliver Smithies and I had supper with Jan and Judith Rapacz who worked in the Department of Meat and Animal Science at the University of Wisconsin. For many years Jan and Judith had been pursuing a relationship between plasma cholesterol, atherosclerosis and immunotypes of apolipoprotein B (apoB) that they had discovered in the plasma of domestic pigs.9 Conversation at the table was lively, and Jan commented that farmers in the US had unknowingly been selecting pigs with particular immunotypes associated with high plasma levels of cholesterol and a high incidence of atherosclerosis because these pigs tended to produce piglets with larger body size and therefore better market values than lean piglets. I asked “Isn’t it about time to clone and sequence the pig Apob gene and determine the changes that cause these phenotypes?” This simple question led us to start a collaboration which also included Alan Attie.10 Alan specialized in lipoprotein metabolism and patiently taught me how it works; I remember being particularly struck by the complexity of the systems that underlie human dyslipidemia which involve multiple genetic and environmental factors.
Animal model of atherosclerosis
When a disease condition is complex, and can only be reproduced in a living animal, then animal models are of vital importance in dissecting its pathogenesis. The best model of inherited atherosclerosis in the 1980’s was the Watanabe Heritable Hyperlipidemic rabbit that has a LDLR gene defect analogous to familial hypercholesterolemia in humans.11,12 Despite their obvious usefulness for biomedical studies and for developing therapeutic devices, Jan’s mutant pigs were large, expensive to maintain, and difficult to handle, particularly when they developed heart disease. Models involving naturally occurring mutations are usually identified because they have a phenotype of interest, and the genetic basis for their condition is unknown and often complex. Furthermore, as is the case in human studies, association between a genotype and a phenotype does not easily translate into causality. A small animal model for atherosclerosis caused by a defined genetic modification would clearly be useful for laboratory experiments.
I became excited about the possibility of introducing human gene variants into small experimental animals via gene targeting in ES cells to determine causal relationships between the variants and complex disease conditions, such as dyslipidemia and atherosclerosis, which can only be replicated in living animals. However, because the majority of humans with hyperlipidemia do not have family histories suggestive of a simple mode of inheritance, I was quite convinced that multiple genetic mutations would have to be combined to create a small animal model for atherosclerosis. Mice are the best experimental laboratory animal for genetic study because multiple, well defined inbred strains are available, and because their gestation period is short. However, despite the occurrence in mice of many models of diseases, no mouse strains had been identified that develop robust atherosclerosis. Indeed, for several reasons, workers in the lipoprotein field at that time generally regarded the mouse as unsuitable to model atherosclerosis. First, unlike humans, mice have low plasma cholesterol and their plasma lipoprotein profile is “protective”, namely high in HDL and low in VLDL/LDL ratios.13 Secondly, their life span of 2-3 years was thought to be too short to develop atherosclerotic plaques, which takes scores of years in humans. Nevertheless, earlier studies by Roberts and Thompson14 and later studies by Paigen et al.15 had shown that some strains of mice develop early fatty streaks after prolonged feeding of a diet high in cholesterol and saturated fat together with sodium cholate. Although the molecular basis of this susceptibility to diet-induced atherosclerosis was not known, these studies suggested that, given a specific genetic constitution, mice could be made susceptible to atherosclerosis. This was good enough for me to initiate a gene targeting approach to generate animal models of this complex human disease.
Embryonic stem cells were only available from mice at that time, but it was still not clear that mice would be a suitable species in which to model atherosclerosis. Alan Attie suggested to me that the plasma lipoprotein profiles of guinea pigs and hamsters are more similar to humans than those of mice.16 I chose hamsters because the early embryology of hamsters is better understood and the collection of timed embryos is easier in hamsters than in guinea pigs. In addition, laboratory hamsters are nearly inbred, since the colony was established from one male and two females, all littermates, captured in Syria in the 1930’s.17 So I began to derive embryonic stem cells of golden hamsters with guidance from Thomas Doetchman who had extensive experience in establishing and culturing mouse ES cells. Together, we made hamster ES cells that have similar characteristics to mouse ES cells.18 But the hamster ES cells proved to be less stable in culture than mouse ES cells, and we stopped pursuing this line of work once we found that our genetically altered mice could indeed provide excellent models for atherosclerosis studies.
Applying for a grant
In order to begin, I applied for an NIH grant in February 1987 with the goal of understanding the relationships between atherogenesis and genetically controlled molecular variability by using targeted modification of the apoB gene in mice. This proposal was submitted eight months before anyone was able to demonstrate targeted modification in ES cells, and two years before anyone had obtained germline transmission of a modified allele in mice. Although theoretically possible, applying the ES cell procedure to a complex disease was likely to be considered too much a jump. Being aware of this, I toned down the application so that the primary aim was identifying mutations in the apoB gene in Jan Rapacz’s pigs, with the secondary aim being to introduce the same mutations into the mouse. The reviewers were quite kind and encouraged me to continue the mutation analyses of the pig gene, but they were unconvinced that the gene targeting portion of the project was useful. The pink sheet (which was actually pink in those days) stated that “it is unclear whether these sophisticated manipulations will actually provide biologic information on the role of apolipoprotein, ------. It would seem reasonable to perform [in vitro] studies before proceeding with the very complex gene modification studies [in animals]”. Criticisms well-taken, but the project was not funded.
When I revised and resubmitted the proposal the next year, the situation was different. Both we19 and Capecchi’s group20 had achieved targeted modification of the HPRT gene in ES cells; we had established hamster ES cells18; and I was more optimistic about the potential application of gene targeting to the study of lipoprotein metabolism. So, and rather boldly I now realize, I completely dropped my studies of pig genes, the part that the previous reviewers had encouraged me to pursue, and concentrated on the targeted modification of apolipoprotein genes (apoB and apoE) in mice and/or hamsters. This time I was able to convince reviewers that my proposal was realistic although clearly difficult, and I was funded. In this regard, the field of lipoprotein metabolism and atherosclerosis was far ahead of other fields in recognizing the potential of gene targeting for the study of common but complex genetic disorders.
Modification of the apolipoprotein genes
Plasma lipoprotein metabolism in mice was not well characterized when I began planning my experiments, although several fundamental publications were available21,15 which suggested that many factors were likely to be involved. So, I hypothesized that combining modifications in multiple genes would be necessary to develop hyperlipidemic mice. The first target that I set for myself was to increase plasma LDL cholesterol levels or the ratios of non-HDL/HDL cholesterol to obtain human-like lipoprotein profiles. I planned three experiments: (1) modify the Apob gene so that the ApoB100 protein would be produced normally but would have altered amino acid residues in the putative LDL receptor binding domains, with the expectation that LDL particles having this defective apoB100 would accumulate in plasma; (2) delete the Apoa1 gene, since apoA1 is the major component of HDL particles, with the expectation that the non-HDL/HDL ratio would increase; and (3) eliminate apolipoprotein E, which is essential for the clearance of chylomicrons and VLDL, with the expectation that cholesterol-rich remnants similar to LDL would accumulate in plasma. When I moved to the University of North Carolina in the summer of 1988, two postdoctoral fellows, Jorge Piedrahita and Terry Smith, and a visiting professor from the University of Iowa, Roger Williams, joined me to tackle all three projects in parallel. By that time, enough DNA sequences for apolipoprotein genes in humans and some in mice were available for us to clone the mouse genomic DNA fragments necessary to assemble individual targeting constructs. Three to four years later, with skilled assistance in embryo manipulation from Paula Oliver (a technician then), we achieved germline transmission of mice having these modifications. Characterization of the resulting mice took a long time and required the efforts of a series of postdocs and graduate students in my laboratory.
For plasma lipid analysis of the newly made mutant mice, I hoped to have help from the clinical laboratories in our Pathology Department. But this did not materialize because the clinical laboratories could only work on human samples and were not allowed to work with samples from experimental animals. So Sunny Zhang, a graduate student, energetically tackled lipoprotein analyses of our mutants by scaling down the various assays available for humans to accommodate the small plasma volumes we could get from mice. We also received help from investigators at Gladstone Laboratory, particularly from Harshini deSilva, for methodologies and antibodies needed for characterizing mouse lipoprotein.
Not all the resulting mutants had the expected plasma lipoprotein profiles. Mice with the modified apoB100 had decreased, not increased, LDL particles due in part to a reduced secretion of modified-apoB100 particles.22, 23 Plasma HDL cholesterol in mice lacking apoA1 was reduced to 20% normal levels, but the non-HDL cholesterol levels remained very low.24 Neither mutants showed increased susceptibility to atherosclerosis.
ApoE-deficient mice
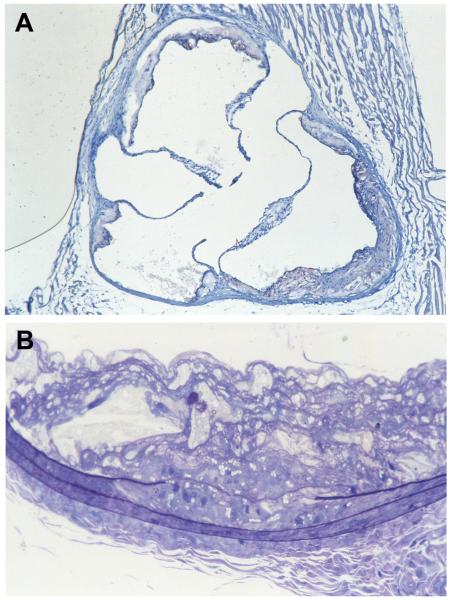
Remarkably, it turned out that lack of apoE is by itself sufficient to cause severe hyperlipidemia in mice. Their average plasma cholesterol levels on a regular mouse chow diet (4.5% fat and 0.02% cholesterol) are about 400 mg/dl compared to about 80 mg/dl in wild type mice. This increase is due to a marked accumulation in plasma of cholesterol-enriched chylomicron remnants and VLDL remnants. LDL particles in these mice are not increased (so much for my earlier prediction!) but HDL cholesterol is low. With such high plasma cholesterol levels and non-HDL/HDL cholesterol ratios, we thought that these mutants might develop atherosclerotic plaques even without additional risk factors such as extra dietary cholesterol. But how long would it take? We decided to examine a 6 months old apoE-deficient mouse, although this age, being equivalent to early adulthood in humans, might be too early. My long time collaborator, Bob Reddick, who is currently Chairman of Pathology at the Texas Health Science Center at San Antonio, sectioned the aorta and examined the plaques from our first apoE deficient mouse. There they were! As he showed me the slides, he pointed to a fold at the corner of an aortic section on the slide and said “I was so excited that I could not steady my hands!”. Indeed, when apoE-deficient mice are maintained on regular mouse chow, plaques become visible as clusters of foam cells near the aortic root as early as two months, and mature with age into raised complex lesions much like the plaques that occur in humans (Figure 1).25 Also with time, there is a progression of lesions to other sites in the aortic tree and associated vessels.
Figure 1.
Atherosclerotic plaques in one of our first apoE deficient mice sectioned by Bob Reddick. Panel A is a frozen section at the aortic root of a 9 month old male mouse stained with Sudan IVB and counterstained with hematoxylin showing plaques adherent to the valve attachment sites and vessel wall. Some areas of the plaque contain cholesterol clefts and fibrous caps while other areas are largely acellular. Panel B is a 1-μm thick section of a carotid artery from the same animal embedded in plastic and stained with Toluidine Blue, showing a plaque composed of foamy macrophages, cholesterol clefts, and fibrous protein deposits. A disruption of internal elastic lamina and the presence of smooth muscle cells with lipid droplets in both the plaque and media indicate smooth muscle cell proliferation, migration and lipid uptake. The mouse was on regular mouse chow.
ApoE deficient mice were produced independently in the laboratories of Jan Breslow26 at Rockefeller University and of Marten Hofker27 at Leiden University, Netherlands, using gene targeting. We and the Breslow group reported our findings essentially simultaneously in 1992. All three independent lines of mutants lack apoE completely, and have the same phenotype. Mice lacking LDLR were produced in a similar way by Joachim Herz and his colleagues28 at the University of Texas Southwestern Medical Center and provided another model. The LDLR-null mice accumulate a large excess of LDL particles in plasma, like humans deficient for LDLR. They have modestly increased VLDL remnants but retain high levels of HDL-cholesterol of about 80 mg/dl. With an average plasma cholesterol level of about 230 mg/dl, they demonstrate less overt vascular disease than do the apoE-null mice on regular chow. However, when fed a diet high in fat and cholesterol, their plasma cholesterol levels increase to more than 1,500 mg/dl and severe atherosclerotic lesions develop throughout their aortas.28 Once these genetically well-defined hyperlipidemic mice were shown to develop mature atherosclerotic plaques in a reasonably short time, the mouse quickly became the prime model for the study of atherosclerosis, and exemplified how to study complex human diseases in general. There are, of course, species differences, such as the finding that plaque ruptures that trigger fatal vascular events in humans are very rare in mice. Nevertheless, apoE deficient mice provide an excellent starting point to study both genetic and environmental factors that influence the early atherogenic processes.29
From early on, it was very important to me that these animal models should be distributed freely to the scientific community. I also worried that this unique mutant might be lost by fire, flood or by infections if they were kept in one colony. So we sent the apoE-deficient mice through Bev Paigen to the Jackson Laboratory for strain preservation and distribution; and this effort led to an establishment of induced mutant resources there a couple of years later. Meanwhile, one of our technicians spent almost full time arranging shipments of mice to other investigators. Our apoE deficient mice (B6.129P2-Apoetm1UNC), available from the Jackson Laboratory and also from Taconics, continue to be widely used.
Strain background and plaque localization
The B6.129P2-Apoetm1UNC mice carry a deletion mutation that we originally made using ES cells derived from a strain 129P2 (129/Ola) mouse. Because 129P2 inbred mice do not reproduce well and because C57BL/6 is a strain susceptible to diet-induced atherosclerosis15, the mutation was subsequently moved into a C57BL/6J (B6) genetic background by backcrossing to B6 for at least 12 generations. Although this extent of backcrossing ensures that the majority of the genome of the resulting mice is now of B6 origin, a relatively large segment of genetic material surrounding the Apoe locus on chromosome 7 is still derived from 129P2. One, therefore, need to be cautious in interpreting traits of apoE- null mice that are quantitatively different from wild type controls but not directly explainable by the function of apoE in lipid metabolism. The observed phenotypes could be the consequence of yet-to-be defined functions of apoE, but the effects could be the consequence of strain differences in genes tightly linked to the Apoe locus, which is derived from 129P2 in the apoE−/− mice but from C57BL/6 in the apoE+/+ mice. Similarly, when other lines of mice carrying additional genetic alterations of interest are crossed with B6.129P2-Apoetm1UNC mice to discover genes and pathways that modify atherogenesis, care has to be taken to eliminate the possibility that the observed effects are due to strain differences in closely linked loci rather than to the mutation of interest itself. The simplest solution to these problems is of course to study the mutation effects in mice having a pure inbred background. This thought prompted me to re-make the apoE-deletion mutation in strain 129S6/SvEvTac ES cells and maintain it on the same 129S6 stain background, since the majority of new mutations have been made with strain 129 ES cells and129S6 inbred mice reproduce well.
There are distinct differences in the phenotypes of apoE-null mutants on the two different genetic backgrounds, B6-apoE and 129-apoE.30 The plasma levels of total cholesterol (800 mg/dl) in the 129-apoE mice are higher than in B6-apoE mice (400 mg/dl). Both VLDL remnants and HDL are two fold increased in the plasma of 129-apoE. Despite having higher plasma cholesterol, however, 129-apoE mice begin to develop plaques at the aortic root later than B6-apoE mice; the plaques becoming visible only at 4 months of age rather than 2 month of age. Unexpectedly, and in a marked contrast, the 129-apoE mice develop much more extensive lesions in the aortic arch than the B6-apoE mice.30 Since circulating plasma components are the same in the aortic root and in the aortic arch, the difference in the susceptibility to atherosclerosis at the two locations of the vasculature suggests a differential vessel response to local hemodynamic forces. There also are distinct and easily recognizable differences in the geometry of the aortic arch between the two strains. For example, 129 mice have narrower vessel diameter and broader curvature of aortic arch than B6 mice. We therefore tested the hypothesis that genetic factors affecting vascular geometry also affect the location and extent of atherosclerotic plaque development using the F2 progeny from a cross between 129-apoE and B6-apoE mice. Quantitative trait locus (QTL) mapping identified two significant QTLs on chromosome 1 for plaque size at the aortic arch, while a single QTL locus for plaque size at the aortic root was found on chromosome 9. Importantly, one of the QTL (Chr1 at 105Mb) for aortic arch atherosclerosis overlapped completely with a significant QTL for the curvature of the aortic arch.31 There are over hundred genes within the overlapping interval and co-localization of QTLs does not establish that a gene affecting atherosclerosis susceptibility in the aortic arch is identical to one leading to wider aortic arch curvature. Nevertheless, the possibility that the ontogeny of the aortic arch formation is a potential risk factor for atherosclerosis is attractive, and the search for a causative gene(s) is a worthwhile effort, although clearly difficult.
Mice with humanized apoE
In closing, I return to my original goal -- to determine causal relationships between human gene variants and complex disease conditions such as atherosclerosis. Recent efforts of my group have been focused on studying mice that express the human apoE isoforms, E2, E3 or E4, in place of mouse apoE. These apoE isoforms are very common in humans and are strongly but differently associated with cardiovascular diseases. Yet the mechanisms underlying these associations have not been determined. One very puzzling observation in humans is that apoE4, which binds to the LDL receptor better than the other isoforms, is associated with increased plasma LDL cholesterol and cardiovascular incidents, while humans with apoE2, which binds poorly to the receptor, are protected from these conditions. Remarkably, the only experimental condition under which the apoE isoform-associated plasma lipid phenotype is replicated in our mice with humanized apoE is when these mice also express high levels of LDL receptor and are fed a Western-type diet high in fat and cholesterol.32 Why too much of a “good” thing (LDL receptor) is harmful has yet to be determined. But high LDL receptor expression and a Western-type diet probably brings the mice a little closer to humans who depend more on LDLR-mediated clearance of plasma lipoproteins than do mice, since various other observations have suggested that humans and mice differ in their relative use of apoE and LDL receptor in lipid metabolism.
Although mice do not always model all aspects of human diseases precisely, this is not usually because there is a fundamental difference in the biology of the gene products in the two species. Rather it is due to differences in the overall physiologic set point of the system. As exemplified in mice with humanized apoE, minimizing species differences can be effective in bringing out the phenotype. The steps needed to achieve this end can, in turn, lead us to new insights and previously unsuspected gene-gene and gene-environmental interactions that underlie the risks for complex human diseases.
Acknowledgements
Our apolipoprotein E-deficient mice and other mutants would not have been made without many colleagues, students, postdoctoral fellows and technicians who shared their passion, excitement, and the fun of science with me. I also acknowledge an NIH grant, HL042630, for funding my research.
Footnotes
Disclosure
None
References
- 1.Maeda N, Smithies O. The evolution of multigene families: human haptoglobin genes. Annu Rev Genet. 1986;20:81–108. doi: 10.1146/annurev.ge.20.120186.000501. [DOI] [PubMed] [Google Scholar]
- 2.Smithies O, Gregg RG, Boggs SS, Koralewski MA, Kucherlapati RS. Insertion of DNA sequences into the human chromosomal beta-globin locus by homologous recombination. Nature. 1985;317(6034):230–234. doi: 10.1038/317230a0. [DOI] [PubMed] [Google Scholar]
- 3.Thomas KR, Folger KR, Capecchi MR. High frequency targeting of genes to specific sites in the mammalian genome. Cell. 1986;44(3):419–428. doi: 10.1016/0092-8674(86)90463-0. [DOI] [PubMed] [Google Scholar]
- 4.Gordon JW, Ruddle FH. Integration and stable germ line transmission of genes injected into mouse pronuclei. Science. 1981;214(4526):1244–1246. doi: 10.1126/science.6272397. [DOI] [PubMed] [Google Scholar]
- 5.Evans MJ, Kaufman MH. Establishment in culture of pluripotential cells from mouse embryos. Nature. 1981;292(5819):154–156. doi: 10.1038/292154a0. [DOI] [PubMed] [Google Scholar]
- 6.Bradley A, Evans M, Kaufman MH, Robertson E. Formation of germ-line chimaeras from embryo-derived teratocarcinoma cell lines. Nature. 1984;309(5965):255–256. doi: 10.1038/309255a0. [DOI] [PubMed] [Google Scholar]
- 7.Breslow JL, McPherson J, Nussbaum AL, Williams HW, Lofquist-Kahl F, Karathanasis SK, Zannis VI. Identification and DNA sequence of a human apolipoprotein E cDNA clone. J Biol Chem. 1982;257(24):14639–14641. [PubMed] [Google Scholar]
- 8.Law SW, Lackner KJ, Fojo SS, Hospattankar A, Monge JC, Brewer HB., Jr. The molecular biology of human apoA-I, apoA-II, apoC-II and apoB. Adv Exp Med Biol. 1986;201:151–162. doi: 10.1007/978-1-4684-1262-8_14. [DOI] [PubMed] [Google Scholar]
- 9.Rapacz J, Elson CE, Lalich JJ. Correlation of an immunogenetically defined lipoprotein type with aortic intimal lipidosis in swine. Exp Mol Pathol. 1977;27(2):249–261. doi: 10.1016/0014-4800(77)90034-x. [DOI] [PubMed] [Google Scholar]
- 10.Maeda N, Ebert DL, Doers TM, Newman M, Hasler-Rapacz J, Attie AD, Rapacz J, Smithies O. Molecular genetics of the apolipoprotein B gene in pigs in relation to atherosclerosis. Gene. 1988;70(2):213–229. doi: 10.1016/0378-1119(88)90194-1. [DOI] [PubMed] [Google Scholar]
- 11.Watanabe Y. Serial inbreeding of rabbits with hereditary hyperlipidemia (WHHL-rabbit) Atherosclerosis. 1980;36(2):261–268. doi: 10.1016/0021-9150(80)90234-8. [DOI] [PubMed] [Google Scholar]
- 12.Kita T, Brown MS, Watanabe Y, Goldstein JL. Deficiency of low density lipoprotein receptors in liver and adrenal gland of the WHHL rabbit, an animal model of familial hypercholesterolemia. Proc Natl Acad Sci U S A. 1981;78(4):2268–2272. doi: 10.1073/pnas.78.4.2268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mills GL, Taylaur CE. The distribution and composition of serum lipoproteins in eighteen animals. Comp Biochem Physiol B. 1971;40(2):489–501. doi: 10.1016/0305-0491(71)90234-3. [DOI] [PubMed] [Google Scholar]
- 14.Roberts A, Thompson JS. Inbred mice and their hybrids as an animal model for atherosclerosis research. Adv Exp Med Biol. 1976;67(00):313–327. doi: 10.1007/978-1-4614-4618-7_18. [DOI] [PubMed] [Google Scholar]
- 15.Paigen B, Morrow A, Brandon C, Mitchell D, Holmes P. Variation in susceptibility to atherosclerosis among inbred strains of mice. Atherosclerosis. 1985;57(1):65–73. doi: 10.1016/0021-9150(85)90138-8. [DOI] [PubMed] [Google Scholar]
- 16.Spady DK, Dietschy JM. Sterol synthesis in vivo in 18 tissues of the squirrel monkey, guinea pig, rabbit, hamster, and rat. J Lipid Res. 1983;24(3):303–315. [PubMed] [Google Scholar]
- 17.Adler JH. The origin of the golden hamster as a laboratory animal. Isr J Med Sci. 1989;25(4):206–209. [PubMed] [Google Scholar]
- 18.Doetschman T, Williams P, Maeda N. Establishment of hamster blastocyst-derived embryonic stem (ES) cells. Dev Biol. 1988;127(1):224–227. doi: 10.1016/0012-1606(88)90204-7. [DOI] [PubMed] [Google Scholar]
- 19.Doetschman T, Gregg RG, Maeda N, Hooper ML, Melton DW, Thompson S, Smithies O. Targetted correction of a mutant HPRT gene in mouse embryonic stem cells. Nature. 1987;330(6148):576–578. doi: 10.1038/330576a0. [DOI] [PubMed] [Google Scholar]
- 20.Thomas KR, Capecchi MR. Site-directed mutagenesis by gene targeting in mouse embryo-derived stem cells. Cell. 1987;51(3):503–512. doi: 10.1016/0092-8674(87)90646-5. [DOI] [PubMed] [Google Scholar]
- 21.LeBoeuf RC, Puppione DL, Schumaker VN, Lusis AJ. Genetic control of lipid transport in mice. I. Structural properties and polymorphisms of plasma lipoproteins. J Biol Chem. 1983;258(8):5063–5070. [PubMed] [Google Scholar]
- 22.Toth LR, Smith TJ, Jones C, de Silva HV, Smithies O, Maeda N. Two distinct apolipoprotein B alleles in mice generated by a single ‘in-out’ targeting. Gene. 1996;178(1-2):161–168. doi: 10.1016/0378-1119(96)00360-5. [DOI] [PubMed] [Google Scholar]
- 23.Johnson LA, Altenburg MK, Walzem RL, Scanga LT, Maeda N. Absence of hyperlipidemia in LDL receptor-deficient mice having apolipoprotein B100 without the putative receptor-binding sequences. Arterioscler Thromb Vasc Biol. 2008;28(10):1745–1752. doi: 10.1161/ATVBAHA.108.169680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li H, Reddick RL, Maeda N. Lack of apoA-I is not associated with increased susceptibility to atherosclerosis in mice. Arterioscler Thromb. 1993;13(12):1814–1821. doi: 10.1161/01.atv.13.12.1814. [DOI] [PubMed] [Google Scholar]
- 25.Zhang SH, Reddick RL, Piedrahita JA, Maeda N. Spontaneous hypercholesterolemia and arterial lesions in mice lacking apolipoprotein E. Science. 1992;258(5081):468–471. doi: 10.1126/science.1411543. [DOI] [PubMed] [Google Scholar]
- 26.Plump AS, Smith JD, Hayek T, Aalto-Setälä K, Walsh A, Verstuyft JG, Rubin EM, Breslow JL. Severe hypercholesterolemia and atherosclerosis in apolipoprotein E-deficient mice created by homologous recombination in ES cells. Cell. 1992;71(2):343–353. doi: 10.1016/0092-8674(92)90362-g. [DOI] [PubMed] [Google Scholar]
- 27.van Ree JH, van den Broek WJ, Dahlmans VE, Groot PH, Vidgeon-Hart M, Frants RR, Wieringa B, Havekes LM, Hofker MH. Diet-induced hypercholesterolemia and atherosclerosis in heterozygous apolipoprotein E-deficient mice. Atherosclerosis. 1994;111(1):25–37. doi: 10.1016/0021-9150(94)90188-0. [DOI] [PubMed] [Google Scholar]
- 28.Ishibashi S, Brown MS, Goldstein JL, Gerard RD, Hammer RE, Herz J. Hypercholesterolemia in low density lipoprotein receptor knockout mice and its reversal by adenovirus-mediated gene delivery. J Clin Invest. 1993;92(2):883–893. doi: 10.1172/JCI116663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Knowles JW, Maeda N. Genetic modifiers of atherosclerosis in mice. Arterioscler Thromb Vasc Biol. 2000;20(11):2336–2345. doi: 10.1161/01.atv.20.11.2336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maeda N, Johnson L, Kim S, Hagaman J, Friedman M, Reddick R. Anatomical differences and atherosclerosis in apolipoprotein E-deficient mice with 129/SvEv and C57BL/6 genetic backgrounds. Atherosclerosis. 2007;195(1):75–82. doi: 10.1016/j.atherosclerosis.2006.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tomita H, Zhilicheva S, Kim S, Maeda N. Aortic arch curvature and atherosclerosis have overlapping quantitative trait loci in a cross between 129S6/SvEvTac and C57BL/6J apolipoprotein E-null mice. Circ Res. 2010;106(6):1052–1060. doi: 10.1161/CIRCRESAHA.109.207175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Malloy SI, Altenburg MK, Knouff C, Lanningham-Foster L, Parks JS, Maeda N. Harmful effects of increased LDLR expression in mice with human APOE*4 but not APOE*3. Arterioscler Thromb Vasc Biol. 2004;24(1):91–97. doi: 10.1161/01.ATV.0000094963.07902.FB. [DOI] [PubMed] [Google Scholar]