Introduction
The circuitry of the human brain is composed of a trillion (1012) neurons and a quadrillion (1015) synapses, whose connectivity underlies all human perception, emotion, thought and behavior. Studies in a range of species have revealed that the overall structure of the nervous system is genetically hard-wired but that neural circuits undergo extensive sculpting and re-wiring in response to a variety of stimuli. This process of experience-dependent changes in synaptic connectivity is called synaptic plasticity.
Studies of synaptic plasticity have begun to detail the molecular mechanisms that underlie these synaptic changes. This research has examined a variety of cell biological processes, including synaptic vesicle release and recycling, neurotransmitter receptor trafficking, cell adhesion and stimulus-induced changes in gene expression within neurons. Taken together, these studies have provided an initial molecular biological understanding of how nature and nurture combine to determine our identities. As such, research on synaptic plasticity promises to provide insight into the biological basis of many neuropsychiatric disorders in which experience-dependent brain rewiring goes awry.
Hippocampal Synaptic Plasticity
The successful study of the cell biology of synaptic plasticity requires a tractable experimental model system. Ideally, such a model should consist of a defined population of identifiable neurons and be amenable to electrophysiological, genetic and molecular cell biological manipulations. A well-studied model system for studying plasticity in the adult vertebrate nervous system is the rodent hippocampus (Figure 1). Critical for memory formation, the anatomy of the hippocampus renders it particularly suitable for electrophysiological investigation. It consists of three sequential synaptic pathways (perforant, mossy fiber and Schaffer collateral pathways), each with discrete cell body layers and axonal and dendritic projections (Figure 1). Synaptic plasticity has been studied in all three hippocampal pathways in in vivo and in vitro preparations. Distinct stimuli elicit changes in synaptic efficacy; high frequency stimuli produce synaptic strengthening called long-term potentiation (LTP) and low frequency stimulation has been shown to produce synaptic weakening, called long-term depression (LTD). Further, different patterns of stimulation elicit changes in synaptic strength that persist over various time domains, with long-lasting forms, but not short-term forms, requiring new RNA and protein synthesis (1).
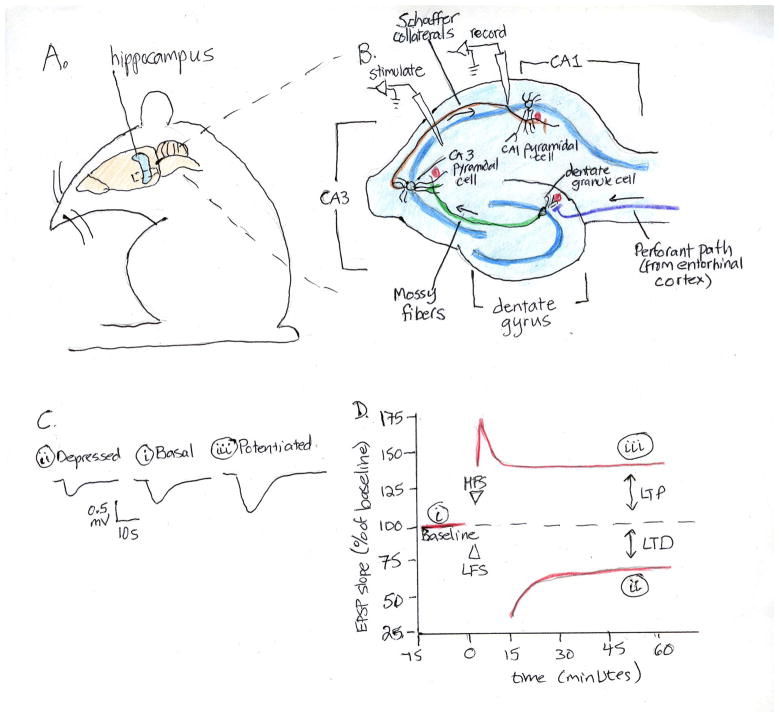
Figure 1. Hippocampal Synaptic Plasticity.
A. The rodent hippocampus can be dissected out and cut into transverse slices (B) preserving all three synaptic pathways. In the perforant pathway (purple), axons from the entorhinal cortex project to form synapses (red circles) on dendrites of dentate granule cells; in the mossy fiber pathway (green), dentate granule axons synapse on CA3 pyramidal neuron dendrites; and in the Schaffer collateral pathway (brown), CA3 axons synapse on CA1 dendrites. The dentate, CA3 and CA1 cell bodies form discrete somatic layers (dark blue lines), projecting axons and dendrites into defined regions. Electrodes can be used to stimulate axonal afferents and record from postsynaptic follower cells, as illustrated for the Schaffer collateral (CA3-CA1) pathway. Test stimuli elicit a stable synaptic response in the follower cell, measured as a excitatory post-synaptic potentials (EPSP, Ci). Trains of high frequency stimulation (HFS) or low frequency stimulation (LFS) to the axonal fibers produce sustained increases or decreases, respectively of the EPSP amplitude in response to subsequent test stimuli (C and D). These forms of plasticity are known as long-term potentiation (LTP) and long-term depression (LTD). Figure adapted from (56).
Hippocampal plasticity can be studied in in vivo and in vitro preparations. Implanted electrodes can be used to stimulate and record from hippocampal pathways in living animals. The hippocampus can be dissected out of the brain and cut into 300–500 micron thick transverse slices that can be maintained and recorded from for hours (Figure 1). Slices can also be kept as organotypic slice cultures for weeks, preserving many aspects of their architecture. Finally, hippocampal neurons can be studied in dissociated cultures, which are particularly amenable to manipulation and dynamic imaging of individual neurons and synapses. The development of genetically modified mice and vectors for acute manipulation of gene expression complete a rich tool-kit for studies of the cell and molecular biology of hippocampal synaptic plasticity.
This review focuses on long-lasting forms of plasticity that underlie learning and memory. We will consider, in turn, each component of the synapse: the presynaptic compartment, the postsynaptic compartment, and the synaptic cleft. In each case, we will discuss processes that undergo activity-dependent modifications to alter synaptic efficacy. Long-lasting changes in synaptic connectivity require new RNA and/or protein synthesis and so we then turn our attention to how gene expression is regulated within neurons. We will concentrate on studies of learning-related plasticity in the rodent hippocampus since these provide the most extensive evidence for the cell biological mechanisms of plasticity in the vertebrate brain. We will also limit our discussion to plastic changes at excitatory chemical synapses (though neurons also communicate at inhibitory and modulatory chemical synapses, and at electrical synapses, all of which show plasticity).
Presynaptic Mechanisms of Plasticity
Communication at chemical synapses involves the release of neurotransmitter from the presynaptic terminal, diffusion across the cleft, and binding to post-synaptic receptors (Figures 2 and 3). Chemical neurotransmission is rapid (occurring in milliseconds) and highly regulated. The presynaptic terminal contains synaptic vesicles filled with neurotransmitter and a dense matrix of cytoskeleton and scaffolding proteins at the site of release, called the active zone. Varying the probability of neurotransmitter release, thereby varying the amount of transmitter released, provides one mechanism for altering synaptic strength during neuronal plasticity.
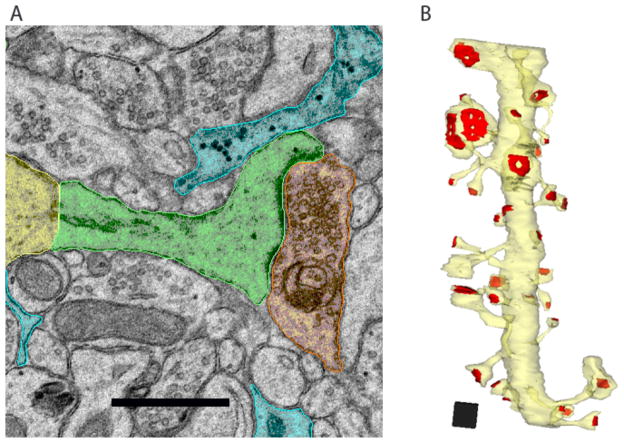
Figure 2. The Ultrastructure of the Synapse.
Neurons communicate with one another at chemical synapses. A) In this electron micrograph from area CA1 in adult rat hippocampus, the CA1 dendritic shaft is colorized in yellow, the spine neck and head in green, the presynaptic terminal in orange, and astroglial processes in blue. Scale bar, 0.5 μm. B) Three-dimensional reconstruction of an 8.5 μm long dendrite (yellow) with the PSDs labeled in red. Note the variation in spine and PSD size and shape. Scale cube, 0.5 μm3. Reproduced with permission from Bourne and Harris (57).
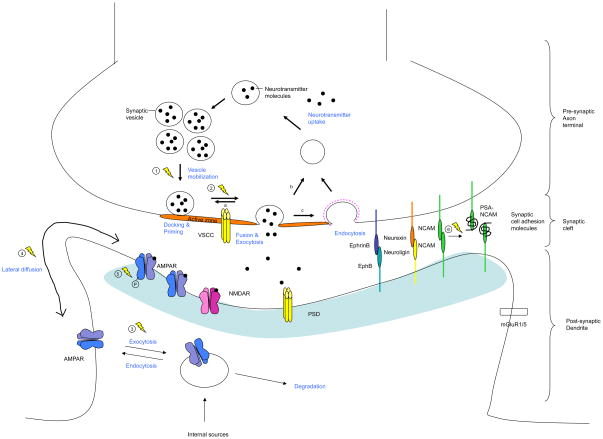
Figure 3. Activity-dependent modulation of pre-, post- and trans-synaptic components.
Presynaptic: Neurotransmitter vesicle cycling. Neurotransmitter release starts with the filling of synaptic vesicles. Filled vesicles then dock and undergo priming at the active zone. Arrival of an action potential induces calcium influx through voltage-sensitive calcium channel (VSCC), which in turn triggers membrane fusion and exocytosis of the neurotransmitters. The synaptic vesicles are then recycled via local reuse (a; also called kiss and stay), fast recycling (b; also called kiss and run), or clathrin-mediated endocytosis (c). The efficacy of the neurotransmitter release can be regulated during plasticity as exemplified by the regulation of synapsin phosphorylation (1) and the regulation of RIM protein phosphorylation (2). Postsynaptic: AMPA receptor trafficking. Locally and somatically synthesized AMPARs enter a pool of endosomes that cycle constitutively and in an activity-dependent manner. During potentiation, greater receptor insertion (3) increases the concentration of AMPARs at the synapse, where they are anchored by interactions at the PSD. During synaptic depression, AMPARs are endocytosed (3). The preferential location of endocytosis and exocytosis is not precisely known, but probably occurs extrasynaptically. Within the plasma membrane, trafficking of AMPARs between the synapse and the point of insertion/removal occurs by lateral diffusion. Extrasynaptic movement of AMPARs increases with neuronal activity (4). Receptor trafficking is modulated by phosphorylation of AMPAR subunits (5), which influences interactions with scaffolding proteins. Trans-synaptic: Synaptic cell adhesion molecules. PSA-NCAM is increased following neuronal activity (6). Lightning bolts indicate activity-dependent processes.
Synaptic vesicle release can be subdivided into distinct steps, including vesicle mobilization, docking, priming, fusion and recycling. While each of these steps may be regulated in an activity-dependent manner, we will highlight three: vesicle mobilization, docking and priming.
Synapsins and synaptic vesicle mobilization
The population of synaptic vesicles within a presynaptic terminal exist in three states, the readily releasable pool docked at the active zone (~1% of synaptic vesicles); the recycling pool, which can be released with moderate stimulation (~15%); and the reserve pool, which is only released in response to strong stimuli (~85%). A family of phosphoproteins called synapsins tether synaptic vesicles to the actin cytoskeleton and to one another. Neuronal stimulation activates kinases that phosphorylate synapsins to modulate synaptic vesicle tethering (2). In this way, activity-dependent regulation of synapsin phosphorylation alters the number of synaptic vesicles available for release. Synapsin knockout mice have significantly reduced reserve pools of synaptic vesicles, and demonstrate deficits in learning and memory as well as various forms of plasticity (3), indicating that activity-dependent modulation of synaptic vesicle mobilization is critical to neuronal and behavioral plasticity.
RIM proteins and synaptic vesicle docking and priming
For synaptic vesicles to become fusion-competent, they must undergo docking and priming, in which vesicle and plasma membrane Soluble NSF-Attachment Protein Receptor (SNARE) proteins are brought into close contact to allow rapid fusion following calcium influx. The Rab3-interacting molecule (RIM) family of proteins has recently been shown to be critical for this process (4). A large, multi-domain protein, RIM clusters calcium channels in the active zone (5) and interacts with the Munc-13 protein (6). The latter is required for efficient SNARE complex formation and membrane fusion. RIM is a substrate for phosphorylation by Protein Kinase A (PKA). Genetic knockout experiments have shown that RIM is required for mossy fiber LTP (7). Together, these findings suggest that RIM proteins regulate not only the coupling between calcium influx and vesicle release, but also vesicle docking and priming.
Postysynaptic Mechanisms of Plasticity
Following release from the presynaptic terminal, neurotransmitter diffuses across the synaptic cleft to bind to receptors on the post-synaptic side of the synapse. Most postsynaptic principle neurons in the brain are studded with membrane protuberances called dendritic spines, which are the post-synaptic compartments. The shape of spines is somewhat heterogeneous (Figure 2), but consists of a bulbous head and a thinner neck that connects the spine to the dendritic shaft; the size of the spine head and the volume of the spine correlates with synaptic strength (8, 9). Spines serve as compartmentalized signaling units, and the number and shape of spines has been shown to change during synaptic plasticity (10). At the ultrastructural level, the postsynaptic compartment is characterized by an electron-dense post-synaptic density (PSD), which consists of neurotransmitter receptors and an extensive network of scaffolding proteins.
Activation of post-synaptic kinases in the spine: CamKII and PKMζ
LTP and LTD induction are both dependent on postsynaptic elevations in intracellular calcium, which activates multiple downstream signaling enzymes including the phosphatase calcineurin and the kinases calcium/calmodulin-dependent protein kinase II (CaMKII) and protein kinase C (PKC). Activation of these enzymes in the postsynaptic compartment plays a major regulatory role during synaptic plasticity. Here we will focus on studies of CaMKII and the PKC isoform PKMζ during hippocampal LTP and learning.
LTP induction in the CA1 region of the hippocampus requires CaMKII activity (11, 12), and transgenic mice lacking the α isoform have defective LTP and spatial learning (13, 14). CaMKIIα undergoes autophosphorylation in response to elevations in Ca2+ bound calmodulin, and this autophosphorylation renders the kinase autonomously active. This switch-like property of CaMKII enables it to persistently phosphorylate targets. Neuronal activity also translocates CaMKIIα to the PSD, where it can phosphorylate many PSD proteins. CaMKIIα knockout mice were among the first transgenic mice shown to have impairments in hippocampal LTP and learning (13, 14). Later studies showed specifically that the autophosphorylation of CaMKIIα is essential for LTP induction and, perhaps, its maintenance (15). More recent findings suggest, however, that autonomous CamKII activity generated by autophosphorylation may be less important for LTP maintenance and long-term memory than previously thought (16).
The brain-restricted atypical PKC isoform, protein kinase M zeta (PKMζ), is constitutively active and thus can persistently phosphorylate targets. PKMζ mRNA is targeted to dendrites where activity-dependent signaling cascades regulate its local translation (17). Protein concentrations increase or decrease with LTP or LTD inducing stimuli, respectively (17). Furthermore, studies that exogenously apply PKMζ or pharmacologically block its activity have shown that PKMζ is sufficient and necessary for LTP maintenance and for the maintenance of long-term memories (17, 18). These data have led to the proposal that PKMζ activation perpetuates synaptic plasticity and memory.
Activity-dependent modulation of postsynaptic glutamate receptors
The main excitatory neurotransmitter in the brain is glutamate, which activates several post-synaptic receptors. Two types of ionotropic glutamate receptors – α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid (AMPA) and N-methyl D-aspartate (NMDA) – have central roles in hippocampal synaptic plasticity. Both are ligand-gated ion channels and have unique properties that subserve different phases of synaptic plasticity. NMDA-type glutamate receptors (NMDARs) are calcium permeable and when activated, allow an influx of calcium needed for the induction of LTP. However, NMDARs are “coincidence detectors” and require both presynaptic transmitter release and postsynaptic depolarization for activation.
AMPA-type glutamate receptors (AMPARs) are important for the expression and maintenance of LTP. Unlike NMDARs, AMPARs can be activated by ligand binding at resting potentials to allow current flow. Increased conductance through AMPARs is responsible for the increase in synaptic connectivity during NMDAR-dependent LTP at CA1 synapses.
Given the importance of AMPARs in determining synaptic connectivity, much effort has focused on delineating the mechanisms that regulate their function. Activity-regulated phosphorylation can change AMPAR function by changing the open probabilities and conductances of the receptors. However, changes in channel properties are unlikely to account for the drastic changes in AMPAR function seen with LTP (19). Instead, changes in AMPAR function during synaptic plasticity are mostly due to phosphorylation-induced changes in its abundance at the synapse.
AMPARs traffic constitutively to and from the plasma membrane via recycling endosomes (20) (Figure 3). Activity modulates receptor trafficking through changes in actin dynamics and AMPAR interactions with scaffolding proteins. One of these scaffolding proteins, Stargazin, mediates the interaction between AMPARs and the PSD protein PSD-95, and this interaction is important for synaptic localization of AMPARs (21). Activity alters the phosphorylation of Stargazin, with phosphorylated Stargazin enhancing AMPAR function. Blocking Stargazin phosphorylation blocks LTP, while blocking dephosphorylation blocks LTD (22).
AMPARs are tetramers, and the cytoplasmic tails of each subunit contain multiple phosphorylation sites that regulate the trafficking of AMPARs. As one example, PKA phosphorylation of S845 in the long cytoplasmic tail of GluA1 increases GluA1 surface expression due to both enhanced insertion and attenuated internalization (23). Conversely, LTD of dissociated cultures and brain slices results in dephosphorylation of S845 and is correlated with an increase in the rate of AMPAR endocytosis (24). The functional consequences of GluA1 phosphorylation are highlighted by studies in knock-in mice with phosphorylation deficient mutations at both S831A and S845A. These mice display a loss of NMDA-induced AMPAR internalization, deficits in LTP and LTD, and have impaired spatial memory (25).
While studies of post-translational modifications at individual sites have established a role for regulating GluA1 trafficking and channel properties, they do not fully account for the changes in GluA1 function observed with synaptic plasticity (26). Activity-modified residues continue to be discovered, including, for example, the highly conserved T840 phosphorylation site, the phosphorylation of which correlates remarkably well with synaptic strength (27). Additionally, the phosphorylation site at S818 appears to have a crucial role in AMPAR trafficking in LTP (26). It is likely that complex patterns of phosphorylation and of other post-translational modifications (e.g. palmitoylation or ubiquitination) combine to regulate AMPAR localization. These studies underscore the importance of activity-dependent modulation of AMPAR trafficking in regulating synaptic strength.
Trans-synaptic signaling; the synaptic cleft
The synaptic cleft is a remarkably regular junction of approximately 20 nm between the pre- and post-synaptic compartments, consisting of a space through which presynaptically released neurotransmitters diffuses to bind postsynaptic receptors, as well as a network of cell adhesion molecules (CAMs) that keeps the synapse together. These adhesive interactions are so strong that it is impossible to biochemically separate intact pre- from post-synaptic compartments.
Role of CAMs in synaptic plasticty
The CAMs that localize to the synaptic cleft include, among others, members of the cadherin, integrin, immunoglobulin (Ig)-containing CAMs, as well as neurexins and neuroligins. Much research has focused on trying to understand whether and how CAMs mediate synapse specificity during neural circuit formation. Here we will focus on the regulation of synaptic CAMs during experience-dependent synaptic plasticity, limiting our discussion to just two of many examples.
One such example involves the addition of large sialic acid homopolymers to the Neural Cell Adhesion Molecule NCAM to form polysialylated NCAM (PSA-NCAM), which decreases hemophilic adhesion to allow new synaptic remodeling and growth. The ratio of PSA-NCAM to NCAM is increased following hippocampal learning tasks and inactivation of the enzyme that adds the poly-sialic moieties blocks hippocampal learning and plasticity (28). The time course of these changes suggests that the increase in PSA-NCAM is required to promote synaptic remodeling during persistent forms of plasticity.
Another family of CAMs that play a role in hippocampal plasticity includes the synaptically localized receptor tyrosine kinase ephrins and ephrin receptors (Eph receptors), which mediate bidirectional signaling at the synapse. Initially studied in the context of neural development, ephrins and Eph receptors have also been found to be essential for hippocampal LTP and LTD in the adult brain (29). Specific ephrins and Eph receptors interact with and regulate the localization and function of NMDA receptors, and can thereby modulate synaptic strength in response to activity. Experiments using inhibitory ephrin and Eph receptor peptides have revealed that both molecules are required, in a kinase-independent manner, for mossy fiber hippocampal LTP (30).
Trans-synaptic signaling by retrograde messengers
Another means of trans-synaptic signaling involves diffusible, membrane soluble messengers. Here we will briefly review recent work outlining a role for endocannabinoids as trans-synaptic retrograde signals critical to synaptic plasticity (31). The CB1 and CB2 cannabinoid receptors were initially identified as receptors for cannabinoid, the active ingredient of THC/marijuana. This led to the identification of endogenous CB1 and CB2 ligands, called endocannabinoids, specifically the arachidonate-based lipids anandamide (N-arachidonoylethanolamide, AEA) and 2-arachidonoylglycerol (2-AG). Discovered only 10 years ago, endocannabinoids have emerged as important modulators of plasticity at synapses. Depolarization and activation of a variety of receptors (including metabotropic glutamate receptors; mGluRs) have been shown to activate release of endocannabinoids from the postsynaptic compartment and binding to presynaptic CB receptors, resulting in a suppression of neurotransmitter release. This form of plasticity is called endocannabinoid-LTD, or eCB-LTD. Studies using CB1 knockout mice or CB1 receptor antagonists have shown that endocannabinoid signaling is required for the extinction but not the acquisition of spatial memories (32). Future studies are likely to reveal additional functions for endocannabinoids as retrograde messengers that modulate brain plasticity.
The tripartite synapse: glia and synaptic plasticity
Once thought of as the “support cells” of the nervous systems, glial cells are now considered essential partners in synapse formation, synaptic transmission and plasticity (33). Astrocytes surround the synapse (Fig 2), forming a “tripartite synapse,” composed of neuronal pre- and post-synaptic compartments as well as surrounding astrocytes. Synaptically localized glia release neuroactive molecules that influence neuronal communication. For example, release of D-serine (a co-activator of the NMDA receptor) from glia has been shown to be required for LTP of hippocampal Schaffer collateral synapses (34) (although see also (35)). Ephrin and Eph receptor signaling between neurons and glia has been shown to regulate the uptake of glutamate through glial glutamate transporters, and thereby affect neurotransmission and synaptic plasticity (36). The release of lactate from astrocytes and uptake by neurons has also been reported to be required for long-term hippocampal memory and plasticity (37). As these examples illustrate, future research in this relatively new field is likely to uncover a multitude of ways in which glia contribute to synaptic plasticity.
Regulating gene expression within neurons during plasticity
Signaling from synapse to nucleus to regulate transcription
Long-lasting forms of synaptic plasticity, such as those underlying long-term memory, require new RNA synthesis (1). This indicates that synaptic signals must be relayed to the nucleus to regulate transcription. Synapse to nucleus signaling poses a unique set of challenges in neurons, where the distance between the synapse and nucleus can be significant. Neurons are specialized for rapid communication between compartments via electrochemical signaling. In this manner, depolarization at the synaptic terminal leads to depolarization at the soma within milliseconds, followed by calcium influx and calcium-dependent nuclear signaling. Calcium influx can occur through voltage- and ligand-gated ion channels. Cytosolic calcium can also be released from intracellular pools following activation of Gq coupled receptors such as mGluRs. Each route of calcium influx induces different programs of gene induction. For example, transcription of brain-derived neurotrophic factor (BDNF) is highly induced following calcium entry through L-type voltage-sensitive calcium channels (VSCCs) in excitatory neurons, but not following calcium entry through NMDA receptors or other VSCCs (38).
Soluble signals can also be transported from the synapse to the nucleus by slower, microtubule and motor protein-dependent pathways (39). This class of signals includes kinases and transcriptional regulators that can indirectly and directly affect transcription. These slower pathways of signaling to the nucleus may sustain changes in gene expression for time periods extending beyond the initial stimulus.
To obtain a global view of how transcription is altered during activity-dependent plasticity, expression profiling has been used to identify changes in transcription following depolarization of cultured mouse neurons. Such studies have identified several hundred activity-regulated genes (40). Genome-wide analyses of transcription factor binding sites of the activated genes have revealed that the transcription factors CREB, MEF2, and Npas4 control the activity-dependent transcription of a large number of downstream activity-regulated genes (40). A larger number of transcription factors also contribute to activity-driven transcription, including SRF, ELK, NFAT, NFκB, DREAM, NeuroD, SP4, and CREST. These transcription factors regulate the expression of overlapping but distinct subsets of activity-regulated genes, suggesting that the precise temporal, spatial, and stimulus-specific cellular response is achieved by the combinatorial control by different transcription factors.
Local Protein Synthesis
Despite requiring new transcription, LTP and LTD can occur in a spatially-restricted manner, raising the question of how gene expression in neurons can be limited to subsets of synapses and not generalized to the entire cell. One way of locally changing the proteome in neurons is through regulated translation of localized mRNAs (Figure 4).
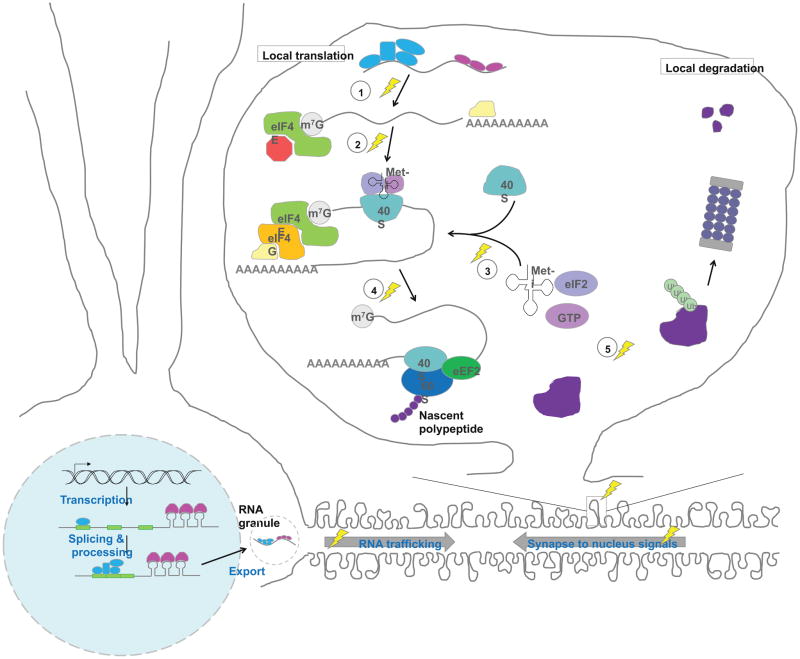
Figure 4. Local regulation of the synaptic proteome.
Synaptic plasticity modifies gene expression at many levels. Strong stimulation of synapses triggers signals that are sent to the nucleus to modify RNA synthesis. Synaptic activity also modifies protein synthesis, and has been found to act at several key steps during translation: (1) Relief of repression, e.g. RISC-mediated repression; (2) Modification of translational initiation to allow 4E–4G interaction and recruitment of 40S; (3) Formation of the preinitiation complex; (4) Dephosphorylation of eEF2 to allow for catalysis of ribosome translocation during translational elongation. To counterbalance local protein synthesis, local protein degradation also occurs at synapses (5). Together, these regulated steps in protein addition and removal allow for rapid, spatially-restricted control of the synaptic proteome. Lightning bolts indicate activity-dependent processes. (Note: while local translation in dendrites is a well-accepted phenomenon, it has not been demonstrated to occur in spines. Also, the precise location of some of the translational machinery and regulatory steps depicted has not been identified.)
Protein synthesis was historically thought to occur exclusively in neuronal cell bodies. The existence of local translation in dendrites of mature neurons was first suggested by electron micrographic identification of polyribosome in hippocampal dendrites (41). Studies in hippocampal slices in which dendrites had been severed from cell bodies were found to retain the ability to express long-lasting LTP and LTD, indicating that local translation can mediate long-term modification of synaptic strength (42, 43).
Studies of mRNA localization have led to the identification of cis-acting RNA elements that bind to RNA-binding proteins to undergo export from the soma into the dendrite (44). Although several dendritic localization elements have been identified, there is to date no consensus on their sequence or structure. Among the best-studied RNA binding proteins involved in dendritic mRNA localization are Staufen, Zipcode binding protein 1 (ZBP1), and hnRNPA2 (44). These proteins bind cis-acting elements and assemble transcripts into larger RNA transport granules, which travel in a kinesin-dependent manner along microtubules to their final destination. Whether localized RNAs undergo directed targeting, anchoring or stabilization at specific sites remains an open question.
In terms of translational regulation, studies have revealed activity-dependent regulation of translation initiation and elongation. A mechanism of translational regulation known to occur at synapses involves the cytoplasmic polyadenylation element binding protein (CPEB). CPEB binding to 3′UTRs indirectly increases the poly(A) tails of mRNAs. Subsequently, poly(A) binding protein (PABP) is recruited to the elongated poly(A) tail, which in turn recruits eIF4G to interact with eIF4E to promote translation initiation (45). CPEB localizes to synapses, and has been shown to regulate translation of dendritically localized CamKIIα mRNA (46, 47).
Another activity-dependent means of regulating translation initiation involves phosphorylation of eIF4E binding proteins (4E-BPs). Hypo-phosphorylated 4E-BPs bind eIF4E and prevent translation initiation; phosphorylated 4E-BP dissociates from eIF4E and relieves translational inhibition. In neurons, activity increases 4E-BP phosphorylation and stimulates translation (48). Studies in 4E-BP2 knockout mice found that E-LTP stimulation protocols could induce L-LTP in brain slices. Recently, two additional 4E-BPs have been identified in neurons: neuroguidin and the cytoplasmic FMRP interacting protein (CYFIP). While 4E-BP1 and 2 are believed to affect general translation, these new 4E-BPs may preferentially affect subgroups of transcripts within dendrites (49, 50).
Activity can also regulate translational elongation during synaptic plasticity. As one example, the elongation factor eEF2 has been shown to undergo activity-dependent changes in phosphorylation. Phosphorylation of eEF2 decreases the rate of translation. Schuman and colleagues have shown that while action potentials decrease eEF2 phosphorylation (thereby increasing translation), spontaneous release of neurotransmitter increases eEF2 phosphorylation and decreases translation (51). These effects occur locally at synapses, indicating that one function of spontaneous release may be to suppress local translation and thereby stabilize synapses.
Translation may also be regulated through the microRNA (miRNA) pathway, where each miRNA can potentially regulate hundreds of transcripts and hence provides a way of coordinating the expression of many genes. Expression profiling has shown that many miRNAs are enriched in, or even restricted to, the brain. While miRNAs can regulate cell-wide levels of translation, their post-transcriptional mode of action makes them especially well-suited to regulating distally localized transcripts. Consistent with a role in local translation, miRNAs have been found in dendrites and at synapses. Further, components of the RNA-induced silencing complex (RISC) machinery itself have been found to be altered by activity (52).
Local protein degradation
The local proteome is regulated not only by local translation but also by protein degradation through the ubiquitin proteasome system (Figure 4). Studies have shown that both protein synthesis and degradation are required for the maintenance of late-phase LTP, suggesting that protein degradation is needed to counterbalance protein synthesis during plasticity (53). Like local translation, protein degradation can be regulated within dendrites. In support of locally regulated degradation, ubiquitin and proteasomal subunits have been found in dendrites and at synapses. Glutamatergic stimulation of cultured hippocampal neurons leads to bidirectional changes in ubiquitin conjugation and to proteasome-dependent changes in protein concentrations of PSD fractions (54). Activity-dependent degradation involves redistribution of proteasomes from dendritic shafts to spines (55). The occurrence of both translation and degradation at the synapse suggests that proper synaptic function requires tight control of the local proteome.
Perspectives
As the above examples indicate, cell biological approaches have provided a detailed understanding of many aspects of activity-dependent plasticity. By focusing on molecular processes occurring within individual neurons and subcellular compartments, we now understand specific processes that are modulated by experience to change synaptic connectivity. These involve alterations in neurotransmitter release, trans-synaptic signaling, post-synaptic receptor dynamics and gene expression within neurons. The results of such studies provide molecular targets for further exploration of the mechanisms of brain plasticity, and potential therapeutic targets for diseases in which brain plasticity is dysfunctional. However, they fall significantly short of elucidating how complex circuits are altered by experience so as to store information and alter behavior. This will require the development of tools for investigating both the dynamic nano-architecture of the synapse and the neural circuit as a whole. A particular challenge in the field is to study plasticity in neural circuits in living animals, and to develop methods to examine how all the components of circuit (excitatory and inhibitory neurons, synapse, glia and vasculature) are regulated to alter circuit function dynamically over various time domains. The development of methodologies for super-resolution time-lapse imaging of synapses, neurons and circuits in live animals promises to move the field forward towards a more nuanced and complete understanding of the experience-dependent plastic changes in the brain that mediate learning and memory.
Acknowledgments
We apologize to those whose primary work could not be cited because of space constraints. We thank J.T. Braslow, T.J. O’Dell and F.E. Schweizer for comments on the manuscript, J. Bourne and K.M. Harris for hippocampal EM images, and all members of the Martin lab for helpful discussions. Support comes from NIH R01 MH077022, R01 NS045324 (to KCM), the Medical Scientist Training Program (NIH T32 GM008042) and the Neurobehavioral Genetics Training Program (NIH T32 MH073526) (to VMH).
References
- 1.Kandel ER. Science. 2001;294:1030. doi: 10.1126/science.1067020. [DOI] [PubMed] [Google Scholar]
- 2.Cesca F, Baldelli P, Valtorta F, Benfenati F. Prog Neurobiol. 2010;91:313. doi: 10.1016/j.pneurobio.2010.04.006. [DOI] [PubMed] [Google Scholar]
- 3.Rosahl TW, et al. Nature. 1995;375:488. doi: 10.1038/375488a0. [DOI] [PubMed] [Google Scholar]
- 4.Kaeser PS, et al. Cell. 2011;144:282. doi: 10.1016/j.cell.2010.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Han Y, Kaeser PS, Sudhof TC, Schneggenburger R. Neuron. 2011;69:304. doi: 10.1016/j.neuron.2010.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Deng L, Kaeser PS, Xu W, Sudhof TC. Neuron. 2011;69:317. doi: 10.1016/j.neuron.2011.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Castillo PE, et al. Nature. 2002;415:327. doi: 10.1038/415327a. [DOI] [PubMed] [Google Scholar]
- 8.Kirov SA, Harris KM. Nat Neurosci. 1999;2:878. doi: 10.1038/13178. [DOI] [PubMed] [Google Scholar]
- 9.Holtmaat A, Svoboda K. Nat Rev Neurosci. 2009;10:647. doi: 10.1038/nrn2699. [DOI] [PubMed] [Google Scholar]
- 10.Engert F, Bonhoeffer T. Nature. 1999;399:66. doi: 10.1038/19978. [DOI] [PubMed] [Google Scholar]
- 11.Malenka RC, et al. Nature. 1989;340:554. doi: 10.1038/340554a0. [DOI] [PubMed] [Google Scholar]
- 12.Malinow R, Schulman H, Tsien RW. Science. 1989;245:862. doi: 10.1126/science.2549638. [DOI] [PubMed] [Google Scholar]
- 13.Silva AJ, Paylor R, Wehner JM, Tonegawa S. Science. 1992;257:206. doi: 10.1126/science.1321493. [DOI] [PubMed] [Google Scholar]
- 14.Silva AJ, Stevens CF, Tonegawa S, Wang Y. Science. 1992;257:201. doi: 10.1126/science.1378648. [DOI] [PubMed] [Google Scholar]
- 15.Lisman J, Schulman H, Cline H. Nat Rev Neurosci. 2002;3:175. doi: 10.1038/nrn753. [DOI] [PubMed] [Google Scholar]
- 16.Buard I, et al. J Neurosci. 2010;30:8214. doi: 10.1523/JNEUROSCI.1469-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sacktor TC. Nat Rev Neurosci. 2011;12:9. doi: 10.1038/nrn2949. [DOI] [PubMed] [Google Scholar]
- 18.Shema R, Sacktor TC, Dudai Y. Science. 2007;317:951. doi: 10.1126/science.1144334. [DOI] [PubMed] [Google Scholar]
- 19.Andrasfalvy BK, Magee JC. J Physiol. 2004;559:543. doi: 10.1113/jphysiol.2004.065219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Park M, et al. Science. 2004;305:1972. doi: 10.1126/science.1102026. [DOI] [PubMed] [Google Scholar]
- 21.Schnell E, et al. Proc Natl Acad Sci U S A. 2002;99:13902. doi: 10.1073/pnas.172511199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tomita S, et al. Neuron. 2005;45:269. doi: 10.1016/j.neuron.2005.01.009. [DOI] [PubMed] [Google Scholar]
- 23.Man HY, Sekine-Aizawa Y, Huganir RL. Proc Natl Acad Sci U S A. 2007;104:3579. doi: 10.1073/pnas.0611698104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ehlers MD. Neuron. 2000;28:511. doi: 10.1016/s0896-6273(00)00129-x. [DOI] [PubMed] [Google Scholar]
- 25.Lee HK, et al. Cell. 2003;112:631. doi: 10.1016/s0092-8674(03)00122-3. [DOI] [PubMed] [Google Scholar]
- 26.Boehm J, et al. Neuron. 2006;51:213. doi: 10.1016/j.neuron.2006.06.013. [DOI] [PubMed] [Google Scholar]
- 27.Delgado JY, et al. J Neurosci. 2007;27:13210. doi: 10.1523/JNEUROSCI.3056-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bonfanti L. Prog Neurobiol. 2006;80:129. doi: 10.1016/j.pneurobio.2006.08.003. [DOI] [PubMed] [Google Scholar]
- 29.Lai KO, Ip NY. Curr Opin Neurobiol. 2009;19:275. doi: 10.1016/j.conb.2009.04.009. [DOI] [PubMed] [Google Scholar]
- 30.Contractor A, et al. Science. 2002;296:1864. doi: 10.1126/science.1069081. [DOI] [PubMed] [Google Scholar]
- 31.Heifets BD, Castillo PE. Annu Rev Physiol. 2009;71:283. doi: 10.1146/annurev.physiol.010908.163149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Marsicano G, Lafenetre P. Curr Top Behav Neurosci. 2009;1:201. doi: 10.1007/978-3-540-88955-7_8. [DOI] [PubMed] [Google Scholar]
- 33.Eroglu C, Barres BA. Nature. 2010;468:223. doi: 10.1038/nature09612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Henneberger C, Papouin T, Oliet SH, Rusakov DA. Nature. 2010;463:232. doi: 10.1038/nature08673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Agulhon C, Fiacco TA, McCarthy KD. Science. 2010;327:1250. doi: 10.1126/science.1184821. [DOI] [PubMed] [Google Scholar]
- 36.Filosa A, et al. Nat Neurosci. 2009;12:1285. doi: 10.1038/nn.2394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Suzuki A, et al. Cell. 2011;144:810. doi: 10.1016/j.cell.2011.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ghosh A, Carnahan J, Greenberg ME. Science. 1994;263:1618. doi: 10.1126/science.7907431. [DOI] [PubMed] [Google Scholar]
- 39.Ch’ng TH, Martin KC. Curr Opin Neurobiol. 2011;21:345. doi: 10.1016/j.conb.2011.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Greer PL, Greenberg ME. Neuron. 2008;59:846. doi: 10.1016/j.neuron.2008.09.002. [DOI] [PubMed] [Google Scholar]
- 41.Steward O, Levy WB. J Neurosci. 1982;2:284. doi: 10.1523/JNEUROSCI.02-03-00284.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kang H, Schuman EM. Science. 1996;273:1402. doi: 10.1126/science.273.5280.1402. [DOI] [PubMed] [Google Scholar]
- 43.Huber KM, Kayser MS, Bear MF. Science. 2000;288:1254. doi: 10.1126/science.288.5469.1254. [DOI] [PubMed] [Google Scholar]
- 44.Kiebler MA, Bassell GJ. Neuron. 2006;51:685. doi: 10.1016/j.neuron.2006.08.021. [DOI] [PubMed] [Google Scholar]
- 45.Richter JD. Trends Biochem Sci. 2007;32:279. doi: 10.1016/j.tibs.2007.04.004. [DOI] [PubMed] [Google Scholar]
- 46.Wu L, et al. Neuron. 1998;21:1129. doi: 10.1016/s0896-6273(00)80630-3. [DOI] [PubMed] [Google Scholar]
- 47.Wells DG, et al. J Neurosci. 2001;21:9541. doi: 10.1523/JNEUROSCI.21-24-09541.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Costa-Mattioli M, Sossin WS, Klann E, Sonenberg N. Neuron. 2009;61:10. doi: 10.1016/j.neuron.2008.10.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jung MY, Lorenz L, Richter JD. Mol Cell Biol. 2006;26:4277. doi: 10.1128/MCB.02470-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Napoli I, et al. Cell. 2008;134:1042. doi: 10.1016/j.cell.2008.07.031. [DOI] [PubMed] [Google Scholar]
- 51.Sutton MA, et al. Cell. 2006;125:785. doi: 10.1016/j.cell.2006.03.040. [DOI] [PubMed] [Google Scholar]
- 52.Banerjee S, Neveu P, Kosik KS. Neuron. 2009;64:871. doi: 10.1016/j.neuron.2009.11.023. [DOI] [PubMed] [Google Scholar]
- 53.Fonseca R, et al. Neuron. 2006;52:239. doi: 10.1016/j.neuron.2006.08.015. [DOI] [PubMed] [Google Scholar]
- 54.Ehlers MD. Nat Neurosci. 2003;6:231. doi: 10.1038/nn1013. [DOI] [PubMed] [Google Scholar]
- 55.Bingol B, Schuman EM. Nature. 2006;441:1144. doi: 10.1038/nature04769. [DOI] [PubMed] [Google Scholar]
- 56.Fleming JJ, England PM. Nature Chem Bio. 2010;6:89. doi: 10.1038/nchembio.298. [DOI] [PubMed] [Google Scholar]
- 57.Bourne J, Harris KM. Curr Opin Neurobiol. 2007;17:381. doi: 10.1016/j.conb.2007.04.009. [DOI] [PubMed] [Google Scholar]