Abstract
The heart is a highly plastic organ. In response to the physiological stress of normal life, as well as the pathological stress of disease, the myocardium manifests robust and rapid changes in mass. In the context of disease-associated stress, this myocardial remodeling response can culminate in ventricular thinning, mechanical dysfunction, and a clinical syndrome of heart failure. Recently, autophagy, a process of cellular cannibalization, has been implicated in many of these remodeling reactions. In some settings, the autophagic response is beneficial and pro-survival; in other contexts, it is maladaptive and promotes disease progression. Together, these observations raise the intriguing prospect of targeting maladaptive autophagy and advancing cell survival-promoting, adaptive autophagy to benefit patients with heart disease.
Keywords: Hypertrophy, Heart failure, Autophagy
Throughout the course of life, the myocardium is faced with ever-changing workload demands. Growth during development is associated with cardiac enlargement in proportion to the growing body. Increases in physiological demand during exercise and pregnancy provoke heart growth as well. Pathological stress on the heart, such as hypertension, valvular disease, myocardial infarction, or excessive neuroendocrine activation, triggers a cardiac growth response that is similarly rapid and robust. At the other end of the spectrum, deconditioning, prolonged bed rest, cancer, and weightlessness each lead to substantial decreases in myocardial mass [17].
These remodeling reactions are common, and in the context of pathological triggers, they represent a significant pathogenetic milestone in disease progression. Indeed, this is one reason why hypertension, with its effects on myocardial architecture, is a leading risk factor for mortality worldwide [50]. Astonishingly, in 2000, >25% of the world's adult population was hypertensive, and this population is projected to reach 1.56 billion in 2025, a 60% increase during 25 years [50]. Mounting epidemiological evidence demonstrates a linear and independent relationship between pathological stress, such as hypertension, and cardiovascular disease [50].
Alterations in ventricular demand, including changes in preload, afterload, and rate of contraction, are capable of altering myocardial wall stress; according to Laplace's law, ventricular wall stress is directly proportional to pressure and chamber size (radius of the cavity) and inversely proportional to wall thickness. As a consequence, structural and functional alterations ensue that normalize wall stress, at least temporarily. For example, left-ventricular wall thickness increases in the setting of increased blood pressure, resulting in decreases in, and even normalization of, wall stress. The majority of this increase in wall thickness derives from myocyte growth (hypertrophy) because cardiomyocytes manifest only modest potential for proliferation [1].
Left-ventricular hypertrophy (LVH) is thought to be adaptive during its initial phases, serving as a response that offsets wall stress-induced increases in oxygen demand. However, analogous to many other instances in biology, the long-term consequences of a short-term compensatory process can be maladaptive; chronic LVH is a major risk factor for systolic dysfunction and clinical heart failure [35]. Despite a great deal of effort being devoted to elucidating molecular events underlying the transition from LVH to heart failure, they still remain elusive. That said, numerous preclinical studies have demonstrated that abrogation of the afterload-induced hypertrophic response is well tolerated and even beneficial [12, 18].
In the setting of increased afterload, such as occurs in hypertension and aortic stenosis, short-axis growth of the myocyte occurs via parallel addition of new sarcomeres; this lateral myocyte growth results in wall thickening with relatively less increase in chamber volume, a process termed “concentric hypertrophy” [17]. Under conditions of increased preload, such as aortic insufficiency, aortovenous shunting, or aortic insufficiency, sarcomeres are added in series, resulting in length-wise growth of the myocyte. The result is ventricular wall thickening accompanied by chamber dilation, a process termed “eccentric hypertrophy.”
Proteostasis in Myocardial Remodeling
Remodeling of any tissue requires alterations in the steady-state equilibrium between protein synthesis and protein degradation. In post-mitotic cells, such as cardiomyocytes, which are largely incapable of re-entering the cell cycle, the fidelity of protein quality control is of paramount importance because accumulation of misfolded proteins and protein aggregates is toxic, thus triggering adverse cellular responses and cell death. The major cellular mechanism for clearing toxic protein aggregates, along with long-lived proteins and dysfunctional organelles, is lysosome dependent.
In the context of cardiovascular disease, activation of lysosomal pathways of protein clearance has been recognized for many years in both human heart failure and in animal models of heart disease [6, 7, 11, 16, 29, 30, 36, 54, 65]. Recent work from our laboratory and those of others has focused on pathways upstream of the lysosome, viz. cardiomyocyte autophagy. Whereas multiple forms of autophagy exist, macroautophagy (hereafter termed “autophagy”) is the most important pathway in the turnover of long-lived proteins and the only means of clearing dysfunctional organelles.
Autophagy is an evolutionarily conserved, ubiquitous mechanism required for cellular homeostasis in multiple contexts [20, 28, 31, 41]. Under resting conditions, for example, autophagy is required for the constitutive turnover of long-lived proteins and dysfunctional organelles. Under conditions of stress, such as nutrient deprivation or hypoxia, autophagic flux is activated, promoting cell survival by way of replenishment of substrates from degraded cellular constituents and by eliminating defective or damaged organelles. However, taken too far, excessive autophagic activation can lead to critical depletion of essential molecules and organelles, thus triggering cell death (autophagic programmed cell death [PCD], type II PCD) [34].
Autophagy: A Process of Cellular Cannibalization
Autophagy is a catabolic process whereby recycling of intracellular components positions a cell to respond to energetic stress [42]. In the presence of ample nutrient supply, growth factors, such as insulin and insulin-like growth factor 1, stimulate glucose uptake and promote anabolic reactions. In this context, autophagy is suppressed. In response to starvation, however, triggered either by a shortage of nutrients or by defects in growth factor-dependent signaling, autophagy is rapidly activated, thus restoring intracellular stores of critical elements of intermediary metabolism and thereby promoting survival. In addition to conditions of nutrient deprivation, enhanced levels of autophagy are observed in a variety of other circumstances, such as microbial invasion, accumulation of misfolded proteins, cancer, neurodegenerative disorders, and cardiac diseases [42].
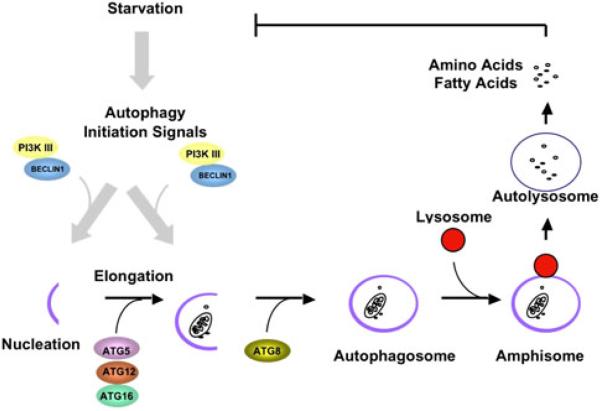
The autophagic cascade involves a complex series of stepwise events, which have been divided into four stages: induction, nucleation, expansion, and maturation/retrieval (Fig. 1) [26, 64]. To date, genetic studies in yeast have identified 32 autophagy-related (ATG) genes involved in the cascade [23, 24, 26]. First, an isolated membrane (phagophore) is localized to a site termed the “phagophore assembly site,” where induction and nucleation take place. Expansion of the phagophore involves addition of several regulatory elements and engulfment of cytoplasmic constituents. The morphologically distinctive autophagosome, the hallmark of autophagy, is a unique, double-membrane vesicle 0.5 to 1.5 μm in diameter.
Fig. 1.
The autophagic process. In response to a variety of cellular stresses a class III PI3 kinase/Beclin 1 complex initiates formation of an isolated double membrane (the membrane nucleation process) and subsequent membrane elongation. The membrane then fuses on itself, forming the distinctive double-membrane autophagosome. Autophagosomes ultimately fuse with lysosomes, and their cargo is degraded, providing fuel and elemental building blocks to preserve vital cellular functions
Next, the autophagosome fuses with a lysosome forming an autolysosome. There, the engulfed cargo and the autophagosome's inner membrane are degraded by acid hydrolases. The resulting small molecules, including amino acids, sugars, and lipids, are released into the cytosol through permeases to serve as cellular building blocks and energy sources. After maturation of the autophagosome, most of the ATG proteins are ultimately recycled through a pathway involving ATG2, ATG9, and ATG18 [64]. Eventually, mTOR (mammalian TOR) is reactivated to attenuate the autophagic response and to restore the cellular complement of lysosomes [68].
In addition to its critical role in cellular responsiveness to nutrient deprivation, autophagy mediates turnover of intracellular organelles. For example, in cardiac myocytes and other highly oxidative tissues, there is constitutive autophagic turnover of mitochondria. Also, opening of the mitochondrial permeability transition and loss of mitochondrial membrane potential triggers their autophagic scavenging [27]. Similarly, fission and subsequent depolarization of mitochondria elicit autophagic sequestration [59]. Thus, autophagy can protect from mitochondria that might otherwise trigger apoptotic cell death. Finally, autophagy contributes importantly to tissue remodeling during development [28, 31, 32, 41].
Regulation of Autophagy
A major upstream negative regulator of autophagy is the protein TOR (target of rapamycin) [24]. In the presence of plentiful nutrients and intact insulin signaling, class I phosphatidylinositol-3-kinase (PI3K) is activated to phosphorylate its downstream target AKT. AKT in turn relays the signal to activate TOR. Active TOR then phosphorylates Atg13, thus inhibiting its interaction with Atg1, a critical step during autophagic induction in budding yeast [23]. By contrast, during starvation, Atg13 binds Atg1 and Atg17, thus promoting induction of autophagy at the phagophore assembly site.
The Atg1–Atg13–Atg17 complex is best characterized in yeast; the mammalian counterpart differs slightly [3]. Under nutrient-rich conditions, mTOR interacts with a complex comprising ULK1 (mammalian ATG1), mATG13, and FIP200, and phosphorylates ULK1 and mATG13 to suppress autophagy. When nutrient supply is low, dissociation of mTOR promotes activation of ULK1, which phosphorylates mATG13 and FIP200 to trigger autophagy. A class III PI3K complex is then recruited to the assembly site to stimulate nucleation, and the lipid kinase Vps34 binds to the phagophore membrane through Vps15. Additionally, Beclin1/ATG6 and ATG14 within this complex regulate Vps34 kinase protein. Activity of this lipid kinase complex is crucial for recruitment of additional ATG proteins, which ultimately complete formation of the autophagosome.
In addition, two cascades with features similar to the ubiquitin-conjugation cascade contribute to expansion of the phagophore and formation of the autophagosome. ATG12 is activated by a ubiquitin E1-like enzyme, ATG7, and transferred in turn to a ubiquitin E2-like enzyme, ATG10. ATG12 is then covalently conjugated to ATG5, and the resulting ATG12-ATG5 complex interacts with ATG16. In the other ubiquitin-like system, LC3 (mammalian homolog of ATG8) is cleaved by ATG4 to expose a carboxyl terminal glycine. This processed LC3-I is then activated by ATG7, an E1-like enzyme. After being transferred by the E2-like enzyme, ATG3, LC3-I is cleaved and subsequently attached to a phosphatidylethanolamine molecule and localized to the phagophore membrane. This lipidated isoform, termed “LC3-II,” migrates faster than LC3-I on SDS-PAGE, and its levels correlate with the abundance of autophagosomes.
Thus, two kinase systems (ATG1–ATG13 and class III PI3K), two ubiquitin-like systems (ATG5–ATG12 and LC3-II-PE), and a retrieval/maturation system—all working in concert—complete the autophagic flux pathway.
Constitutive Autophagy in the Heart
We have known for 3 decades that lysosomal pathways are activated in virtually all forms of heart disease [6, 7, 11, 16, 29, 30, 36, 54, 65]. Until recently, however, it has not been possible to manipulate these pathways to explore their functional significance or to consider manipulating them for therapeutic gain. Now, thanks to insights derived largely from the field of yeast genetics [20], it has become possible to upregulate or downregulate autophagic flux pathways to dissect their molecular circuitry, to explore their role(s) in health and disease, and to begin to envision therapeutic interventions in disease-related autophagy.
During cell growth and repair, regulation of proteolysis is critical. The requirement of maintenance of proteostasis is especially true in long-lived post-mitotic cells, such as cardiac myocytes, where cell replacement is limited. Short-lived proteins are targeted for degradation by way of ubiquitination and proteasomal processing. In contrast, degradation of longer-lived proteins, protein complexes, aggregates of misfolded proteins, and organelles occurs in lysosomes. Also, as a pathway to rid the cell of long-lived proteins, and the only mechanism capable of eliminating dysfunctional organelles, autophagy is critical to the maintenance of cellular homeostasis in response to fluctuating environmental conditions. Indeed, the vital importance of autophagy under basal conditions is highlighted by cell death in the absence of autophagy. Using siRNA technology, Nakai et al. depleted ATG5, the ubiquitin E1-like enzyme, from neonatal rat ventricular myocytes (NRVM) [43]. With decreased autophagic activity, NRVM manifested a classic hypertrophic response, including increased cell size, activation of a fetal gene program, processing of caspase 12, and decreased viability. Consistent with these in vitro results, the investigators found that loss of autophagy in the heart in vivo triggered hypertrophic growth, cardiac dysfunction, and ultimately heart failure [43].
The importance of autophagy under basal conditions is highlighted further by reports addressing mechanisms of Danon disease, a cardioskeletal myopathy that develops due to deficiency of LAMP2, a lysosomal membrane protein [38, 45, 56]. In the absence of LAMP2, fusion of autophagosomes with lysosomes is blocked, a defect that in turn leads to accumulation of long-lived proteins and consequent cardiac and skeletal myopathies. Together, these results point to the crucial housekeeping role of constitutive autophagy in muscle.
Autophagy in Cardiac Stress Responsiveness
During fasting, progressive decreases in heart weight are observed, consistent with activation of catabolic pathways, including autophagy, which is known to be induced rapidly [25]. Although partial inhibition of autophagy does not alter cardiac function under fed conditions, suppression of autophagy during starvation results in decreased cardiac adenosine triphosphate content and impaired heart performance. Similar findings have been reported for NRVMs in culture [40]. Collectively, these results are consistent with autophagy playing a critical, protective role in cardiomyocytes in the setting of nutrient deprivation.
Growth of myocytes is accomplished by increases in protein synthesis, construction of new sarcomeres, and remodeling of existing cellular elements. In initial phases, anabolic processes predominate, resulting in new protein synthesis and increases in cell size. Some evidence suggests that catabolic processes are suppressed during the early-phase response [43, 48]. We have reported robust activation of cardiomyocyte autophagy in the setting of pressure overload, a scenario that is common clinically and leads to heart failure [70]. Nakai et al. reported that autophagy was increased in mice 4 weeks after thoracic aortic constriction (TAC) [43]. Eventually, a new, steady-state equilibrium between anabolism and catabolism is achieved, albeit at a higher level, with balance achieved between synthetic and degradative processes.
At first glance, it seems paradoxical that a mechanism of protein degradation is activated in the setting of cell growth. However, hypertrophic remodeling involves more than just addition of proteins; as just one example, substantial alterations in the content of numerous sarcomeric components also occur. Thus, although the overall result is an increase in cell size, activation of protein degradative and quality control pathways, such as the proteasome and autophagy, are required in the remodeling of existing and newly synthesized cellular elements.
Autophagy in Cardiac Proteostasis
In neurons, protein aggregates are capable of inducing autophagy [21]. We have observed robust accumulation of polyubiquitinated proteins co-localized to autophagic active sites in pressure-stressed left ventricle [58]. We went on to show that polyubiquitinated proteins are sufficient to induce autophagy in NRVMs maintained in culture. Consistent with our findings, Depre et al. reported increased proteasome expression and activity in load-stressed heart [9]. Collectively, accumulation of protein aggregates in response to pressure overload elicits an upregulated protein quality-control response, which includes both lysosomal and proteasomal mechanisms.
The importance of protein aggregation in cardiac myocytes is highlighted further in the desmin-related myopathies (DRCM), where mutations in genes encoding desmin or the chaperone protein alpha-B-crystallin (CryAB) lead to accumulation of protein aggregates and profound heart failure [5, 46, 47, 49, 61, 63]. In this disorder, mutant CryAB confers a dominant-negative action to disrupt the chaperone function of CryAB, thus leading to protein misfolding, protein aggregation, and sarcomeric disarray. Severe cardiomyopathy ensues due to toxic protein (and protein aggregate) accumulation along with disruption of the myocyte's desmin architecture.
CryABR120G-associated DRCM has been replicated in two independently-derived transgenic mouse lines [49, 62]. The CryABR120G mutation results in protein aggregation and aggresome formation [53], mitochondrial toxicity [37], disruption of proteasome function [4], and a state of “reductive stress” [49]. We have reported robust activation of autophagy in DRCM, a response that clears toxic aggregates and ameliorates disease progression [57]. Thus, we found, similar to analogous disorders in brain, that autophagic activity is enhanced and confers an adaptive function to facilitate clearance of aggregates [57]. This finding contrasts with our finding of protein aggregation— and a maladaptive, autophagic response—in load-induced heart failure [58, 70].
Cardiomyocyte Autophagy in Load-Induced Heart Disease
Dysregulation of autophagy contributes to the pathogenesis of numerous diseases, including neurodegenerative disorders, cancer, skeletal myopathy, and microbial infection [33]. In the case of the heart, multiple forms of stress, including pressure overload, chronic ischemia, and ischemia-reperfusion, provoke an increase in cardiomyocyte autophagic activity [39, 44, 51, 52, 70]. However, despite considerable evidence linking autophagic cell death to heart failure progression, uncertainty remains regarding whether increased autophagy is an epiphenomenon or a causative factor in the dying myocyte. In recent years, a number of theories have been promoted in an attempt to define mechanisms governing the transition from stable, compensated hypertrophy to systolic dysfunction and decompensated heart failure. A common element among these theories is myocyte dropout by cell death. In tissues from patients with end-stage heart failure, evidence for each of the three major types of cell death has been reported, with autophagic cell death being a consistently prominent one [29, 30].
Our group reported that pressure overload induces autophagy as early as 24 h after TAC [70]. We found that partial suppression of autophagic activity in Beclin haploinsufficient mice (beclin 1+/–), a model where levels of autophagy are decreased, blunted load-induced pathological remodeling. Conversely, cardiomyocyte-restricted overexpression of Beclin 1, with associated increases in autophagy, substantially amplified adverse remodeling [70]. In other words, using gain- and loss-of-function approaches, we found that amplifying the autophagic response exacerbates load-induced pathological remodeling and that decreasing the autophagic response blunts pathological remodeling [70]. Thus, our results suggest that autophagy can be a maladaptive response to hemodynamic stress. Our studies went on to demonstrate that protein aggregation is a proximal trigger of load-induced cardiomyocyte autophagy [58].
Consensus is emerging that cardiomyocyte autophagy triggered by increases in afterload has both adaptive and maladaptive features. Complete abrogation of autophagic flux is incompatible with cell survival. Autophagic activation occurring in the setting of pressure stress may be beneficial up to a point, and overactivation of autophagic flux is maladaptive. At one level, this is not surprising, because the dual nature of autophagy is a recurring theme in other organ systems and disease states [32]. Indeed, we have postulated that the physiological impact of autophagy exists as a continuum and that a window of optimal autophagic activation (“Goldilocks zone” of autophagy) is critical to the maintenance of cellular homeostasis and function (Fig. 2).
Fig. 2.
Dual nature of autophagy in heart disease. Basal activation of autophagy is required for normal cellular function, and too little or too much autophagy can each be maladaptive. Hemodynamic stress activates autophagy, which, depending on the amplitude and duration of autophagic response and the initial level of basal autophagic flux, may elicit either harmful or beneficial effects [52] (figure used with permission of the American Heart Association)
Autophagy in Myocardial Ischemia
Autophagy is activated in response to myocardial ischemia, a fact first demonstrated 30 years ago in rabbit models [6, 8]. More recently, Matsui et al. reported dramatic upregulation of LC3-II and autophagosome formation 30 min after ischemia-inducing surgery in mice [40]. Going further, these investigators reported that pharmacological suppression of autophagy enhanced myocyte death triggered by glucose deprivation, an in vitro condition that mimics aspects of tissue ischemia [40]. These findings, then, suggest that ischemia-induced autophagy is protective, serving to replenish depleted energy stores and rid the cell of dysfunctional, potentially toxic, organelles.
Some evidence points to stabilization of hypoxiainducible factor 1α (HIF1α) as a trigger of autophagy in ischemia; inactivation of HIF1α blunts hypoxia-induced autophagy in fibroblasts [69]. In this setting, the cell survival-promoting activity of autophagy may derive from its ability to clear dysfunctional mitochondria, which would otherwise release reactive oxygen species and pro-apoptotic molecules. In addition, AMP-activated protein kinase (AMPK) participates in the regulation of autophagy during ischemia [40]. Together, these data suggest that both HIF1α and AMPK participate in the activation of autophagy in myocardial ischemia, which in turn serves to eliminate dysfunctional organelles and provide energy sources required for cellular homeostasis.
Ischemia may play a role in the autophagic response observed in hypertensive heart disease. For example, coronary angiogenesis is induced during hypertrophic growth of the myocardium to meet the increased metabolic and oxygen demands of enlarged cardiomyocytes. Indeed, precise coordination of the processes of cellular growth and angiogenesis is critical to the remodeling process elicited by changes in functional demand [22, 55]. At some point, however, continued growth of the heart ultimately depletes the capacity for angiogenesis, resulting in limited nutrient and oxygen supply; these latter events may in turn induce autophagy. Consistent with this model, decreased capillary density has been reported in failing human hearts, suggesting again that angiogenesis inadequate to meet the metabolic demands of the hypertrophic myocardium contributes to cardiac decompensation [19].
Autophagy in Ischemia/Reperfusion
Reoxygenation after ischemia triggers a second wave of lysosomal activation [6, 8]. Increased expression of elements of the autophagic pathway, including Beclin 1 and LC3, have been reported in a swine model of chronic ischemia/reperfusion (I/R) [66]. Similar findings have been reported in the cardiac myoblast cell line H9c2, HL-1 cells, NRVMs, and isolated rat hearts [13, 14, 60].
However, divergent results have emerged regarding whether autophagy is adaptive or maladaptive in I/R injury, a fact that may stem from the inherent complexity of this process and/or differences in experimental systems employed. In mice exposed to I/R injury, genetic loss-of-autophagic function models manifested attenuated infarction [40]. Consistent findings have been reported in both NRVM and adult cardiomyocytes [60]; I/R activated cell death and autophagy, and pharmacological suppression of autophagy promoted cell viability. Collectively, these data lend support to the notion that autophagy induced by I/R is detrimental.
Different results have been reported in the HL-1 cell line, however, where there was augmentation of simulated I/R injury with rapamycin or Beclin 1 overexpression was protective [14]. Brief episodes of ischemia activate myocyte autophagy, and treatment with wortmannin to block autophagic induction abolishes the cell-protective effects [13]. Autophagic activation in simulated I/R injury elicited by an adenosine receptor agonist is protective in both HL-1 cells and in NRVM [67]. Together, these data suggest that autophagy activated during simulated I/R in these cells is a prosurvival, adaptive response. Clearly, more work is warranted to elucidate the role of autophagy as protective or detrimental during I/R.
Bnip3, a downstream target of HIF1α, is expressed at negligible levels under basal conditions, but its abundance increases substantially after I/R [10], and some evidence implicates it in I/R-induced cell death. Ablation of Bnip3 does not elicit an obvious cardiac phenotype. However, left-ventricular systolic performance and diminished I/R-induced cardiac dilatation are seen in Bnip3-deficient mice, even though infarct size was similar to wild-type animals [10]. Conversely, cardiomyocyte-specific overexpression of Bnip3 induces progressive ventricular dilation and impaired systolic performance, possibly due to increased apoptosis. These results suggest that Bnip3 promotes I/R-induced cell death. Consistent with this, Bnip3 induces mitochondrial fragmentation and autophagy in cardiac myocytes, and suppression of Bnip3 using a dominant-negative mutant protected against I/R injury [15]. Similarly, hypoxia is capable of inducing Bnip3, which in turn is required for mitochondrial autophagy [69]. Collectively, these findings suggest that Bnip3 contributes to cell death during I/R injury, positioning Bnip3 as a potential therapeutic target.
Perspective
The expanding worldwide epidemic of cardiovascular disease poses an enormous public health challenge. Despite the complexity of the myriad manifestations of these disorders, activation of cardiomyocyte autophagy has emerged as a near-ubiquitous feature. Whereas basal, constitutive autophagy is indispensible to maintain cellular homeostasis and normal cardiac function, autophagy is activated in response to the cellular stresses occurring in virtually all forms of heart disease. In some contexts, autophagy is adaptive and protective, providing energy resources and molecular building blocks to promote cell function and survival. Under other circumstances, however, autophagic activity is maladaptive, promoting disease pathogenesis and cell death.
Not only is cardiomyocyte autophagy remarkably prevalent in cardiovascular physiology and pathology, the process itself can be manipulated at several crucial nodal points. Indeed, a number of small molecule modulators of autophagy are available (e.g., PI3K inhibitors) or in development (e.g., histone deaceytylase inhibitors) [2]. Thus, it is easy to envision a day when therapeutic titration of cardiomyocyte autophagy will emerge as a strategy to prevent, slow, or even reverse, heart disease.
Precedent exists in other areas, such as oncology, for a requirement of finely tuned autophagic activation; too little autophagy and too much autophagy can each be detrimental. Despite considerable progress in recent years in our understanding of the molecular machinery of autophagy, our understanding of its adaptive-versus-maladaptive consequences is lagging. Looking to the future, detailed elucidation of the mechanisms and effects of autophagy in heart disease will be essential to our long-term goal of targeting pathological autophagy for therapeutic gain.
Acknowledgments
We are grateful to members of the Hill laboratory for valuable suggestions and comments. This work was supported by grants from the National Institutes of Health NIH (Grants No. HL-075173, HL-080144, and HL-090842), The American Heart Association (AHA; Grant No. 0640084N), and the AHA—Jon Holden DeHaan Foundation.
Footnotes
Conflicts of interest None.
References
- 1.Bergmann O, Bhardwaj RD, Bernard S, Zdunek S, Barnabe-Heider F, Walsh S, et al. Evidence for cardiomyocyte renewal in humans. Science. 2009;324:98–102. doi: 10.1126/science.1164680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cao DJ, Wang ZV, Battiprolu PK, Jiang N, Morales C, Kong Y, et al. HDAC inhibitors attenuate cardiac hypertrophy by suppressing autophagy. Proc Natl Acad Sci USA. 2011 doi: 10.1073/pnas.1015081108. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chan EY. mTORC1 phosphorylates the ULK1-mAtg13-FIP200 autophagy regulatory complex. Sci Signal. 2009;2:pe51. doi: 10.1126/scisignal.284pe51. [DOI] [PubMed] [Google Scholar]
- 4.Chen Q, Liu JB, Horak KM, Zheng H, Kumarapeli AR, Li J, et al. Intrasarcoplasmic amyloidosis impairs proteolytic function of proteasomes in cardiomyocytes by compromising substrate uptake. Circ Res. 2005;97:1018–1026. doi: 10.1161/01.RES.0000189262.92896.0b. [DOI] [PubMed] [Google Scholar]
- 5.Dalakas MC, Park KY, Semino-Mora C, Lee HS, Sivakumar K, Goldfarb LG. Desmin myopathy, a skeletal myopathy with cardiomyopathy caused by mutations in the desmin gene. N Engl J Med. 2000;342:770–780. doi: 10.1056/NEJM200003163421104. [DOI] [PubMed] [Google Scholar]
- 6.Decker RS, Wildenthal K. Lysosomal alterations in hypoxic and reoxygenated hearts. I. Ultrastructural and cytochemical changes. Am J Pathol. 1980;98:425–444. [PMC free article] [PubMed] [Google Scholar]
- 7.Decker RS, Decker ML, Herring GH, Morton PC, Wildenthal K. Lysosomal vacuolar apparatus of cardiac myocytes in heart of starved and re-fed rabbits. J Mol Cell Cardiol. 1980;12:1175–1189. doi: 10.1016/0022-2828(80)90064-4. [DOI] [PubMed] [Google Scholar]
- 8.Decker RS, Poole AR, Crie JS, Dingle JT, Wildenthal K. Lysosomal alterations in hypoxic and reoxygenated hearts. II. Immunohistochemical and biochemical changes in cathepsin D. Am J Pathol. 1980;98:445–456. [PMC free article] [PubMed] [Google Scholar]
- 9.Depre C, Wang Q, Yan L, Hedhli N, Peter P, Chen L, et al. Activation of the cardiac proteasome during pressure overload promotes ventricular hypertrophy. Circulation. 2006;114:1821–1828. doi: 10.1161/CIRCULATIONAHA.106.637827. [DOI] [PubMed] [Google Scholar]
- 10.Diwan A, Krenz M, Syed FM, Wansapura J, Ren X, Koesters AG, et al. Inhibition of ischemic cardiomyocyte apoptosis through targeted ablation of Bnip3 restrains postinfarction remodeling in mice. J Clin Invest. 2007;117:2825–2833. doi: 10.1172/JCI32490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Elsasser A, Vogt AM, Nef H, Kostin S, Mollmann H, Skwara W, et al. Human hibernating myocardium is jeopardized by apoptotic and autophagic cell death. J Am Coll Cardiol. 2004;43:2191–2199. doi: 10.1016/j.jacc.2004.02.053. [DOI] [PubMed] [Google Scholar]
- 12.Frey N, Katus HA, Olson EN, Hill JA. Hypertrophy of the heart: a new therapeutic target? Circulation. 2004;109:1580–1589. doi: 10.1161/01.CIR.0000120390.68287.BB. [DOI] [PubMed] [Google Scholar]
- 13.Gurusamy N, Lekli I, Gorbunov NV, Gherghiceanu M, Popescu LM, Das DK. Cardioprotection by adaptation to ischaemia augments autophagy in association with BAG-1 protein. J Cell Mol Med. 2009;13:373–387. doi: 10.1111/j.1582-4934.2008.00495.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hamacher-Brady A, Brady NR, Gottlieb RA. Enhancing macroautophagy protects against ischemia/reperfusion injury in cardiac myocytes. J Biol Chem. 2006;281:29776–29787. doi: 10.1074/jbc.M603783200. [DOI] [PubMed] [Google Scholar]
- 15.Hamacher-Brady A, Brady NR, Logue SE, Sayen MR, Jinno M, Kirshenbaum LA, et al. Response to myocardial ischemia/reperfusion injury involves Bnip3 and autophagy. Cell Death Differ. 2007;14:146–157. doi: 10.1038/sj.cdd.4401936. [DOI] [PubMed] [Google Scholar]
- 16.Hein S, Arnon E, Kostin S, Schonburg M, Elsasser A, Polyakova V, et al. Progression from compensated hypertrophy to failure in the pressure-overloaded human heart: structural deterioration and compensatory mechanisms. Circulation. 2003;107:984–991. doi: 10.1161/01.cir.0000051865.66123.b7. [DOI] [PubMed] [Google Scholar]
- 17.Hill JA, Olson EN. Cardiac plasticity. N Engl J Med. 2008;358:1370–1380. doi: 10.1056/NEJMra072139. [DOI] [PubMed] [Google Scholar]
- 18.Hill JA, Karimi M, Kutschke W, Davisson RL, Zimmerman K, Wang Z, et al. Cardiac hypertrophy is not a required compensatory response to short-term pressure overload. Circulation. 2000;101:2863–2869. doi: 10.1161/01.cir.101.24.2863. [DOI] [PubMed] [Google Scholar]
- 19.Hudlicka O, Brown M, Egginton S. Angiogenesis in skeletal and cardiac muscle. Physiol Rev. 1992;72:369–417. doi: 10.1152/physrev.1992.72.2.369. [DOI] [PubMed] [Google Scholar]
- 20.Inoue Y, Klionsky DJ. Regulation of macroautophagy in Saccharomyces cerevisiae. Semin Cell Dev Biol. 2010;21:664–670. doi: 10.1016/j.semcdb.2010.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Iwata A, Riley BE, Johnston JA, Kopito RR. HDAC6 and microtubules are required for autophagic degradation of aggregated huntingtin. J Biol Chem. 2005;280:40282–40292. doi: 10.1074/jbc.M508786200. [DOI] [PubMed] [Google Scholar]
- 22.Izumiya Y, Shiojima I, Sato K, Sawyer DB, Colucci WS, Walsh K. Vascular endothelial growth factor blockade promotes the transition from compensatory cardiac hypertrophy to failure in response to pressure overload. Hypertension. 2006;47:887–893. doi: 10.1161/01.HYP.0000215207.54689.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kamada Y, Funakoshi T, Shintani T, Nagano K, Ohsumi M, Ohsumi Y. Tor-mediated induction of autophagy via an Apg1 protein kinase complex. J Cell Biol. 2000;150:1507–1513. doi: 10.1083/jcb.150.6.1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kamada Y, Sekito T, Ohsumi Y. Autophagy in yeast: a TOR-mediated response to nutrient starvation. Curr Top Microbiol Immunol. 2004;279:73–84. doi: 10.1007/978-3-642-18930-2_5. [DOI] [PubMed] [Google Scholar]
- 25.Kanamori H, Takemura G, Maruyama R, Goto K, Tsujimoto A, Ogino A, et al. Functional significance and morphological characterization of starvation-induced autophagy in the adult heart. Am J Pathol. 2009;174:1705–1714. doi: 10.2353/ajpath.2009.080875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kanki T, Wang K, Cao Y, Baba M, Klionsky DJ. Atg32 is a mitochondrial protein that confers selectivity during mitophagy. Dev Cell. 2009;17:98–109. doi: 10.1016/j.devcel.2009.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim I, Rodriguez-Enriquez S, Lemasters JJ. Selective degradation of mitochondria by mitophagy. Arch Biochem Biophys. 2007;462:245–253. doi: 10.1016/j.abb.2007.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Klionsky DJ. Autophagy: from phenomenology to molecular understanding in less than a decade. Nat Rev Mol Cell Biol. 2007;8:931–937. doi: 10.1038/nrm2245. [DOI] [PubMed] [Google Scholar]
- 29.Knaapen MW, Davies MJ, De Bie M, Haven AJ, Martinet W, Kockx MM. Apoptotic versus autophagic cell death in heart failure. Cardiovasc Res. 2001;51:304–312. doi: 10.1016/s0008-6363(01)00290-5. [DOI] [PubMed] [Google Scholar]
- 30.Kostin S, Pool L, Elsasser A, Hein S, Drexler HC, Arnon E, et al. Myocytes die by multiple mechanisms in failing human hearts. Circ Res. 2003;92:715–724. doi: 10.1161/01.RES.0000067471.95890.5C. [DOI] [PubMed] [Google Scholar]
- 31.Kundu M, Thompson CB. Autophagy: basic principles and relevance to disease. Annu Rev Pathol. 2008;3:427–455. doi: 10.1146/annurev.pathmechdis.2.010506.091842. [DOI] [PubMed] [Google Scholar]
- 32.Levine B, Klionsky DJ. Development by self-digestion: molecular mechanisms and biological functions of autophagy. Dev Cell. 2004;6:463–477. doi: 10.1016/s1534-5807(04)00099-1. [DOI] [PubMed] [Google Scholar]
- 33.Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132:27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Levine B, Yuan J. Autophagy in cell death: an innocent convict? J Clin Invest. 2005;115:2679–2688. doi: 10.1172/JCI26390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Levy D, Garrison RJ, Savage DD, Kannel WB, Castelli WP. Prognostic implications of echocardiographically determined left ventricular mass in the Framingham Heart Study. N Engl J Med. 1990;322:1561–1566. doi: 10.1056/NEJM199005313222203. [DOI] [PubMed] [Google Scholar]
- 36.Lockshin RA, Zakeri Z. Caspase-independent cell deaths. Curr Opin Cell Biol. 2002;14:727–733. doi: 10.1016/s0955-0674(02)00383-6. [DOI] [PubMed] [Google Scholar]
- 37.Maloyan A, Sanbe A, Osinska H, Westfall M, Robinson D, Imahashi K, et al. Mitochondrial dysfunction and apoptosis underlie the pathogenic process in alpha-B-crystallin desmin-related cardiomyopathy. Circulation. 2005;112:3451–3461. doi: 10.1161/CIRCULATIONAHA.105.572552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Maron BJ, Roberts WC, Arad M, Haas TS, Spirito P, Wright GB, et al. Clinical outcome and phenotypic expression in LAMP2 cardiomyopathy. JAMA. 2009;301:1253–1259. doi: 10.1001/jama.2009.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Martinet W, Knaapen MW, Kockx MM, De Meyer GR. Autophagy in cardiovascular disease. Trends Mol Med. 2007;13:482–491. doi: 10.1016/j.molmed.2007.08.004. [DOI] [PubMed] [Google Scholar]
- 40.Matsui Y, Takagi H, Qu X, Abdellatif M, Sakoda H, Asano T, et al. Distinct roles of autophagy in the heart during ischemia and reperfusion: roles of AMP-activated protein kinase and Beclin 1 in mediating autophagy. Circ Res. 2007;100:914–922. doi: 10.1161/01.RES.0000261924.76669.36. [DOI] [PubMed] [Google Scholar]
- 41.Mizushima N. Autophagy: process and function. Genes Dev. 2007;21:2861–2873. doi: 10.1101/gad.1599207. [DOI] [PubMed] [Google Scholar]
- 42.Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature. 2008;451:1069–1075. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nakai A, Yamaguchi O, Takeda T, Higuchi Y, Hikoso S, Taniike M, et al. The role of autophagy in cardiomyocytes in the basal state and in response to hemodynamic stress. Nat Med. 2007;13:619–624. doi: 10.1038/nm1574. [DOI] [PubMed] [Google Scholar]
- 44.Nishida K, Kyoi S, Yamaguchi O, Sadoshima J, Otsu K. The role of autophagy in the heart. Cell Death Differ. 2009;16:31–38. doi: 10.1038/cdd.2008.163. [DOI] [PubMed] [Google Scholar]
- 45.Nishino I, Fu J, Tanji K, Yamada T, Shimojo S, Koori T, et al. Primary LAMP-2 deficiency causes X-linked vacuolar cardiomyopathy and myopathy (Danon disease). Nature. 2000;406:906–910. doi: 10.1038/35022604. [DOI] [PubMed] [Google Scholar]
- 46.Omary MB, Coulombe PA, McLean WH. Intermediate filament proteins and their associated diseases. N Engl J Med. 2004;351:2087–2100. doi: 10.1056/NEJMra040319. [DOI] [PubMed] [Google Scholar]
- 47.Perng MD, Wen SF, van den IJssel P, Prescott AR, Quinlan RA. Desmin aggregate formation by R120G alphaB-crystallin is caused by altered filament interactions and is dependent upon network status in cells. Mol Biol Cell. 2004;15:2335–2346. doi: 10.1091/mbc.E03-12-0893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pfeifer U, Fohr J, Wilhelm W, Dammrich J. Short-term inhibition of cardiac cellular autophagy by isoproterenol. J Mol Cell Cardiol. 1987;19:1179–1184. doi: 10.1016/s0022-2828(87)80528-x. [DOI] [PubMed] [Google Scholar]
- 49.Rajasekaran NS, Connell P, Christians ES, Yan LJ, Taylor RP, Orosz A, et al. Human alpha B-crystallin mutation causes oxido-reductive stress and protein aggregation cardiomyopathy in mice. Cell. 2007;130:427–439. doi: 10.1016/j.cell.2007.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Roger VL, Go AS, Lloyd-Jones DM, Adams RJ, Berry JD, Brown TM, et al. Heart disease and stroke statistics̄2011 update: a report from the American Heart Association. Circulation. 2010 [Google Scholar]
- 51.Rothermel BA, Hill JA. Myocyte autophagy in heart disease: friend or foe? Autophagy. 2007;3:632–634. doi: 10.4161/auto.4913. [DOI] [PubMed] [Google Scholar]
- 52.Rothermel BA, Hill JA. Autophagy in load-induced heart disease. Circ Res. 2008;103:1363–1369. doi: 10.1161/CIRCRESAHA.108.186551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sanbe A, Osinska H, Saffitz JE, Glabe CG, Kayed R, Maloyan A, et al. Desmin-related cardiomyopathy in transgenic mice: a cardiac amyloidosis. Proc Natl Acad Sci USA. 2004;101:10132–10136. doi: 10.1073/pnas.0401900101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shimomura H, Terasaki F, Hayashi T, Kitaura Y, Isomura T, Suma H. Autophagic degeneration as a possible mechanism of myocardial cell death in dilated cardiomyopathy. Jpn Circ J. 2001;65:965–968. doi: 10.1253/jcj.65.965. [DOI] [PubMed] [Google Scholar]
- 55.Shiojima I, Sato K, Izumiya Y, Schiekofer S, Ito M, Liao R, et al. Disruption of coordinated cardiac hypertrophy and angiogenesis contributes to the transition to heart failure. J Clin Invest. 2005;115:2108–2118. doi: 10.1172/JCI24682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tanaka Y, Guhde G, Suter A, Eskelinen EL, Hartmann D, Lullmann-Rauch R, et al. Accumulation of autophagic vacuoles and cardiomyopathy in LAMP-2-deficient mice. Nature. 2000;406:902–906. doi: 10.1038/35022595. [DOI] [PubMed] [Google Scholar]
- 57.Tannous P, Zhu H, Johnstone JL, Shelton JM, Rajasekaran NS, Benjamin IJ, et al. Autophagy is an adaptive response in desmin-related cardiomyopathy. Proc Natl Acad Sci USA. 2008;105:9745–9750. doi: 10.1073/pnas.0706802105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tannous P, Zhu H, Nemchenko A, Berry JM, Johnstone JL, Shelton JM, et al. Intracellular protein aggregation is a proximal trigger of cardiomyocyte autophagy. Circulation. 2008;117:3070–3078. doi: 10.1161/CIRCULATIONAHA.107.763870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Twig G, Elorza A, Molina AJ, Mohamed H, Wikstrom JD, Walzer G, et al. Fission and selective fusion govern mitochondrial segregation and elimination by autophagy. EMBO J. 2008;27:433–446. doi: 10.1038/sj.emboj.7601963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Valentim L, Laurence KM, Townsend PA, Carroll CJ, Soond S, Scarabelli TM, et al. Urocortin inhibits Beclin1-mediated autophagic cell death in cardiac myocytes exposed to ischaemia/reperfusion injury. J Mol Cell Cardiol. 2006;40:846–852. doi: 10.1016/j.yjmcc.2006.03.428. [DOI] [PubMed] [Google Scholar]
- 61.Vicart P, Caron A, Guicheney P, Li Z, Prevost MC, Faure A, et al. A missense mutation in the alphaB-crystallin chaperone gene causes a desmin-related myopathy. Nat Genet. 1998;20:92–95. doi: 10.1038/1765. [DOI] [PubMed] [Google Scholar]
- 62.Wang X, Osinska H, Dorn GW, 2nd, Nieman M, Lorenz JN, Gerdes AM, et al. Mouse model of desmin-related cardiomyopathy. Circulation. 2001;103:2402–2407. doi: 10.1161/01.cir.103.19.2402. [DOI] [PubMed] [Google Scholar]
- 63.Wang X, Osinska H, Klevitsky R, Gerdes AM, Nieman M, Lorenz J, et al. Expression of R120G-alphaB-crystallin causes aberrant desmin and alphaB-crystallin aggregation and cardiomyopathy in mice. Circ Res. 2001;89:84–91. doi: 10.1161/hh1301.092688. [DOI] [PubMed] [Google Scholar]
- 64.Xie Z, Klionsky DJ. Autophagosome formation: core machinery and adaptations. Nat Cell Biol. 2007;9:1102–1109. doi: 10.1038/ncb1007-1102. [DOI] [PubMed] [Google Scholar]
- 65.Yamamoto S, Sawada K, Shimomura H, Kawamura K, James TN. On the nature of cell death during remodeling of hypertrophied human myocardium. J Mol Cell Cardiol. 2000;32:161–175. doi: 10.1006/jmcc.1999.1064. [DOI] [PubMed] [Google Scholar]
- 66.Yan L, Vatner DE, Kim SJ, Ge H, Masurekar M, Massover WH, et al. Autophagy in chronically ischemic myocardium. Proc Natl Acad Sci USA. 2005;102:13807–13812. doi: 10.1073/pnas.0506843102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yitzhaki S, Huang C, Liu W, Lee Y, Gustafsson AB, Mentzer RM, Jr, et al. Autophagy is required for preconditioning by the adenosine A1 receptor-selective agonist CCPA. Basic Res Cardiol. 2009;104:157–167. doi: 10.1007/s00395-009-0006-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yu L, McPhee CK, Zheng L, Mardones GA, Rong Y, Peng J, et al. Termination of autophagy and reformation of lysosomes regulated by mTOR. Nature. 2010;465:942–946. doi: 10.1038/nature09076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhang H, Bosch-Marce M, Shimoda LA, Tan YS, Baek JH, Wesley JB, et al. Mitochondrial autophagy is an HIF-1-dependent adaptive metabolic response to hypoxia. J Biol Chem. 2008;283:10892–10903. doi: 10.1074/jbc.M800102200. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 70.Zhu H, Tannous P, Johnstone JL, Kong Y, Shelton JM, Richardson JA, et al. Cardiac autophagy is a maladaptive response to hemodynamic stress. J Clin Invest. 2007;117:1782–1793. doi: 10.1172/JCI27523. [DOI] [PMC free article] [PubMed] [Google Scholar]