Abstract
Genomic alterations have been linked to the development and progression of cancer. The technique of comparative genomic hybridization (CGH) yields data consisting of fluorescence intensity ratios of test and reference DNA samples. The intensity ratios provide information about the number of copies in DNA. Practical issues such as the contamination of tumor cells in tissue specimens and normalization errors necessitate the use of statistics for learning about the genomic alterations from array CGH data. As increasing amounts of array CGH data become available, there is a growing need for automated algorithms for characterizing genomic profiles. Specifically, there is a need for algorithms that can identify gains and losses in the number of copies based on statistical considerations, rather than merely detect trends in the data.
We adopt a Bayesian approach, relying on the hidden Markov model to account for the inherent dependence in the intensity ratios. Posterior inferences are made about gains and losses in copy number. Localized amplifications (associated with oncogene mutations) and deletions (associated with mutations of tumor suppressors) are identified using posterior probabilities. Global trends such as extended regions of altered copy number are detected. Because the posterior distribution is analytically intractable, we implement a Metropolis-within-Gibbs algorithm for efficient simulation-based inference. Publicly available data on pancreatic adenocarcinoma, glioblastoma multiforme, and breast cancer are analyzed, and comparisons are made with some widely used algorithms to illustrate the reliability and success of the technique.
Keywords: Amplifications, Cancer, Copy number, Deletions, DNA, Genomic alterations, Intensity ratios, MCMC, Tumor
1. INTRODUCTION
The Genomics of Cancer
The normal DNA of human females has two copies of the genomic code because there are 23 matched pairs of chromosomes. Human males have 22 matched pairs of nonsex (or autosomal) chromosomes and an unmatched pair of sex chromosomes. Hence, the copy number of normal male DNA is two for the autosomal chromosomes. The ends of the chromosomes are called the telomeres. The telomere corresponding to the short arm of a chromosome is called the p telomere, whereas the one corresponding to the long arm is called the q telomere.
Human cells can be classified into somatic (or body) cells and germ cells. Barring a few exceptions like red blood cells, muscle cells, and brain cells, the life cycle of somatic cells consists of a period of growth followed by cell division through mitosis. The cells must satisfy certain “quality control checks” before they can progress to a subsequent stage of the cycle. These checks ensure that the cells develop normally, that defects are repaired, and that DNA is correctly copied during mitosis. Two kinds of genes play very important roles in the regulation procedure: proto-oncogenes and tumor suppressors. Proto-oncogenes encourage the body cells to grow and divide, pushing them through the quality control check points. Tumor suppressors tend to hold the cells back, inhibiting mitosis when there are cell defects and signaling the cells to die when their lifespans have ended or when there are cell defects that cannot be repaired. Further details about the relevant biology for this problem are given in Pasternak (1999).
Occasionally, proto-oncogenes may mutate into oncogenes. The mutations are propagated to new cells through mitosis. Oncogenes duplicate themselves through several stages of mitosis so that cells end up with multiple copies of oncogenes. Oncogenes have a dominant effect on the cell function, causing the cells to divide at a rapid rate and resulting in the development of tumors. Tumors may also develop due to mutations in tumor suppressors that cause them to become nonfunctional and allow the proto-oncogenes to play a dominant role. Tumor-suppressor mutations eventually result in the loss of one or both copies of the gene. A deletion is the loss of both copies in a genomic region.
A single mutation is usually not enough to trigger cancer. A number of complex biological events occur before a person acquires the phenotype of cancer. An example, but not a necessary condition, is the ability of tumor cells to metastasize, making the tumor malignant. Not all the cells in a tumor specimen necessarily exhibit the same kind of genomic alteration. Additionally, there is a lot of variation among individuals. As the disease progresses, there are larger scale changes in tumor DNA because of the breakdown of quality control in cell division.
Copy number changes, or alterations in the number of copies in tumor DNA, are, therefore, closely associated with the development and progression of cancer. A number of methods are currently available to detect genomic changes. Karyotyping views the chromosomes through a microscope during the metaphase stage of the cell cycle. This technique covers the entire genome but has low resolution because only changes spanning large regions of the DNA, such as missing chromosomes, monosomies (loss of single copies), and trisomies (gain of additional copies of chromosomes), can be detected by this method. At the other end of the spectrum, molecular genetic studies are capable of single-base-pair resolution. Because the genome consists of approximately 3 billion bases, this technique cannot be used in the absence of prior knowledge to identify the DNA regions associated with a disease. Researchers must rely on other methods to first identify candidate loci involved in the disease pathogenesis.
Array CGH
Comparative genomic hybridization (CGH) has emerged as a powerful technique because it combines relatively high resolution of a few million bases with the ability to span the entire genome in a single experiment (Kallioniemi et al. 1992). Fragmented DNA from a test sample is labeled with fluorochrome (typically Cy3) and is mixed with normal DNA that is identically fragmented but labeled using a different fluorochrome (typically Cy5). The normal and tumor DNA fragments are simultaneously hybridized to a normal metaphase spread. Image analysis yields data consisting of fluorescence intensity ratios along the genomes of the test and reference DNA samples. The more recently developed array CGH techniques (Solinas-Toldo et al. 1997; Pinkel et al. 1998; Snijders et al. 2001; Pinkel and Albertson 2005) hybridize the DNA fragments or “clones” to mapped array fragments rather than metaphase chromosomes. CGH arrays that rely on BAC (bacterial artificial chromosome) clones have a resolution of the order of 1 Mb (1 million base pairs). Oligonucleotide and cDNA arrays (Pollack et al. 1999; Brennan et al. 2004) provide a higher resolution of 50–100 kb (1 kb = 1,000 base pairs). As with all hybridization-based techniques, the fluorescence intensity ratios have to be normalized as part of a preprocessing step to correct for nonbiological sources of error such as intensity fluctuations, background noise, and fabrication artifacts (Brown, Goodwin, and Sorger 2001; McLachlan, Do, and Ambroise 2004). Refer to Khojasteh, Lam, Ward, and MacAulay (2005) for a comparison of different normalization methods for array CGH data.
Array CGH intensity ratios (equivalently, their transformation on the log2 scale) provide much useful information about genome-wide changes in copy number. Imagine an idealized situation where all the cells in a tumor specimen have identical genomic alterations and are uncontaminated by cells from surrounding normal tissue. In the absence of normalization or measurement errors, the normal (or copy-neutral) clones would correspond to a log2 ratio of 0 because the normal and tumor DNA fragments both have two copies. The log-intensity ratios of single-copy losses would be exactly log2 1/2 = −1 and those of single-copy gains would be log2 3/2 = .58. Multiple-copy gains or amplifications, often associated with oncogenes, would correspond to data belonging to the sequence log2 4/2, log2 5/2, …. Losses of both copies or deletions, often associated with tumor-suppressor mutations, would correspond to a value of −∞. In this hypothetical situation, the genomic alterations can be easily deduced from the data without statistical techniques.
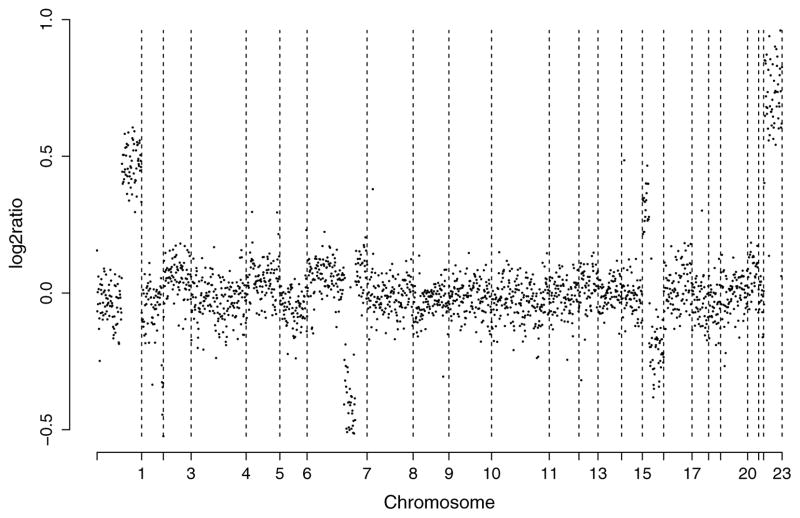
For comparison with the preceding idealized scenario, Figure 1 plots the normalized log2 ratios of breast cancer specimen S0034 analyzed by Snijders et al. (2001). The data are available from table J at http://www.nature.com/ng/journal/v29/n3/suppinfo/ng754_S1.html. Although relatively clean by array CGH standards, the data highlight some of the issues that necessitate the use of statistical methods. For example, even after accounting for measurement error, the log2 ratios differ considerably from the theoretical values. In particular, the numbers are typically shrunk toward 0. This is caused by several factors, including contamination of the tumor sample with normal cells. There is a more subtle effect of the 0 varying slightly from chromosome to chromosome. There is also an obvious dependence among the intensity ratios of neighboring clones.
Figure 1.
Normalized copy number ratios of a comparison of DNA from cell strain S0034 (Snijders et al. 2001) with normal DNA. The BACs are ordered by position in the genome beginning at 1p and ending at Xq. The vertical bars indicate borders between chromosomes.
As increasing amounts of array CGH data become available, there is a need for automated algorithms for characterizing the genomic profiles. A number of well-known methods strive to fulfill this need. For example, Pollack et al. (2002) proposed a threshold method for identifying clones having extreme value of emissions. Cheng, Kimmel, Neiman, and Zhao (2003) discussed a regression-based test for altered copy numbers. Hodgson et al. (2001) used a normal mixture of three components to model the observed emissions. Olshen, Venkatraman, Lucito, and Wigler (2004) developed a variation of binary segmentation to identify chromosomal segments with altered copy numbers. Fridlyand, Snijders, Pinkel, Albertson, and Jain (2004) applied an unsupervised hidden Markov model. Wang, Kim, Pollack, Balasubramanian, and Tibshirani (2005) built hierarchical clustering-style trees along each chromosome and selected interesting clusters by controlling the false discovery rate. Jong et al. (2003) proposed a break-point model to segment the clones. Eilers and de Menezes (2005) applied a quantile smoothing method, whereas Huang, Wu, Lizardi, and Zhao (2005) used penalized least squares regression and Hsu et al. (2005) applied wavelets. Hupe, Stransky, Thiery, Radvanyi, and Barillot (2004) relied on a likelihood function with adaptively determined weights using a smoothed version of the data. Picard, Robin, Lavielle, Vaisse, and Daudin (2005) used a penalized likelihood function. Myers, Dunham, Kung, and Troyanskaya (2004) applied an edge filter to detect the segments. Lingjaerde, Baumbusch, Liestol, Glad, and Borresen-Dale (2005) performed smoothing using the signs of neighboring data values and inspecting the width and magnitude of the segments to detect regions of copy number change.
A recent article by Lai, Johnson, Kucherlapati, and Park (2005) made comparisons of some of the preceding algorithms using real and simulated data. In evaluating the algorithms, Lai and co-authors commented that “a particularly helpful feature for future implementations of some algorithms would be to estimate the statistical significance of the detected copy number changes and then rank them accordingly.” They pointed out that only two algorithms (those of Wang et al. 2005 and Lingjaerde et al. 2005) could actually detect copy number changes based on statistical significance. Both methods rely on false discovery rates.
In Section 2 we develop a statistical framework for detecting copy number gains and losses, identifying localized amplifications and deletions, and partitioning tumor DNA into regions of relatively stable copy number. We rely on the hidden Markov model (HMM) to account for the dependence between neighboring clones. We adopt a Bayesian approach, assuming informative priors for the model parameters that are flexible enough to allow Bayesian learning. Because the posterior distribution is analytically intractable, Section 3 develops a framework for simulation-based posterior inference. In Section 4 we demonstrate the success of the technique using publicly available data. Section 4.4 compares the proposed Bayesian HMM with some of the existing algorithms using the framework of Lai et al. (2005).
Unlike the HMM of Fridlyand et al. (2004), which is purely a segmentation method, the likelihood function of Section 2.1 allows the use of objective decision rules based on posterior probabilities to detect copy number alterations. Unlike most of the existing array CGH methods, the biologist is not required to subjectively decide, after the algorithm’s output has been obtained, plausible thresholds for identifying changes in the number of DNA copies. The proposed framework allows the use of the simple classification scheme of Section 3.1, which is motivated by biological considerations and which makes the algorithm output easy to interpret. Section 5 uses simulation studies to compare our Bayesian HMM with alternative techniques for analyzing array CGH data.
2. BAYESIAN HIDDEN MARKOV MODEL
2.1 Likelihood Function
Because the propensity for genomic alterations varies across the chromosomes, we allow each chromosome to have a distinct set of parameters. For a given chromosome, let L1, …, Ln represent the mapped clones or DNA fragments arranged from the p telomere to the q telomere. Let Yk denote the normalized log2 ratio observed at clone Lk.
As mentioned earlier, the aim of the analysis is to learn about genome-wide changes in copy number from the data. A key innovation that directly achieves this goal is a latent variable called the copy number state sk associated with each clone Lk, where k = 1, …, n. The variable sk takes values in the set {1, 2, 3, 4}. The value sk = 1 represents a copy number loss at Lk that could be either a single-copy loss or a deletion; sk = 2 represents the copy-neutral state; sk = 3 represents a single-copy gain; sk = 4 represents an amplification (i.e., multiple copy gain) at Lk. The parameters of interest that summarize the copy number changes on the chromosome are s1, …, sn.
For j = 1, …, 4, we define μj as the expected log2 ratio of all clones Lk for which sk = j. For example, the expected log2 ratio of single-copy gains is μ3. The theoretical value of μ3 is .58, but, as mentioned earlier, the actual value could be different for many reasons (e.g., contamination of tumor samples with normal tissue). Although the μj ’s are unknown parameters, the biological interpretation associated with the state space of sk allows us to assume the ordering: μ1 < μ2 < μ3 < μ4. Conditional on the copy number states, the normalized log2 ratios are assumed to be distributed as , where k = 1, …, n.
We model the dependence of the neighboring clones using a hidden Markov model (Rabiner 1989; MacDonald and Zucchini 1997; Durbin, Eddy, Krogh, and Michison 1998). For any m indices for which 1 ≤ k1 ≤ · · · ≤ km ≤ n, a Markov model for the copy number states assumes that Pr[skm| s1, …, skm−1] = Pr[skm| skm−1]. The hidden Markov model assumes that the conditional probabilities of neighboring clones is Pr[sk+1 | sk ] = asksk+1, where A = ((aij )) is the matrix of stationary transition probabilities. We assume that the elements of A are strictly positive. The hidden Markov process is then aperiodic and irreducible, and its four states are positive recurrent. Transition matrix A has a unique stationary distribution, denoted by πA = (πA(1), πA(2), πA(3), πA(4)), where πA(i) is strictly positive for state i = 1, …, 4 (Karlin and Taylor 1975). We also assume that s1, the copy number state of the first clone, is distributed as πA. Together with the hidden Markov assumption, this uniquely determines the joint likelihood of a given sequence s1, …, sn. The chromosome-specific hyperparameters are, therefore, the transition probability matrix A, means {μ1, μ2, μ3, μ4}, and error variances { }.
2.2 Priors
The Bayesian approach assumes priors for all unknown parameters. Because the copy number states defined in Section 2.1 have a well-defined meaning, this facilitates the use of informative priors based on our knowledge of array CGH data. For example, we know that the mean μ1 of copy number losses cannot be a positive number, although individual log2 ratios that correspond to copy number losses could be. Independent priors are assumed for the chromosome-specific parameters. This results in independent posteriors for all the chromosomes. The marginal posterior [s1, …, sn | Y1, …, Yn] is of interest. As with many Bayesian applications, the marginal posterior cannot be analytically computed, and so simulation-based techniques are necessary. While analyzing HMMs, a key issue is label switching (refer to Scott 2002 for a discussion). This is an identifiability issue where the likelihood is invariant under arbitrary permutations of the state space labels, resulting in inefficient exploration of the posterior by simulation. The likelihood of Section 2.1 avoids this problem by assuming order constraints. Specifically, the constraint μ1 < μ2 < μ3 < μ4 is violated on permutating the labels.
Let X ~ F · I (c < X < d) imply that X has the distribution F restricted to the interval (c, d) with the density suitably rescaled to make it a random variable. For the mean μ1 corresponding to copy number losses, we assume the prior where ε > 0. We comment later on the choice of ε. For the copy-neutral state, we assume . For single-copy gains, we assume , and for multiple-copy gains, we assume . These informative priors were chosen as follows. For μ2 and μ3, the means of the untruncated distributions are set equal to the theoretical values for pure samples. For μ1 (μ4), the untruncated distribution is centered at the theoretical value for a loss (gain) of one copy. The lower endpoint of the support of μ4 is chosen to be 3σ3 units away from μ3 so that a small fraction of single-copy gains are erroneously classified as multiple-copy gains. The results are not sensitive to choices of τ1, τ2, and τ3 belonging to the interval [.5, 2]. Setting τ4 ≤ 2 guarantees sufficiently high prior probability to large values of μ4 associated with high-level amplifications. We set τ1 = τ2 = τ3 = 1 and τ4 = 2 in Sections 4 and 5.
Unlike a threshold-based approach for detecting changes in copy number, the constant ε determines the boundaries for the means μj rather than for the log2 ratios. These boundaries are not the same as threshold levels for detecting gains and losses. In fact, our assumptions allow positive log-intensity ratios for copy number losses, especially with large measurement errors, although μ1 itself cannot exceed −ε. In our analyses of actual array CGH data, we have found the results to be robust to choices of ε in the range [.05, .15]. This is shown in Section 5.2. For all our analyses, we set ε = .1.
For the measurement error precisions, we assume the priors for j = 1, 2, 3, and . For the states j = 1, 2, 3, the assumption is equivalent to σj < .41. This assumption is mild because typical array CGH data suggest much lower within-group variability for the states 1, 2, and 3. The support of is not bounded below because state 4 is an aggregation of multiple-copy gains, which usually results in a higher within-group variability (i.e., smaller precision).
We assume independent Dirichlet priors on ℜ4 for the rows of the stochastic matrix A, because this distribution has the set of all probability 4-tuples as its support. That is, with ai denoting the ith row of matrix A, we assume that , where i = 1, …, 4 and the constants {θij } are positive. As shown in Section 5.2, the results are not affected by the choices of θij that are small in comparison to n. We fixed the θij ’s equal to 1 in Sections 4 and 5.
The preceding priors are found to work consistently well for array CGH data. They are flexible enough to allow Bayesian learning and information sharing across the clones. We find in Sections 4 and 5 that the posterior inference is reliable and sensitive to the characteristics of the data.
3. CHARACTERIZING ARRAY CGH PROFILES
We rely on simulation-based methods for inference because the posterior distribution cannot be investigated by mathematical analysis or numerical integration. An efficient Metropolis-within-Gibbs algorithm for generating posterior samples of the parameters is given in the Appendix. The algorithm generates the parameters in blocks conditional on the remaining parameters and the data. The transition matrix A is generated using an independent-proposal Metropolis–Hastings algorithm. The copy number states are simulated by a stochastic version of the forward–backward algorithm (Chib 1996; Robert, Ryden, and Titterington 1999) that mixes faster than a Gibbs sampler (refer to Scott 2002). The remaining model parameters are generated by Gibbs sampling. The algorithm has been implemented using R and will soon be publicly available.
3.1 Classification Scheme
The generated copy number states represent draws from the marginal posterior of interest, [s1, …, sn | Y1, …, Yn]. For each Markov chain Monte Carlo (MCMC) draw, the generated states are inspected and, possibly nonexclusively, classified as focal aberrations, transition points, amplifications, outliers, and whole chromosomal changes. In the following discussion, altered state refers to a copy number state that is different from 2.
Focal aberrations (Fridlyand et al. 2004). Focal aberrations represent localized regions of altered copy number: (i) a single clone not belonging to a telomere having an altered state different from its neighbors; (ii) two clones belonging to a telomere sharing a common altered state, different from that of the adjacent clone to the telomere; or (iii) two or more adjacent clones mapped within 5 Mb (or any threshold representing a small region of the genome) and having a common altered state different from their neighbors. Focal aberrations are used to detect transition points and outliers (defined later).
Transition points. Transition points can be regarded as a property of the n − 1 interclonal spaces on the chromosome. An interclonal space is a transition point if it borders on two large regions associated with different copy number states. In contrast, focal aberrations represent small regions of altered copy number. A transition point is an interclonal space for which both of these conditions hold: (i) It is not adjacent to a telomere; and (ii) after excluding all focal aberrations on the chromosome, the neighboring clones on both sides of the interclonal space have different copy number states. Transition points differ from “segments” defined by the CBS (circular binary segmentation) algorithm of Olshen et al. (2004), an outstanding algorithm (refer to Lai et al. 2005) for analyzing array CGH data. The CBS algorithm segments clones regardless of their spacing on the chromosome. A transition point, on the other hand, is associated with large-scale regions of gains and losses and is declared only when the width of the altered region exceeds 5 Mb. For example, five contiguous clones that are highly amplified would generally be identified as a segment by the CBS algorithm (although there are examples in Sec. 4 where the procedure ignores obvious amplifications and deletions to control the false-positive rate). In contrast, if these five clones are located within 5 Mb, the proposed classification scheme labels them as focal aberrations rather than identify them as a separate region.
High-level amplifications. A clone for which sk = 4.
Outliers. An outlier is a focal aberration satisfying (i) sk = 1 and (Yk − μ1)/σ1 < −2, or (ii) sk = 3 and (Yk − μ3)/σ3 > 2. The first type of outliers could be associated with mutations on tumor suppressors and are labeled as deletions. The second type of outliers may be associated with oncogene mutations.
Whole chromosomal changes. The entire chromosome is identified as gained or lost if all the clones except the focal aberrations have altered copy number states.
3.2 Posterior Inference
For a given clone, the classification scheme of Section 3.1 results in a Bernoulli variable for each MCMC iterate and type of genomic alteration. For example, the kth clone is classified as a focal aberration (“1”) for some MCMC draws and as “0” for the remaining draws. The probability that this Bernoulli variable equals 1 is the posterior probability that clone Lk is a focal aberration. For a sufficiently large number of MCMC samples, the average of these binary outcomes is a simulation-consistent estimate of the posterior probability. Therefore, we declare clone Lk to be a focal aberration if this posterior probability exceeds .5. A similar method is used to identify deletions. Whole chromosomal changes correspond to a common Bernoulli outcome for all n clones. A chromosomal alteration is declared if the posterior probability of a chromosome-wide alteration exceeds .5.
High-level amplifications could be detected by a similar method. However, a more efficient method is available as a byproduct of the forward–backward algorithm, which computes the conditional probability that sk = 4 given the hyperparameters and the data. Averaging these conditional probabilities over the MCMC sample gives a simulation-consistent estimate of the posterior probability that clone Lk is a high-level amplification.
We have noticed a potential problem with identifying transition points based on the marginal posterior probabilities of the interclonal gaps. We recommend detecting the change points based on the configuration of change points having the highest joint posterior probability. Formally, let us write the configuration of change points as ν(s) = (g1, …, gn−1), where gj equals 1 if the j th interclonal gap is a change point, and equals 0 otherwise. Notice that the mapping from s to ν(s) is many–one. The posterior distribution of ν(s) is maximized to compute ν*, the configuration having the highest posterior probability. A simulation-consistent estimate of ν* is computed using the MCMC sample and is used to detect the transition points.
Summary tables and plots that are of direct interest to the biologist can now be constructed. Large-scale and localized regions of copy number change identified by the Bayesian HMM algorithm can be important tools for identifying candidate genes associated with cancer.
4. ILLUSTRATIONS
4.1 Pancreatic Adenocarcinoma Data
Pancreatic adenocarcinoma is among the most lethal of cancers. The disease is characterized by a high level of genomic instability from the earliest stages of the disease (Gisselsson et al. 2000, 2001; van Heek et al. 2002). Genomic changes identified in the progression of the disease include early-stage mutations in the oncogene KRAS and later-stage losses of the tumor suppressors p16INK4A, p53, and SMAD4 (Bardeesy and DePinho 2002). Using a variety of techniques ranging from karyotype analyses, CGH, and loss of heterozygosity mapping, frequent gains and losses have been mapped to regions on chromosomes 3–13, 17, 18, 21, and 22 (Johansson et al. 1992; Solinas-Toldo et al. 1997; Mahlamaki et al. 1997, 2002; Seymour et al. 1994; among many others).
Aguirre et al. (2004) studied the array CGH profiles of 24 pancreatic adenocarcinoma cell lines and 13 primary tumor specimens. In that article the profiles were individually analyzed using the CBS algorithm of Olshen et al. (2004), which segments the data and computes the within-segment means but does not detect gains or losses. The CBS algorithm was first run on the unnormalized log2 ratios to obtain the distribution of the within-segment means. The tallest mode of the distribution was subtracted from the data to compute the normalized log2 ratios, which are available at http://genomic.dfci.harvard.edu/array_cgh.htm. Setting thresholds in an ad hoc manner, Aguirre et al. (2004) declared normalized log2 ratios greater than .13 in magnitude as copy number changes (gains or losses), greater than .52 as high-level amplifications, and less than −.58 as deletions. They also defined objective criteria for comparing the copy number alterations of the 37 array CGH profiles. These criteria were applied to identify 54 frequently altered minimal common regions (MCRs) associated with pancreatic adenocarcinoma. In a subsequent study, candidate genes located within the MCRs were confirmed by the analysis of expression profiles.
We applied the Bayesian HMM algorithm to analyze these data and made comparisons with the CBS procedure. The complete set of results are presented in the supplementary materials. Throughout, the Bayesian HMM is found to perform reliably and compare favorably with the CBS procedure. We discuss a few examples here. Our primary reference for the MCRs associated with pancreatic cancer is Aguirre et al. (2004).
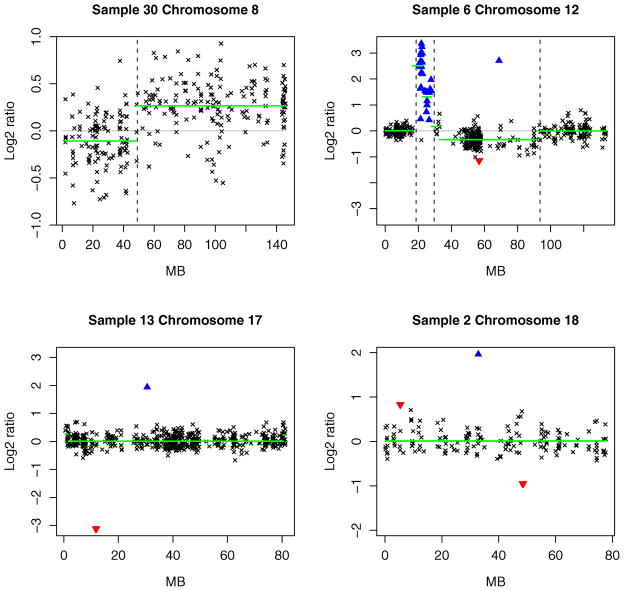
The upper left panel of Figure 2 displays the result for chromosome 8 of specimen 30. The bold horizontal lines represent the within-segment means computed by the CBS algorithm. The vertical lines correspond to the transition points identified by the Bayesian HMM. We find that both algorithms picked up the overall trend in the data. However, while the end user (often a biologist with relatively little statistical training) decides whether or not the CBS algorithm’s within-segment means correspond to copy number changes, the Bayesian HMM automatically identified the first region as primarily copy neutral and the second region as consisting of mainly single-copy gains.
Figure 2.
Array CGH profiles of some pancreatic cancer specimens. In each panel, the clonal distance (in Mb) from the p telomere has been plotted on the x axis. High-level amplifications and outliers are, respectively, indicated by ▲ and ▼. The broken vertical lines represent transition points. For comparison, the bold horizontal lines display the segment means computed by the CBS algorithm. See Section 4.1 for further discussion.
In the upper right panel of Figure 2, the CBS procedure declared the first set of high-intensity ratios on chromosome 12 of specimen 6 as two separate segments. This is because the CBS procedure identifies trends in the data. The Bayesian HMM, on the other hand, is motivated from the perspective of copy number change. It declared these clones as high-level amplifications and, therefore, as a single region. The next set of clones having lower log2 ratios were identified as focal aberrations because they are localized changes less than 2 Mb in width. The two amplified regions detected by the Bayesian HMM correspond to the two minimal common regions (MCRs) on chromosome 12 associated with copy number gains (see table 1 of Aguirre et al. 2004). The first MCR contains the KRAS2 gene, point mutations of which occur in more than 75% of pancreatic cancer cases (Almoguera et al. 1988). The CBS algorithm failed to detect the second MCR. This MCR has been biologically verified by Aguirre et al. (2004) using quantitative polymerase chain reaction (PCR) techniques.
The bottom left panel of Figure 2 displays the profile for chromosome 17 of specimen 13. The region from 17p13.3 to 17q11.1 (10.36–12.8 Mb) contains the tumor suppressors p53 and MKK4. Mutations on the gene p53 are found in at least 50% of pancreatic adenocarcinoma cases (Caldas et al. 1994). The single probe corresponding to this region was easily detected by the Bayesian HMM as a deletion. In contrast, the CBS algorithm effectively declared the entire chromosome as copy neutral.
The bottom right panel presents the array CGH profile of chromosome 18 of specimen 2. The Bayesian HMM algorithm detected an outlier associated with a copy number loss around 48 Mb. The outlier corresponds to the SMAD4 tumor suppressor gene located at 18q21, a mutation on which is associated with pancreatic cancer (Bardeesy and DePinho 2002). Aguirre and co-authors mentioned that the CBS procedure completely missed the well-established association with the SMAD4 gene, even though it was clearly visible in several specimens of the dataset.
The CBS procedure often ignores obvious single-probe aberrations to control the false discovery rate. Such errors can be misleading, because subsequent gene validation involves experimental techniques that are much more expensive than CGH. For this reason, single-probe aberrations that are frequently observed across tumor specimens provide one of the most cost-effective avenues for further research about the underlying causes of cancer. There are many other instances of the differences between the CBS and Bayesian HMM algorithms. For example, the MCR from 68.27 to 68.85 Mb on chromosome 12 maps to highly amplified clones in 34 out of 37 specimens (see the supplementary materials). In every case, the Bayesian HMM declared them as high-level amplifications, but the CBS procedure detected only the amplification in specimen 8. The Bayesian HMM also outperformed the CBS algorithm in detecting the mutation on gene FEZ1 in specimen 26 and on the genes OZF and AKT2 in specimen 6.
The results demonstrate that the Bayesian HMM is effective in detecting not only global trends but also highly localized changes in copy number. This feature is important in identifying genes associated with cancer (e.g., SMAD4 in the foregoing example) on which the point mutations do not become large-scale genomic changes as the disease progresses. Compared with other algorithms for analyzing array CGH data, the Bayesian HMM has potential as a diagnostic tool during the early stages of disease, when genomic alterations remain localized to relatively smaller regions of the genome.
4.2 Corriel Cell Lines
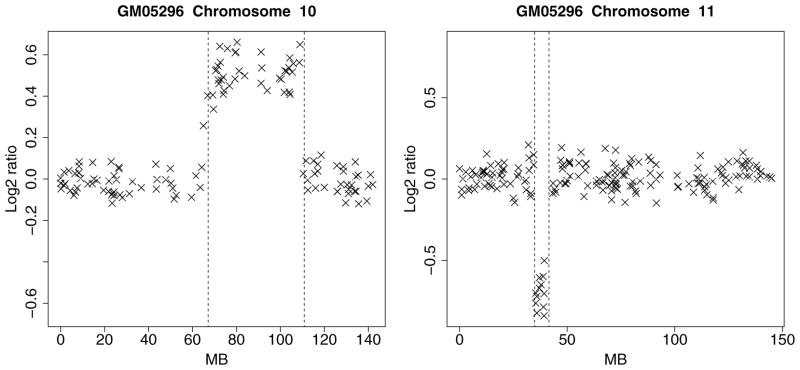
The Corriel cell line is widely regarded a “gold standard” dataset and analyzed in Snijders et al. (2001). The data, normalized to the genome-wide median log2 ratio, are available in tables E–H at http://www.nature.com/ng/journal/v29/n3/suppinfo/ng754_S1.html. A table of known karyotypes is presented in table I on the same website. We compared these cytogenically mapped alterations with the profiles produced by our algorithm and verified that the results match in all the specimens. For example, for cell line GM05296, table I reports a trisomy at 10q21–10q24 and a monosomy at 11p12–11p13. The array CGH profile for chromosomes 10 and 11 of cell line GM05296 are displayed in Figure 3. The regions of gain and loss identified by the Bayesian HMM match the karyotypes presented in table I. We omit the results for the other cell lines for brevity.
Figure 3.
Array CGH profile of chromosomes 10 and 11 of Corriel cell strain GM05296. The x axis displays the clonal distance (in Mb) from the p telomere. The broken vertical lines represent transition points.
4.3 Breast Cancer Data
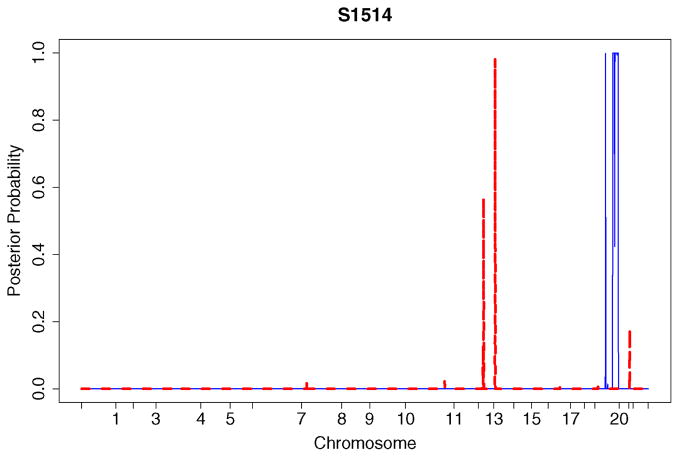
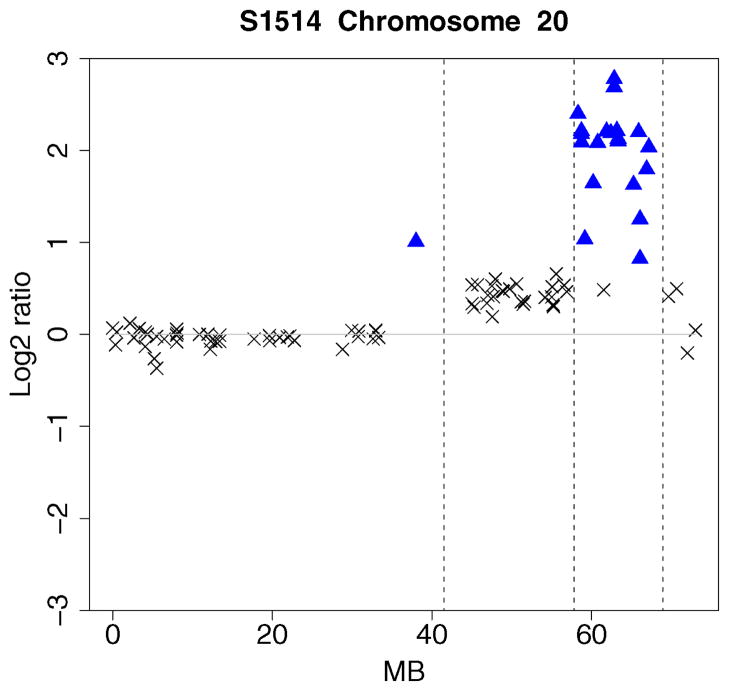
A useful feature of the Bayesian approach is that posterior probability plots can be created for the different kinds of genomic alterations. These plots provide a “bird’s eye view” of the copy number alterations. They are useful in identifying genomic regions associated with the disease. The procedure can be easily automated for a large number of genomic profiles. To illustrate, we analyzed the breast cancer data given in Snijders et al. (2001). The data were normalized by centering to the genome-wide median log2 ratios. The posterior probability plot for specimen S1514 is displayed in Figure 4. There are several high-level amplifications on chromosome 20 and deletions on chromosomes 13 and 14. Consistent with Figure 4, a region of high-level amplifications is seen on the array CGH profile of chromosome 20 in Figure 5.
Figure 4.
Posterior probabilities of genomic alterations for specimen S1514. The solid line represents high-level amplifications, whereas the dashed line corresponds to deletions. The numbers on the horizontal axis represent the q telomere of the chromosomes. The BACs are ordered by position in the genome beginning at 1p and ending at Xq.
Figure 5.
Array CGH profile of chromosome 20 of S1514. The x axis represents clonal distance (in Mb) from the p telomere. The broken vertical lines represent transition points. High-level amplifications are shown using ▲.
4.4 Comparisons With Some Existing Methods
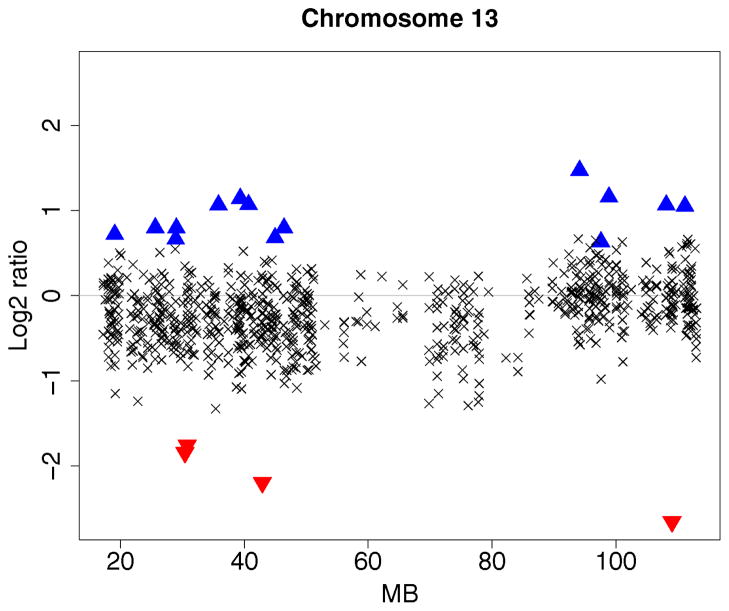
Using the glioblastoma multiforme data of Bredel el al. (2005), Lai et al. (2005) evaluated 11 array CGH algorithms based on segment detection as well as smoothing. The data were normalized using the Limma package (Smyth 2004) and are available at http://www.chip.org/~ppark/Supplements/Bioinformatics05b.html. Graphical summaries of the results are presented in that article as figures 3 and 4. Sample GBM31 (fig. 3 of Lai et al. 2005) exhibits a low signal-to-noise ratio. There is a large region of losses on chromosome 13. Lai and co-authors found that the algorithms CGHseg of Picard et al. (2005), GLAD of Hupe et al. (2004), CBS of Olshen et al. (2004), and GA of Jong et al. (2003) segmented chromosome 13 into two regions and detected the region of copy number loss. Smoothing-based methods like lowess, the quantreg algorithm of Eilers and de Menezes (2005), and the wavelet algorithm of Hsu et al. (2005) were sensitive to local trends but were less effective in detecting global trends. The HMM algorithm of Fridlyand et al. (2004) did not find any segments.
We followed an identical evaluation procedure to compare the Bayesian HMM with the aforementioned methods. Figure 6 displays the result for sample GBM31. The partitioned regions are the same as those identified by the CGHseg, CBS, GLAD, and GA algorithms. Local changes in the number of copies, identical to those collectively detected by the GLAD and CGH-seg algorithms, are marked as high-level amplifications (▲) and deletions (▼).
Figure 6.
Array CGH profile of chromosome 13 of GBM31. The clonal distance (in Mb) from the p telomere is plotted on the x axis. High-level amplifications and outliers are, respectively, indicated using ▲ and ▼. The broken vertical line represents a transition point.
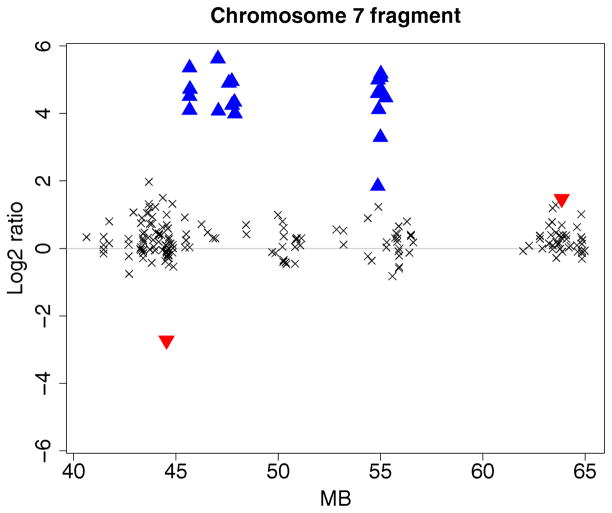
The second dataset investigated in Lai et al. (2005) is a fragment of chromosome 7 from sample GBM29 (refer to fig. 4 of that article). The data show some high log2 intensity ratios around the EGFR locus. The algorithms CGHseg, quantreg, GLAD, wavelet, and GA separated the data into three distinct amplification regions. The algorithms CBS, CLAC (Wang et al. 2005), and ACE (Lingjaerde et al. 2005) detected two distinct regions instead of three. ChARM (Myers et al. 2004) grouped all the high log2 intensity ratios into a single region. The HMM algorithm of Fridlyand et al. (2004) did not detect the amplifications.
Figure 7 displays the results for the Bayesian HMM algorithm. The high log2 ratios are identified as high-level amplifications (▲). Unlike the algorithms investigated in Lai et al. (2005), the single clone having a highly negative value is detected by the algorithm and marked as a deletion. The amplifications are identified as focal aberrations, rather than as separate regions, because both clusters are less than 5 Mb in width.
Figure 7.
Partial array CGH profile of chromosome 7 of GBM29. The clonal distance (in Mb) from the p telomere is plotted on the x axis. High-level amplifications and outliers are, respectively, indicated using ▲ and ▼.
We find that the Bayesian HMM algorithm combines the strength of the smoothing-based algorithms in detecting local features with the strength of the segmentation-based methods in detecting global trends. The reliability of the procedure is especially impressive with noisy data.
5. SIMULATION STUDIES
5.1 Comparison With Non-Bayesian HMM and CBS Algorithms
The frequentist analysis matching the foregoing Bayesian procedure estimates the hyperparameters of the likelihood using the Baum–Welch expectation-maximization (EM) algorithm, iteratively incrementing the likelihood until relative changes in the hyperparameters become sufficiently small. Conditional on the estimated hyperparameters, the Viterbi algorithm then computes the a posteriori most likely sequence of states s1, …, sn. Notice that this technique is different from the non-Bayesian HMM of Fridlyand et al. (2004). In particular, the latter method does not assign biological meanings to the latent states and cannot directly detect changes in copy number.
To find the global maximum in the 20-dimensional hyper-parameter space, the EM algorithm has to be run from several starting points. For typical array CGH data, each run often requires hundreds of iterations to converge. Because of this, the computational costs associated with the frequentist and Bayesian analyses are often comparable. When R is used as the computing platform, the CBS algorithm is considerably faster than either method. However, all three approaches are computationally feasible and have negligible costs compared to the many months of experimental effort required to process the tumor specimens.
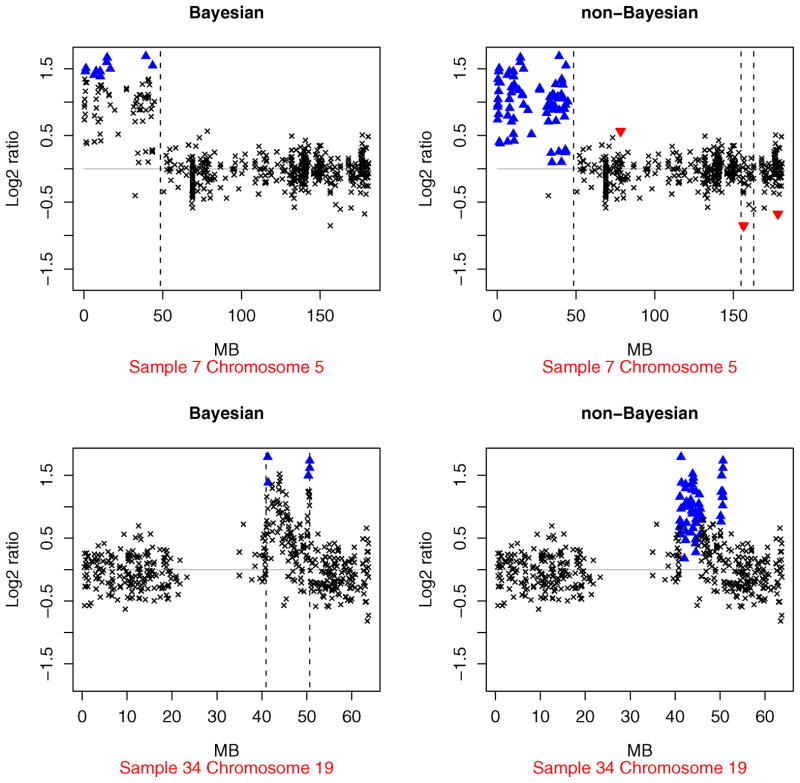
The non-Bayesian array CGH profiles for the Section 4.1 data are presented in the supplementary materials. A detailed comparison with the Bayesian profiles reveals that the two procedures often gave similar results. However, there are many profiles for which the answers are noticeably different. Examples of such chromosome–specimen pairs include (5, 2), (5, 7), (12, 10), (7, 13), (15, 13), (5, 19), (18, 31), and (19, 34). Two of the profiles are displayed in Figure 8. The non-Bayesian hyperparameter estimates correspond to a greater value of the likelihood function than the Bayes estimates in all these examples. However, the Bayesian profiles look more reasonable when we compare the smallest log2 ratios that are labeled as amplifications by the two methods.
Figure 8.
Examples from Section 4.1 where the Bayesian and non-Bayesian array CGH profiles are different. The upper panels correspond to chromosome 5 of sample 7, and the lower panels correspond to chromosome 19 of sample 34. The clonal distance (in Mb) from the p telomere has been plotted on the x axis. High-level amplifications and outliers are indicated using ▲ and ▼, respectively. The broken vertical lines represents transition points.
We performed a simulation study of the differences between the methods. For each of the aforementioned chromosome–specimen pairs, we obtained signal-to-noise ratios that were typical of array CGH data by setting the hyperparameters equal to their estimated values. Using the model described in Section 2.1, we then generated the underlying copy number states and log ratios for n = 200 clones. The Bayesian and non-Bayesian HMMs were applied to infer the latent copy number states. The procedure was independently replicated 100 times. Table 1 displays the percentage of correctly labeled copy number states for the two methods. The Bayesian HMM outperforms the non-Bayesian HMM in all the cases.
Table 1.
Estimated percentages of correctly discovered copy number states for the Bayesian and non-Bayesian methods, along with the estimated standard errors
| Source
|
Bayesian HMM
|
Non-Bayesian HMM
|
|||
|---|---|---|---|---|---|
| Chromosome | Specimen | % accuracy | SE | % accuracy | SE |
| 5 | 2 | 94.81 | .789 | 86.89 | 1.685 |
| 5 | 7 | 91.99 | 1.188 | 81.44 | 1.942 |
| 12 | 10 | 95.22 | .390 | 89.08 | 1.378 |
| 7 | 13 | 92.41 | 1.019 | 80.09 | 2.333 |
| 15 | 13 | 92.42 | 1.322 | 82.55 | 1.649 |
| 5 | 19 | 88.02 | 2.189 | 73.09 | 2.873 |
| 18 | 31 | 84.95 | 2.512 | 71.17 | 2.448 |
| 19 | 34 | 88.13 | 2.000 | 72.10 | 2.124 |
NOTE: The estimates were based on 100 independently generated datasets. The first two columns specify the chromosome and specimen numbers of the Section 4.1 dataset whose estimated hyperparameters were used to generate the data. See the text for an explanation.
Using eight randomly selected chromosome–specimen pairs, but an otherwise identical simulation strategy, Table 2 compares the CBS algorithm with the Bayesian and non-Bayesian HMMs. The method used by Aguirre et al. (2004) was applied to declare copy number gains and losses for the CBS algorithm. The Bayesian HMM outperforms the CBS algorithm, often substantially, in seven cases. The difference is inconclusive in one case. In six out of eight cases, the Bayesian HMM outperforms the non-Bayesian HMM, with the difference being inconclusive in one case. These results provide significant evidence in favor of the Bayesian HMM. We would like to emphasize here that the simulated data were generated from the Section 2.1 model, a strategy that is likely to favor the HMM-based methods. There may be alternative simulation procedures where the reliability of the CBS algorithm is greater than the proposed Bayesian HMM.
Table 2.
Estimated percentages of correctly discovered copy number states for the Bayesian and non-Bayesian methods, along with the estimated standard errors
| Source
|
Bayesian HMM
|
Non-Bayesian HMM
|
CBS
|
||||
|---|---|---|---|---|---|---|---|
| Chromosome | Specimen | % accuracy | SE | % accuracy | SE | % accuracy | SE |
| 13 | 33 | 94.38 | 1.203 | 72.01 | 2.634 | 67.72 | 3.512 |
| 19 | 4 | 88.20 | 1.129 | 87.94 | .534 | 75.36 | 1.726 |
| 14 | 1 | 87.35 | 1.893 | 76.47 | 1.834 | 86.70 | .426 |
| 12 | 17 | 80.84 | 1.736 | 76.11 | 1.453 | 44.12 | 1.791 |
| 1 | 24 | 40.64 | 2.512 | 54.31 | 1.460 | 35.37 | 2.470 |
| 3 | 35 | 96.03 | .239 | 72.06 | 2.509 | 92.43 | .488 |
| 23 | 12 | 74.31 | 3.417 | 65.2 | 2.420 | 58.08 | 3.311 |
| 15 | 34 | 90.79 | 2.164 | 68.3 | 2.798 | 55.22 | 4.175 |
NOTE: The estimates were based on 100 independently generated data sets. The first two columns specify the chromosome and specimen numbers of the Section 4.1 dataset whose estimated hyperparameters were used to generate the data. See the text for an explanation.
The proposed Bayesian HMM is found to benefit from the informative priors of Section 2.2. Prior knowledge about array CGH helps the procedure distinguish between competing sets of hyperparameter values that are almost equally plausible under the likelihood but not under the posterior. For example, consider the frequently encountered situation where very few log2 ratios are assigned to one or more copy number states. In such a situation, the likelihood alone may be unable to distinguish between the matching non-Bayesian HMM and a model having fewer than four states. This results in likelihood-based estimates where one or more of the μj ’s are approximately equal. Because of the well-defined meanings assigned to the four states of the HMM, the sequence of copy number states assigned by the non-Bayesian model often seem incorrect in such cases. The Bayesian approach is more robust in such situations. The informative priors prevent even states having very few probes and log2 ratios having a considerable amount of overlap due to high measurement error from being classified as a common state. For some data, a model having fewer states than four may be better fitting than the proposed model. However, the states might not have a simple biological interpretation in terms of copy number change. The detection of copy number gains and losses, which is one of the main goals of the analysis, may also be less straightforward.
Several examples in Section 4.1 suggest that our Bayesian HMM is better than the CBS algorithm in detecting amplifications that are localized to a small number of probes. This advantage is of practical importance, because single-probe amplifications frequently occurring across specimens are often the focus of future, more expensive gene validation studies. To investigate the difference by a controlled simulation, we independently generated 25 datasets using the following procedure: (i) Fifty out of n = 200 clones were randomly chosen to be amplifications having a mean signal of 2 on the log2 scale. (ii) The remaining clones were assumed to be copy neutral with a mean signal of 0. (iii) The data were generated by adding Gaussian noise with a standard deviation of .1 to these means.
The high signal-to-noise ratio (SNR) of 20 is atypical of array CGH data. The percentage of amplified probes (25%) is also very high. However, in spite of these features that simplify the detection of copy number change, the CBS algorithm failed to detect any amplification. The Bayesian HMM, on the other hand, correctly identified all the amplifications. Moreover, the false discovery rate of our Bayesian HMM was 0 for all the datasets and the average true discovery rate exceeded 99%.
5.2 Prior Sensitivity
The preceding analyses assumed that ε = .1 for the supports of the μj ’s (refer to Sec. 2.2) and that θij = 1 for the priors of the transition matrix rows, where i = 1, …, 4 and j = 1, …, 4. To alleviate concerns that the results are sensitive to the choice of ε, we generated 100 datasets with n = 500 clones each. For each dataset, the true means μ1, …, μ4 were uniformly generated from narrow intervals centered, respectively, at −.5, 0, .5, and 1. The standard deviations σj were uniformly generated in the interval [.2, .25], which is typical of noisy array CGH data. The true transition matrices were simulated as follows. For row 2 corresponding to the copy-neutral state, the off-diagonal elements were uniformly generated in the intervals [.01, .02]; for the remaining rows, the off-diagonal elements were uniformly generated in the intervals [.02, .05]. These nine elements uniquely determined the row-stochastic transition matrix. For k = 1, …, 500, the copy number states sk were then generated and the data were obtained by adding Gaussian noise to the means μsk.
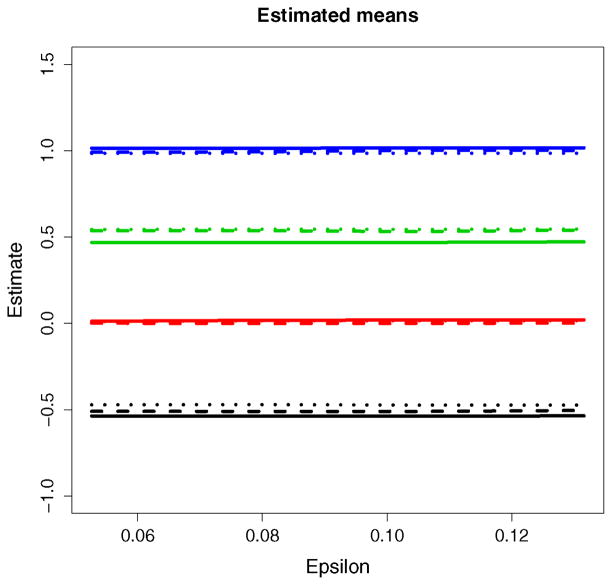
For ε belonging to a grid of points in the interval [.05, .15], the Bayesian HMM was used to analyze each simulated dataset. The posterior expectations of the means μj, the true discovery rates, and the false discovery rates were found to be robust to the choice of ε. Figure 9 plots the estimates of μ1, …, μ4 for three randomly chosen datasets as ε varies. The flatness of the lines provides evidence of the lack of sensitivity to ε ∈ [.05, .15]. The results were also found to be robust to {θij }i,j that were small compared to n.
Figure 9.
Estimated means Ê[μj |Y] for three independently generated datasets (shown by solid, dashed, and dotted lines) plotted against ε.
6. CONCLUSIONS
We propose a Bayesian hierarchical approach relying on a hidden Markov model for analyzing array CGH data. The informative priors allow Bayesian learning from the data. One of the strengths of the fully automated approach is the ability to detect copy number changes like gains, losses, amplifications, outliers, and transition points based on the posterior. Summaries of the array CGH profiles are generated. The profiles can then be compared across individuals to identify the genomic alterations involved in the disease pathogenesis. Recent research (Freeman et al. 2006) suggests that such comparisons must adjust for copy number variation in “normal” human DNA outside of cancer.
The examples of Section 4 demonstrate the reliability of our Bayesian HMM. The sensitivity of the algorithm to individual probes often allows us to find candidate genes that are missed by other algorithms. The performance of the algorithm is impressive not only for the “gold-standard” Corriel cell lines but also for the glioblastoma dataset of Bredel el al. (2005) having high measurement error. Combined with the results presented in Lai et al. (2005), the latter analysis reveals a very favorable comparison with outstanding algorithms like those of Pi-card et al. (2005) and Olshen et al. (2004). Section 5 compares our Bayesian HMM and alternative algorithms using controlled simulations. The results confirm the accuracy of the approach.
A strength of our Bayesian HMM is that it relies on essentially no tuning parameters. Unlike many other algorithms (see Lai et al. 2005), the user is only required to input the normalized log2 ratios. This is a convenient feature for the end user with little or no statistical training. In all our analyses, we have used the default parameterizations specified in Section 2.2. Certain features of the Bayesian HMM may be changed to produce a different result. Possible features include the constant ε in the prior specification of the means μj and the constants θij in the transition matrix priors in Section 2.2. However, the simulation study in Section 5.2 and our own experience with the algorithm indicate that the results are robust to variations in these quantities. The informative priors for the means μj substantially influence the results, as we find in Section 5.1 on comparing our Bayesian HMM with the matching non-Bayesian model. However, the order constraints on the μj ’s and the biological meanings assigned to sk ∈ {1, 2, 3, 4} allow the specification of priors that work consistently well across different datasets. For this reason, we recommend using the default parameterizations of our Bayesian HMM for most array CGH applications.
Acknowledgments
This material is based on work supported by the National Institute of Health under award R01CA95747. The first author thanks Professors Steven MacEachern, Louise Ryan, and David Harrington for many insightful comments that helped improve the focus of the article.
APPENDIX: AN MCMC ALGORITHM
The following algorithm is independently run for each chromosome to generate an MCMC sample for the chromosomal parameters. We group the model parameters into four blocks, namely, B1 = A, B2 = (s1, …, sn), B3 = (μ1, μ3, μ4), and . The starting values of the parameters are generated from the priors. The algorithm iteratively generates each of the four blocks conditional on the remaining blocks and the data. Let denote the values of the blocks at the (v − 1)st iteration. In the next iteration, the blocks are generated as follows.
Updating Block B1
The transition matrix is generated using a Metropolis–Hastings step because the normalizing constant of the full conditional cannot be computed in closed form. This step makes independent proposals from a distribution that closely approximates the full conditional of the transition matrix. The proposal is accepted or rejected with a probability that compensates for the approximation. Typically, most of the Metropolis–Hastings proposals are accepted. Using the copy number states generated at iteration v − 1, we compute the number of transitions from state i to state j, denoted by , where i, j = 1, …, 4. We generate a proposal C for the transition matrix from the distributions , where row i = 1, …, 4 and B−1 denotes the blocks, {B2, B3, B4}. The proposal ignores the marginal distribution of state s1 and so it differs from the full conditional of the transition matrix. To compensate for this, we accept the proposal (in other words, set A(v) = C) with probability β, where , and otherwise reject the proposal (in other words, set A(v) = A(v−1)). As defined earlier, πD(s) denotes the probability of state s under the stationary distribution of a given transition matrix D.
Updating Block B2
The copy number states are generated by a stochastic version of the forward–backward algorithm. We compute the distribution [sn | B−2, Y1, …, Yn] at the beginning of the backward step. We generate sn from this distribution. The backward step is continued to compute and generate a draw from the distribution [sn−1 | sn, B−2, Y1, …, Yn]. The sequence of computing and generating a draw from [sk | sk+1, B−2, Y1, …, Yn] is iterated for k = n − 2 down to k = 1. This produces a sample from the joint distribution [s1, …, sn | B−2, Y1, …, Yn].
Updating Block B3
For s = 1, …, 4, let δ0s be the center of the untruncated normal distribution in the prior specification of μs. Compute the sums , averages , precisions , and weighted means . For s = 1, …, 4, generate , where the intervals Is denote the support of the μs (see prior specification).
Updating Block B4
For j = 1, …, 4, compute and . Generate
Contributor Information
Subharup Guha, Email: GuhaSu@missouri.edu, Department of Statistics, University of Missouri–Columbia, Columbia, MO 65211.
Yi Li, Email: yili@hsph.harvard.edu, Department of Biostatistics, Harvard School of Public Health, Boston, MA 02115.
Donna Neuberg, Email: neuberg@hsph.harvard.edu, Department of Biostatistics, Harvard School of Public Health, Boston, MA 02115.
References
- Aguirre AJ, Brennan C, Bailey G, Sinha R, Feng B, Leo C, Zhang Y, Zhang J, Gans JD, Bardeesy N, Cauwels C, Cordon-Cardo C, Redston MS, DePinho RA, Chin L. High-Resolution Characterization of the Pancreatic Adenocarcinoma Genome. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:9067–9072. doi: 10.1073/pnas.0402932101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almoguera C, Shibata D, Forrester K, Martin J, Arnheim N, Perucho M. Most Human Carcinomas of the Exocrine Pancreas Contain Mutant c-K-ras Genes. Cell. 1988;53:549–554. doi: 10.1016/0092-8674(88)90571-5. [DOI] [PubMed] [Google Scholar]
- Bardeesy N, DePinho RA. Pancreatic Cancer Biology and Genetics. Nature Reviews Cancer. 2002;2:897–909. doi: 10.1038/nrc949. [DOI] [PubMed] [Google Scholar]
- Bredel M, Bredel C, Juric D, Harsh GR, Vogel H, Recht LD, Sikic BI. High-Resoluton Genome-Wide Mapping of Genetic Alternations in Human Glial Brain Tumors. Cancer Research. 2005;65:4088–4096. doi: 10.1158/0008-5472.CAN-04-4229. [DOI] [PubMed] [Google Scholar]
- Brennan C, Zhang Y, Leo C, Feng B, Cauwels C, Aguirre AJ, Kim M, Protopopov A, Chin L. High-Resolution Global Profiling of Genomic Alterations With Long Oligonucleotide Microarray. Cancer Research. 2004;64:4744–4748. doi: 10.1158/0008-5472.CAN-04-1241. [DOI] [PubMed] [Google Scholar]
- Brown CS, Goodwin PC, Sorger PK. Image Metrics in the Statistical Analysis of DNA Microarray Data. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:8944–8949. doi: 10.1073/pnas.161242998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caldas C, Hahn SA, da Costa LT, Redston MS, Schutte M, Seymour AB, Weinstein CL, Hruban RH, Yeo CJ, Kern SE. Frequent Somatic Mutations and Homozygous Deletions of the p16 (MTS1) Gene in Pancreatic Adenocarcinoma. Nature Genetics. 1994;8:27–32. doi: 10.1038/ng0994-27. [DOI] [PubMed] [Google Scholar]
- Cheng C, Kimmel R, Neiman P, Zhao LP. Array Rank Order Regression Analysis for the Detection of Gene Copy-Number Changes in Human Cancer. Genomics. 2003;82:122–129. doi: 10.1016/s0888-7543(03)00122-8. [DOI] [PubMed] [Google Scholar]
- Chib S. Calculating Posterior Distributions and Modal Estimates in Markov Mixture Models. Journal of Econometrics. 1996;75:79–97. [Google Scholar]
- Durbin R, Eddy S, Krogh A, Michison G. Biological Sequence Analysis. New York: Cambridge University Press; 1998. [Google Scholar]
- Eilers PHC, de Menezes RX. Quantile Smoothing of Array CGH Data. Bioinformatics. 2005;21:1146–1153. doi: 10.1093/bioinformatics/bti148. [DOI] [PubMed] [Google Scholar]
- Freeman JL, Perry GH, Feuk L, Redon R, McCarroll SA, Altshuler DM, Hiroyuki A, Jones KW, Tyler-Smith C, Hurles ME, Carter NP, Scherer SW, Lee C. Copy Number Variation: New Insights in Genome Diversity. Genome Research. 2006;16:949–961. doi: 10.1101/gr.3677206. [DOI] [PubMed] [Google Scholar]
- Fridlyand J, Snijders AM, Pinkel D, Albertson DG, Jain AN. Application of Hidden Markov Models to the Analysis of the Array CGH Data. Journal of Multivariate Analysis. 2004;90:132–153. [Google Scholar]
- Gisselsson D, Jonson T, Petersen A, Strombeck B, Dal Cin P, Hoglund M, Mitelman F, Mertens F, Mandahl N. Telomere Dysfunction Triggers Extensive DNA Fragmentation and Evolution of Complex Chromosome Abnormalities in Human Malignant Tumors. Proceedings of the National Academy of Sciences of the United States America. 2001;98:12683–12688. doi: 10.1073/pnas.211357798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gisselsson D, Pettersson L, Hoglund M, Heidenblad M, Gorunova L, Wiegant J, Mertens F, Dal Cin P, Mitelman F, Mandahl N. Chromosomal Breakage-Fusion-Bridge Events Cause Genetic Intratumor Heterogeneity. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:5357–5362. doi: 10.1073/pnas.090013497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgson G, Hager J, Volik S, Hariono S, Wernick M, Moore D, Nowak N, Albertson D, Pinkel D, Collins C, et al. Genome Scanning With Array CGH Delineates Regional Alterations in Mouse Islet Carcinomas. Nature Genetics. 2001;929:459–464. doi: 10.1038/ng771. [DOI] [PubMed] [Google Scholar]
- Huang T, Wu B, Lizardi P, Zhao H. Detection of DNA Copy Number Alterations Using Penalized Least Squares Regression. Bioinformatics. 2005;21:3811–3817. doi: 10.1093/bioinformatics/bti646. [DOI] [PubMed] [Google Scholar]
- Hupe P, Stransky N, Thiery J-P, Radvanyi F, Barillot E. Analysis of Array CGH Data: From Signal Ratio to Gain and Loss of DNA Regions. Bioinformatics. 2004;20:3413–3422. doi: 10.1093/bioinformatics/bth418. [DOI] [PubMed] [Google Scholar]
- Hsu L, Self SG, Grove D, Randolph T, Wang K, Delrow JJ, Loo L, Porter P. Denoising Array-Based Comparative Genomic Hybridization Data Using Wavelets. Biostatistics. 2005;6:211–226. doi: 10.1093/biostatistics/kxi004. [DOI] [PubMed] [Google Scholar]
- Johansson B, Bardi G, Heim S, Mandahl N, Mertens F, Bak-Jensen E, Andren-Sandberg A, Mitelman F. Nonrandom Chromosomal Rearrangements in Pancreatic Carcinomas. Cancer. 1992;69:1674–1681. doi: 10.1002/1097-0142(19920401)69:7<1674::aid-cncr2820690706>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- Jong K, Marchiori E, Vaart A, Ylstra B, Weiss M, Meijer G. Applications of Evolutionary Computing: Evolutionary Computation and Bioinformatics. Vol. 2611. New York: Springer-Verlag; 2003. Chromosomal Breakpoint Detection in Human Cancer; pp. 54–65. [Google Scholar]
- Kallioniemi A, Kallioniemi OP, Sudar D, Rutovitz D, Gray JW, Waldman F, Pinkel D. Comparative Genomic Hybridization for Molecular Cytogenetic Analysis of Solid Tumors. Science. 1992;258:818–821. doi: 10.1126/science.1359641. [DOI] [PubMed] [Google Scholar]
- Karlin S, Taylor HM. A First Course in Stochastic Processes. 2. New York: Academic Press; 1975. [Google Scholar]
- Khojasteh M, Lam WL, Ward RK, MacAulay C. A Stepwise Framework for the Normalization of Array CGH Data. BMC Bioinformatics. 2005;6:274. doi: 10.1186/1471-2105-6-274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai W, Johnson MJ, Kucherlapati R, Park PJ. Comparative Analysis of Algorithms for Identifying Amplifications and Deletions in Array CGH Data. Bioinformatics. 2005;21:3763–3770. doi: 10.1093/bioinformatics/bti611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingjaerde OC, Baumbusch LO, Liestol K, Glad IK, Borresen-Dale A-L. CGH–Explorer: A Program for Analysis of Array-CGH Data. Bioinformatics. 2005;21:821–822. doi: 10.1093/bioinformatics/bti113. [DOI] [PubMed] [Google Scholar]
- MacDonald IL, Zucchini W. Hidden Markov and Other Models for Discrete-Value Time Series. Boca Raton, FL: Chapman & Hall; 1997. [Google Scholar]
- Mahlamaki EH, Barlund M, Tanner M, Gorunova L, Hoglund M, Karhu R, Kallioniemi A. Frequent Amplification of 8q24, 11q, 17q, and 20q-Specific Genes in Pancreatic Cancer. Genes Chromosomes Cancer. 2002;35:353–358. doi: 10.1002/gcc.10122. [DOI] [PubMed] [Google Scholar]
- Mahlamaki EH, Hoglund M, Gorunova L, Karhu R, Dawiskiba S, Andren-Sandberg A, Kallioniemi OP, Johansson B. Comparative Genomic Hybridization Reveals Frequent Gains of 20q, 8q, 11q, 12p, and 17q, and Losses of 18q, 9p, and 15q in Pancreatic Cancer. Genes Chromosomes Cancer. 1997;20:383–391. doi: 10.1002/(sici)1098-2264(199712)20:4<383::aid-gcc10>3.0.co;2-o. [DOI] [PubMed] [Google Scholar]
- McLachlan GJ, Do K-A, Ambroise C. Analyzing Microarray Gene Expression Data. Hoboken, NJ: Wiley; 2004. [Google Scholar]
- Myers CL, Dunham MJ, Kung SY, Troyanskaya OG. Accurate Detection of Aneuploidies in Array CGH and Gene Expression Microarray Data. Bioinformatics. 2004;20:3533–3543. doi: 10.1093/bioinformatics/bth440. [DOI] [PubMed] [Google Scholar]
- Olshen AB, Venkatraman ES, Lucito R, Wigler M. Circular Binary Segmentation for the Analysis of Array-Based DNA Copy Number Data. Biostatistics. 2004;4:557–572. doi: 10.1093/biostatistics/kxh008. [DOI] [PubMed] [Google Scholar]
- Pasternak JJ. An Introduction to Human Molecular Genetics: Mechanism of Inherited Diseases. Bethesda, MD: Fitzgerald Science Press; 1999. [Google Scholar]
- Picard F, Robin S, Lavielle M, Vaisse C, Daudin J-J. A Statistical Approach for Array CGH Data Analysis. BMC Bioinformatics. 2005;6:27. doi: 10.1186/1471-2105-6-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinkel D, Albertson DG. Array Comparative Genomic Hybridization and Its Applications in Cancer. Nature Genetics. 2005;37:11–17. doi: 10.1038/ng1569. [DOI] [PubMed] [Google Scholar]
- Pinkel D, Segraves R, Sudar D, Clark S, Poole I, Kowbel D, Collins C, Kuo W, Chen C, Zhai Y, et al. High Resolution Analysis of DNA Copy Number Variation Using Comparative Genomic Hybridization to Microarrays. Nature Genetics. 1998;20:207–211. doi: 10.1038/2524. [DOI] [PubMed] [Google Scholar]
- Pollack J, Sorlie T, Perou C, Rees C, Jeffrey S, Lonning P, Tibshirani R, Botstein D, Borresen-Dale A, Brown P. Microarray Analysis Reveals a Major Direct Role of DNA Copy Number Alteration in the Transcriptional Program of Human Breast Tumors. Proceedings of the National Academy of Sciences, USA. 2002;99:12963–12968. doi: 10.1073/pnas.162471999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollack JR, Perou CM, Alizadeh AA, Eisen MB, Pergamenschikov A, Williams CF, Jeffrey SS, Botstein D, Brown PO. Genome-Wide Analysis of DNA Copy-Number Changes Using cDNA Microarrays. Nature Genetics. 1999;23:41–46. doi: 10.1038/12640. [DOI] [PubMed] [Google Scholar]
- Rabiner LR. A Tutorial on Hidden Markov Models and Selected Applications in Speech Recognition. Proceedings of the IEEE. 1989;77:257–286. [Google Scholar]
- Robert CP, Ryden T, Titterington DM. Convergence Controls for MCMC Algorithms, With Applications to Hidden Markov Chains. Journal of Statistical Computing and Simulation. 1999;64:327–355. [Google Scholar]
- Scott S. Bayesian Methods for Hidden Markov Models: Recursive Computing in the 21st Century. Journal of the American Statistical Association. 2002;97:337–351. [Google Scholar]
- Seymour AB, Hruban RH, Redston M, Caldas C, Powell SM, Kinzler KW, Yeo CJ, Kern SE. Allelotype of Pancreatic Adenocarcinoma. Cancer Research. 1994;54:2761–2764. [PubMed] [Google Scholar]
- Smyth GK. Linear Models and Empirical Bayes Methods for Assessing Differential Expression in Microarray Experiments. Statistical Applications in Genetics and Molecular Biology. 2004;3:article 3. doi: 10.2202/1544-6115.1027. [DOI] [PubMed] [Google Scholar]
- Snijders AM, Nowak N, Segraves R, Blackwood S, Brown N, Conroy J, Hamilton G, Hindle AK, Huey B, Kimura K, Law S, Myambo K, Palmer J, Ylstra B, Yue JP, Gray JW, Jain AN, Pinkel D, Albertson DG. Assembly of Microarrays for Genome-Wide Measurement of DNA Copy Number. Nature Genetics. 2001;29:4370–4379. doi: 10.1038/ng754. [DOI] [PubMed] [Google Scholar]
- Solinas-Toldo S, et al. Matrix-Based Comparative Genomic Hybridization: Biochips to Screen for Genomic Imbalances. Genes Chromosomes Cancer. 1997;20:399–407. [PubMed] [Google Scholar]
- van Heek NT, Meeker AK, Kern SE, Yeo CJ, Lillemoe KD, Cameron JL, Offerhaus GJ, Hicks JL, Wilentz RE, Goggins MG, et al. Telomere Shortening Is Nearly Universal in Pancreatic Intraepithelial Neoplasia. American Journal of Pathology. 2002;161:1541–1547. doi: 10.1016/S0002-9440(10)64432-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P, Kim Y, Pollack J, Balasubramanian N, Tibshirani R. A Method for Calling Gains and Losses in Array CGH Data. Biostatistics. 2005;6:45–58. doi: 10.1093/biostatistics/kxh017. [DOI] [PubMed] [Google Scholar]