Abstract
Carcinogen-induced oral cavity squamous cell carcinoma (OSCC) incurs significant morbidity and mortality and constitutes a global health challenge. To gain further insight into this disease, we generated cell line models from DMBA-induced murine primary OSCC capable of tumor formation upon transplantation into immunocompetent wild-type mice. While several lines grew rapidly and were capable of metastasis, some grew slowly and did not metastasize. Aggressively growing lines displayed ERK1/2 activation, which stimulated expression of CD44, a marker associated with EMT and putative cancer stem cells. MEK inhibition upstream of ERK1/2 decreased CD44 expression and promoter activity and reduced cell migration and invasion. Conversely, MEK1 activation enhanced CD44 expression and promoter activity, whereas CD44 attenuation reduced in vitro migration and in vivo tumor formation. Extending these findings to freshly resected human OSCC, we confirmed a strict relationship between ERK1/2 phosphorylation and CD44 expression. In summary, our findings identify CD44 as a critical target of ERK1/2 in promoting tumor aggressiveness and offer a preclinical proof of concept to target this pathway as a strategy to treat head and neck cancer.
Keywords: ERK1/2, oral cancer, CD44, metastasis, mouse models
Introduction
Oral cavity squamous cell carcinoma (OSCC) is a prominent subset of head and neck cancers, which are the 6th most common cancer worldwide (1). The major risk factor for developing OSCC is carcinogen exposure, which distinguishes this subset of head and neck cancers from those induced by human papillomavirus (1, 2). Despite advances in detection, surgery, chemotherapy, and radiation, the prognosis for OSCC has remained stable for decades (1–3). Furthermore, at the time of diagnosis, approximately two-thirds of OSCC patients have locoregionally advanced disease resulting in increased morbidity and mortality (1–3).
Multiple mutations and epigenetic alterations of signaling and regulatory proteins have been identified as promoters of OSCC. These include alterations in TP53, CDKN2A, PTEN, PIK3CA, and Notch1 (4, 5). The RAS signaling pathway is also modulated, with 5–50% of OSCC having mutated RAS and 45–80% having overexpressed non-mutated RAS (1, 6). Similarly, overexpression of the epidermal growth factor receptor (EGFR), found in 80–90% of OSCC, heralds a worse prognosis (7). Furthermore, key pro-growth and pro-survival signaling cascades, including signal transducer and activator of transcription 3 (STAT3), protein kinase B (AKT), and nuclear factor-κB (NFκB), are constitutively activated in OSCC (8). The extracellular signal-regulated kinases (ERK1/2) have received less attention in OSCC but do activate IL-8 and VEGF expression in human OSCC cell lines and in the context of PTPN13 loss, promote tumorigenesis in both HPV positive and negative squamous cancers (9, 10). Aberrations in these pathways are common to many OSCC and thus, defining pathways or tumor subtypes that distinguish more aggressive lesions is critical for therapeutic targeting.
CD44+ cells are such targets for three reasons—first, they have enhanced chemoresistance; second, they are proposed to be cancer stem cells (CSCs) as they engraft at higher frequencies in mice; and third, in other systems, they are associated with the epithelial to mesenchymal transition (EMT), a genetic program associated with metastasis (11–13). CD44 functions as a transmembrane protein that, along with RHAMM, is the principal receptor for hyaluronan and also is a co-receptor for several receptor tyrosine kinases (RTKs) including c-MET and EGFR (14, 15). Initial work on its regulation focused on alternative splicing and showed that the v6 CD44 isoform promoted metastatic activity (16). In addition, other levels of regulation have been demonstrated including p53-mediated repression, RAS-mediated alternative splicing or transcription and microRNA-mediated attenuation (17–20). Regulation of CD44 expression remains incompletely understood in head and neck cancers. In OSCC, increased CD44 expression has been associated with decreased survival and increased recurrence, metastasis, and resistance to chemo/radiation therapy (11, 21, 22). Whether these properties are due to a functional contribution of CD44 or whether CD44 is simply a marker of cells with more aggressive behavior has not been fully explored in OSCC (21).
Models for the study of OSCC have relied primarily on immunodeficient xenograft models, which overlook tumor-immune interactions. In contrast to the extensive literature on syngeneic murine transplantable models in other tumor systems, studies in OSCC are limited. Herein, we describe a transplantable syngeneic murine model of OSCC derived from carcinogen-induced primary tumors. Utilizing this model, we identified that increased activation of ERK1/2 was associated with locoregional aggressiveness and enhanced transcription of CD44, a key driving force for in vitro migration and in vivo growth. We then extended these data to human OSCC and identified that increased ERK1/2 phosphorylation and CD44 expression were tightly associated. Thus, these mouse cell lines provide insight into ERK1/2-CD44 contribution to tumor aggressiveness and constitute a syngeneic, transplantable murine model of OSCC that will allow an evaluation of the intrinsic tumor biology and the host immune responses to OSCC development and metastasis.
Materials and Methods
Animals
C57BL/6 mice were from Taconic and CXCR3−/− mice have been described (23). Studies were performed under approved protocols of the Animal Studies Committee of Washington University.
Plasmids
Mouse CD44 shRNA and control scramble plasmids were from Sigma-Aldrich. Mouse CD44 luciferase reporters were provided by Mark Perrella (24). The estrogen regulated constitutively active MEK1/R4F-ER was from Addgene (25).
Antibodies
All primary antibodies were from Cell Signaling Technologies except anti-STAT3 (Santa Cruz Biotechnology) and anti-β-actin (Sigma-Aldrich). Secondary antibodies were conjugated to Alexaflour 680 (Invitrogen) or IRDye 800 (Rockland). PE-anti-mouse/human CD44 (1M7), PE-rat IgG2b-κ isotype control, and APC anti-mouse CD24 were from Biolegend. For immunofluorescence, anti-cytokeratin (DAKO), CD44 (1M7) (BD Biosciences) and p-ERK1/2 (Cell Signaling Technologies) were used. Secondaries were Cy2 and Cy3-conjugated donkey anti-rabbit (Jackson ImmunoResearch), and Alexa-488 goat anti-rat antibodies (Invitrogen).
Cell lines
Primary DMBA induced mouse OSCC were generated as described (26). Single cell suspensions of individual primary oral cavity tumors were made with Collagenase IA (Sigma-Aldrich) and cultured in IMDM/F12 (2:1) with 5% FCS, penicillin/streptomycin, 1% amphotericin, 5 ng/mL EGF (Millipore), 400 ng/mL hydrocortisone, and 5 μg/mL insulin. Sequential differential trypsinization was then used to clear fibroblast contamination. MOC1, 7, 10, 22 and 23 were derived from primary tumors in C57BL/6 WT mice and MOC2 was derived from a chemokine receptor CXCR3deficient mouse on a pure C57BL/6 background (27) (of note, no major differences in the incidence of tumor formation were noted between the different genotypes). CXCR3 is not detectable on oral keratinocytes and does not contribute to MOC2 growth (Figure S3). Immunofluorescence staining for cytokeratin was performed to confirm an epithelial phenotype (Figure 1C and Figure S1C). PCI-13 was obtained from Dr. Theresa Whiteside, UPCI:SCC029B and UPCI:SCC068 were obtained from Dr. Suzanne Gollin, and all were used with minimal passaging. The UM-SCC-1 cell line (from Dr. Tom Carey) was genotyped in May, 2011 and concordance with published data was established (27).
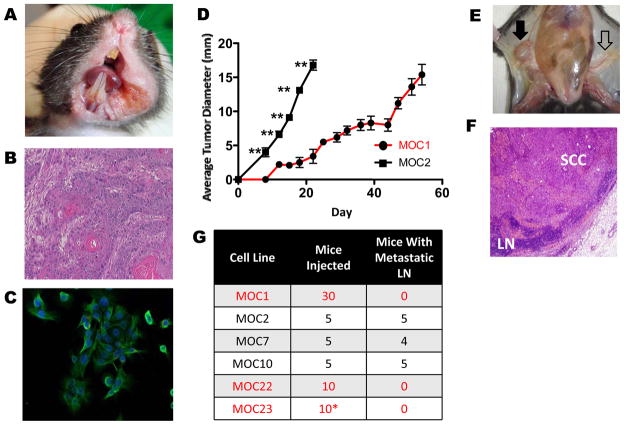
Figure 1. Variant growth patterns and lymphatic metastasis in a C57BL/6 syngeneic model of OSCC.
A. Representative primary OSCC in left floor of mouth/buccal region after 25 weeks of biweekly DMBA treatment (MOC10 parent tumor). B. Representative H&E stained section of primary tumor (MOC10 parent tumor) showing moderately differentiated squamous cell carcinoma (40X magnification). C. Immunofluorescence of MOC10 cell line showing epithelial phenotype with positive cytokeratin staining (green) and DAPI for nuclear staining (40X magnification). D. Representative in vivo growth curves comparing indolent MOC1 and aggressive MOC2 after 1×106 cells were injected into the right flank of C57BL/6 WT mice (**=p<0.01 in all days post day 0). E. Metastatic inguinal draining lymph node (black filled arrow) that is enlarged and discolored compared to contralateral normal appearing lymph node (black open arrow). F. H&E stained metastatic lymph node shows effacement of normal architecture by SCC (LN= lymph node, 20X magnification). G. Summary of LN metastatic capacity of each cell line. (*MOC23 data is shown for RAG2−/− mice, as this line does not form progressive tumors in WT mice)
Transwell Migration Assay
1×105 cells were loaded into the upper chamber and complete media in the lower chamber of Transwell plates (8 μm, BD Biosciences). After incubation for 24 hours, cells in the lower chamber were fixed and stained (DiffQuick, Dade Behring) and counted.
Wound Healing (Scratch) Test
Cells at 80% confluency were wounded with a sterile 200μl pipette. Cell migration was recorded by microscopy at 0 and 24 hours.
Western Blot and immunoprecipitation
Cell Extraction Buffer (Invitrogen) was used to make lysates and westerns were performed as described (28). A Pierce Co-Immunoprecipitation Kit (Thermo Scientific) was used for STAT3 analysis.
FACS
Tumor cells were blocked with rat serum and stained with antibodies at 4°C for 30 minutes. Data were collected on a FACSCalibur (BD Biosciences) and analyzed using FloJo software (Tree Star). Further details are included in the Supplementary Methods.
Immunofluorescence
Cells were grown on cover slips, fixed and permeabilized. Paraffin embedded sections underwent antigen retrieval. Specimens were blocked and then incubated with primary antibodies for 1 hour. Detection was accomplished with secondaries and DAPI (Invitrogen) for the nuclear stain.
Tumor transplantation
Cell lines were harvested, washed twice in D-PBS (Fisher), and injected into the right subcutaneous flank of mice. Tumor growth was recorded as the average of the two largest diameters.
Luciferase assay and infections
Cell lines were transfected (Fugene, Promega) and lysates were analyzed using the Dual Luciferase Reporter System (Promega). For shRNA targeting, cell lines underwent lentiviral transduction and selected using puromycin (2.5μg/ml). For analysis of enforced ERK1/2 activation, MEK1/R4F-ER was retrovirally transduced into MOC1. Cells were then treated with vehicle or 4-OH-tamoxifen (200 nM, referred to as tamoxifen) for 48 hours and analyzed for CD44 expression. CD44 luciferase activity was evaluated 24 hours after co-transfection with MEK1/R4F-ER and CD44 reporter. For studies with U0126 (10 μM), cell lines were transfected and treated with drug for 24 hours and assays performed as above.
Human Tumor Analysis
Under an IRB approved protocol at Stanford University, human primary OSCC tumors were dissociated and analyzed by flow cytometry. CD45+/CD31+ cells were excluded and tumor cells analyzed for CD44 and phospho-ERK1/2 (see Supplementary Methods).
Statistical Analysis
All analyses were performed with oversight from a biostatistician. Heterotopic tumor growth was analyzed by single day comparison analysis using Mann-Whitney U test (Nonparametric equivalent of independent samples t-test). Migration differences (Figure 2D) were analyzed by independent samples t-test, and in addition a two way ANOVA was performed. For comparison of tumor growth in CD44 knockdowns (Figure 3E), a mixed between-within subjects analysis of variance comparing scramble with the indicated CD44 knockdown was performed. Luciferase assays were all analyzed using independent samples t-tests as indicated.
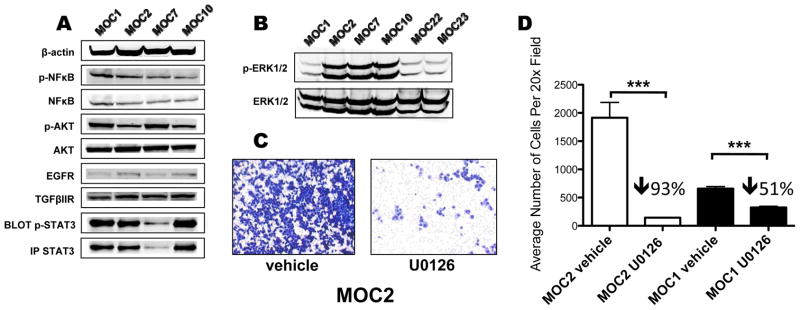
Figure 2. Increased activation of ERK1/2 is associated with increased OSCC aggressiveness.
A. Western blots of MOC1/2/7/10 for phospho-NFκB, NFκB, phospho-AKT, AKT, EGFR, TGFβIIR, and β-actin. STAT3 and phospho-STAT3 were visualized by immunoprecipation and Western blotting. Note in the experiment shown MOC7 had less total STAT3 immunoprecipitated, but in repeat experiments it was found to express similar amounts of STAT3 when compared with the other cell lines (data not shown). B. Western blot of p-ERK1/2 and ERK1/2 for all 6 MOC lines. C. Representative 20× microscopic image of a Transwell migration assay of MOC2 cells treated with vehicle control (DMSO) or U0126 (10μM). D. Quantitation of Transwell migration assay where 4 random sections per filter × 3 filters were counted in a blinded fashion by light microscopy at 20× magnification for both MOC1 and MOC2 cells treated with vehicle or U0126 (***=p<0.001). The percentage decrease relative to vehicle treatment is indicated above the bar graph.
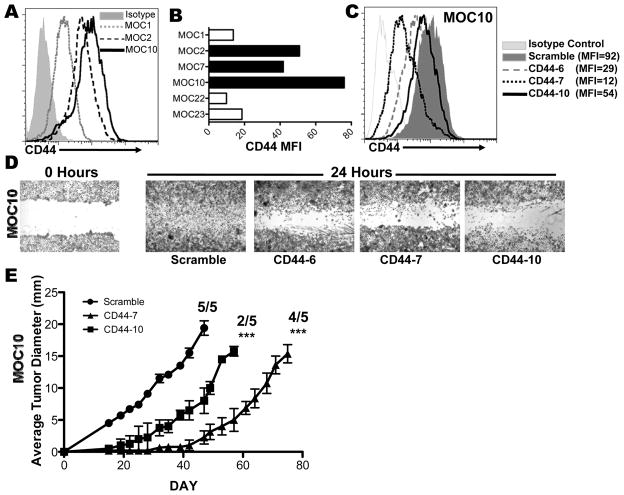
Figure 3. CD44 is associated with and contributes to increased OSCC aggressiveness.
A. FACS analysis of cell surface CD44 expression by pan-CD44 antibody (1M7) and isotype control shown for MOC1, 2 and 10. B. Representative mean fluorescence intensity for cell surface CD44 expression in all 6 cell lines (from one of at least three experiments). C. FACS analysis of cell surface CD44 expression in MOC10 after shRNA knockdown of CD44 with 3 distinct shRNAs (CD44-6, CD44-7, CD44-10) or scramble control shRNA (quantitated by indicated MFI). D. Scratch Test of MOC10 after transduction with indicated CD44 or scramble shRNAs. E. CD44 shRNA knockdown leads to delayed or abrogated growth of MOC10. MOC10 cells (1×104) transduced with scramble, CD44-7, or CD44-10 shRNA were injected into the right flank of C57BL/6 WT mice and monitored. Growth curves only represent average tumor diameter of transplanted tumors that grew out as indicated (***=p<0.001).
Results
A new OSCC model
To model OSCC, we generated six transplantable mouse OSCC cell lines from independent carcinogen-induced tumors (26). After 25 weeks of twice weekly oral cavity DMBA application, mice developed squamous cell carcinomas (Figure 1A, B and Figure S1A, B) and six cell lines, designated mouse oral cancer (MOC1, 2, 7, 10, 22 and 23), were derived.
To assess their in vivo growth, cell lines were transplanted either heterotopically in the flank, where tumors can be easily monitored, or orthotopically in the floor of mouth/buccal region (Figure S2B). Growth was seen at both sites; thus for ease of measurement we utilized heterotopic transplantation for these studies. The cell lines segregated into either an indolent (MOC1, 22) or aggressive (MOC2, 7, and 10) growth phenotypes (Figure 1D and S2A). MOC23 did not form tumors in WT mice and only grew in RAG2−/− immunodeficient mice (data not shown). The aggressive growth of MOC2 and MOC10 was illustrated by their ability to form tumors with injection of as few as 10,000 cells (Figures 3E and S6E). Notably, the cell lines reflected the clinical appearance of the primary tumors—MOC1, 22 and 23 were derived from exophytic lesions (see Figure S1A for MOC1) whereas MOC2, 7 and 10 were invasive appearing (Figure 1A and S1A).
Lymph node metastatic capacity of MOC lines
A key poor prognosticator of human OSCC is the presence of lymph node (LN) metastases. To assess this in the MOC lines, tissues from all mice in the transplant experiments in Figure 1D and S2A were examined for metastases (Figure 1E,F,G). We did not observe any lung or liver metastases in these mice, but interestingly, MOC2/7/10 displayed spontaneous metastasis to regional LNs in 5/5, 4/5 and 5/5 mice, respectively. Orthotopic injection of MOC2 and 10 also showed cervical metastasis at the same rate as in the flank (Figure S2C). In contrast MOC1 and MOC22 did not metastasize regionally or distantly (combined n=30 for MOC1 and n=10 for MOC22). When 1×104 MOC10 cells were injected, slower but progressive primary tumor growth was seen and both lymphatic and lung metastases were found (Figure S2D, E) suggesting that distant metastases require a longer primary tumor growth. Thus, 3 of 6 lines displayed regional LN metastasis recapitulating an important aspect of human OSCC.
ERK 1/2 activation is associated with increased OSCC aggressiveness
Given differences in growth phenotypes, we next addressed possible aberrant intracellular signaling pathways that could mediate these differences. We focused on well-characterized pathways from human OSCC and found minimal correlation in levels of EGFR, TGFβRII, phospho-AKT (Ser473), phospho-NFκB p65 (Ser536) or phopho-STAT3 (Tyr705) with the aggressive phenotype (Figure 2A). Interestingly, the most profound difference was in the level of phosphorylated ERK1/2 (Thr202/Tyr 204). MOC2/7/10, which all grew aggressively and consistently metastasized to draining lymph nodes in vivo, had increased phospho-ERK1/2 compared to the indolent MOC1, 22 or 23 (Figure 2B).
K-RAS mutations are associated with increased phospho-ERK1/2 in MOC lines
Although DMBA is widely mutagenic, DMBA induced RAS-family mutations are key oncogenic drivers in multistage carcinogenesis (29). To this end, we evaluated H-, K-, N-RAS and B-Raf for mutations in the cell lines. We found no N-RAS or B-Raf mutations in any of the cell lines. However, an activating H-RAS mutation in codon 61 was identified in MOC1 and MOC22, while the aggressive lines MOC2/7/10 bore an activating K-RAS mutation in codon 61 (Figure S4A). Evaluation of activated RAS identified that 5/6 cell lines had activated total RAS, and more specifically, the aggressive lines MOC2/7/10 had an approximate 2-fold increase in activated K-RAS compared to the indolent lines MOC1/22 (Figure S4B). Interestingly, the MOC23 cell line that formed tumors only in RAG2−/− immunodeficient mice, had no RAS pathway mutations. This analysis of the MOC lines identified an oncogenic pathway that is upstream of ERK1/2 phosphorylation and potentially plays a primary role in the development and aggressiveness of mouse DMBA-induced OSCC.
Phosphorylated ERK 1/2 regulates OSCC migration and invasion
The contribution of ERK1/2 in tumor cell migration and invasion in OSCC has received limited attention, and therefore we examined the effect of U0126, a selective inhibitor of the MEK1/2 kinases, in our system. Given the toxicity of U0126 (10 μM) beyond 48 hours, we assessed the effect of U0126 on ERK1/2 phosphorylation, cellular proliferation, and migration after 24 hours. We found no effect on proliferation despite significant inhibition of ERK1/2 phosphorylation (Figure S5A and data not shown). Interestingly, the more aggressive MOC2 had nearly four-fold higher basal capacity to migrate in Transwell assays compared to MOC1 (Figure 2C,D), and this was proportionally inhibited to a higher extent by U0126 (93% for MOC2 versus 51% for MOC1, significant interaction effect -Line*vehicle F(1,18)=52.824, p<0.001), suggesting a greater dependence on ERK1/2 for MOC2. Concordantly, U0126 treatment dramatically inhibited MOC2’s migratory ability in a scratch assay whereas MOC1 was less affected (data not shown). Finally, assessment of invasion across a Matrigel matrix showed MOC2 had a greater ability to invade and was more attenuated upon U0126 treatment compared to MOC1 (Figure S5C,D). Thus, ERK1/2 activity is directly associated with migration and invasion in vitro.
CD44 expression is correlated with increased ERK 1/2 activation
We next investigated the downstream targets of ERK1/2 that could be driving OSCC migration and invasion. Others have shown that ERK1/2 is associated with EMT, which itself is associated with CD44 expression (13, 30). Therefore, we assessed the expression of CD44 on the MOC lines and found increased cell surface CD44 on the aggressive MOC2/7/10 cell lines (MFI= 51, 42 and 76, respectively) compared to indolent MOC1/22 and 23 cell lines (MFI=13.7, 9.87 and 18.6, Figure 3A,B). This correlation was also detected in MOC1 and MOC2 cell lines growing in vivo (Figure 4A, S8). Thus, there is an association between tumor cell aggressiveness, ERK1/2 phosphorylation, and CD44 expression in our cell lines.
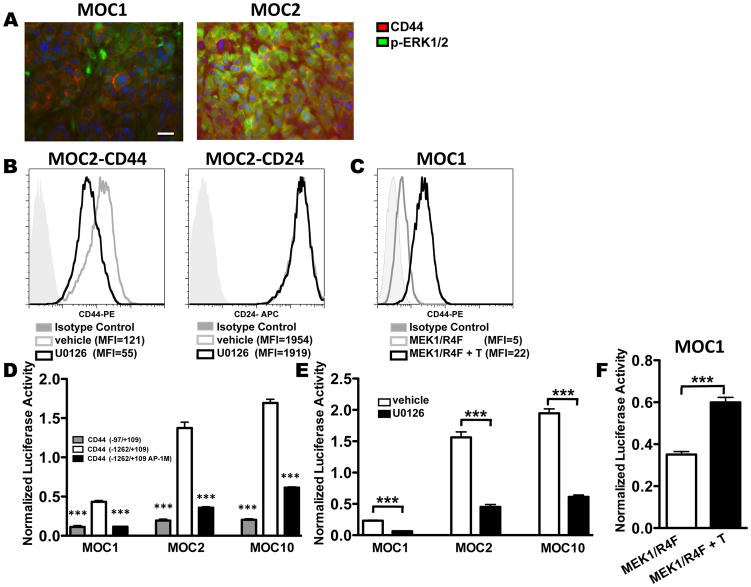
Figure 4. ERK1/2 activation regulates CD44 activity in MOC cells.
A. Dual immunofluorescence of p-ERK1/2 (green) and CD44 (red) was performed on paraffin-embedded sections of transplanted MOC1 and MOC2 tumors. All cell lines have nuclear staining with DAPI (blue). Images are representative of at least 3 sections with the same exact settings for both tumors (40X magnification). Scale bar in MOC1 represents 100 μM—the same scale applies to all images. B. FACS analysis of cell surface CD44 or CD24 expression on MOC2 after 48 hours of treatment with vehicle control (gray line) or U0126 (10μM, black line). The isotype control for each analysis is indicated by gray shaded curve. MFIs are as indicated. C. FACS analysis of cell surface CD44 expression after MOC1 was transduced with tamoxifen regulated MEK1/R4F and treated with (black line) or without (gray line) 200nM tamoxifen for 48 hours. Isotype control is indicated by the gray shaded curve and MFIs are as indicated. D. MOC1, MOC2, or MOC10 cells were co-transfected with indicated CD44 promoter-luciferase and Renilla plasmids in triplicate. CD44 luciferase plasmids utilized were the basal CD44 promoter (CD44 -97+109) (gray), the full length CD44 promoter (CD44 -1262/+109) (white) or the full length CD44 promoter with a mutated AP-1 site (CD44 -1262/+109 AP-1M) (black). Luciferase activity was normalized to Renilla activity to control for transfection efficiency (***=p<0.001 for basal or mutant AP-1 construct vs. full length CD44 promoter). E. MOC1, 2, and 10 were co-transfected with CD44 full length promoter luciferase (CD44 -1262/+109) and Renilla plasmids. Cells in triplicate were then treated with vehicle (DMSO) or U0126 (10μM) for 24 hours (***=p<0.001 for DMSO vs. U0126 treatment only). F. MOC1 cells were co-transfected with tamoxifen regulated MEK1/R4F, CD44 full length promoter luciferase (CD44 -1262/+109), and Renilla plasmids. Cells in triplicate were treated with or without 200nM tamoxifen (***=p<0.001 for vehicle vs. tamoxifen treatment only). All data are representative of at least 2 independent experiments.
CD44 is critical for MOC migration and in vivo growth
We next addressed a causal connection between ERK1/2 activity, CD44, and migration/invasion. Three pan-CD44 shRNAs that target the CD44 coding region and a control scramble shRNA were used to create knockdown cell lines of MOC10 (CD44 knockdown of 70%, 88%, and 42%, Figure 3C) and MOC2 (CD44 knockdown of 63%, 92%, and 29%, Figure S6A). Although the cells with CD44 knockdown morphologically appeared to be more clustered in culture (not shown), there was no effect on cell viability (Figure S6B) or alteration in ERK1/2 phosphorylation (Figure S6C). In contrast, reduced CD44 expression drastically inhibited in vitro cellular migration and invasion (Figure 3D, S6D, and data not shown) and in vivo growth and metastasis (Figure 3E, S6E,F and data not shown). Specifically, when assessed 50 days post-tumor transplant, MOC10 scramble shRNA cells formed 20 mm tumors in 5/5 mice, with 4/5 displaying LN metastases, whereas MOC 10 with silenced CD44 formed smaller (2–7 mm) tumors in fewer (6/10) mice with no evidence of metastasis. Interestingly, when examined at later timepoints, some CD44 knockdown tumors did contain metastatic LN deposits, which are likely escape mutants that had recovered CD44 expression (Figure S7). Similar results were found at a higher dose of tumor cells (Figure S6F), confirming that MOC cell CD44 plays a pivotal role in local aggressiveness and tumor growth in vitro and in vivo.
ERK1/2 regulates CD44 expression
Having established that CD44 contributes to growth in vivo and its surface levels correlated with phospho-ERK1/2 in vitro, we next tested whether this correlation was maintained in vivo. Transplanted MOC1 and MOC2 tumors were analyzed by immunofluorescence, which revealed that intracellular phospho-ERK1/2 staining coincided with cell surface CD44 for MOC2 (Figure 4A, S8). Although low levels of CD44 cell surface staining were detectable for MOC1, minimal phospho-ERK1/2 staining was detected (Figure 4A).
We next employed loss- and gain-of-function approaches to assess whether ERK1/2 regulated CD44 expression. Blockade of ERK1/2 activation by U0126 consistently decreased CD44 surface expression compared to cells treated with vehicle control (Figure 4B, S9A, B). This effect of U0126 on CD44 expression was specific as cell surface CD24 was unchanged after 48 hours of U0126 treatment (Figure 4B).
We then transduced the low phospho-ERK1/2 MOC1 line with a tamoxifen inducible constitutively active MEK1/R4F construct (25). Tamoxifen treatment of these cells induced a four-fold increase in cell surface CD44 compared to vehicle alone; however, tamoxifen treatment of parental MOC1 revealed no change in cell surface CD44 (Figure 4C and S9C). Thus, ERK1/2 activity is strongly linked to cell surface CD44 expression.
To address the mechanism of ERK1/2 regulation of CD44, MOC lines were assessed for differences in alternative splicing of CD44 and no isoform differences were found (data not shown). We next investigated whether the ERK1/2 pathway regulated CD44 gene expression, which has not been studied extensively but was suggested 18 years ago when Ponta and colleagues showed that c-H-RAS transcriptionally induced CD44 message (31). We found using luciferase reporter constructs attached to basal (-97/+105) and full-length (-1262/+105) sequences of the CD44 promoter (24) that cell surface CD44 expression correlated with luciferase activity as MOC2 and MOC10 had more (3- to 4-fold) full-length CD44 promoter driven luciferase activity compared to MOC1 (Figure 4D). Importantly, this activity was reduced by mutating the AP-1 transcription factor binding site (-1262/+105 AP-1M) (Figure 4D) or by addition of U0126 (Figure 4E). Finally, when MOC1 cells were co-transfected with the full-length reporter and a tamoxifen inducible MEK1/R4F construct, treatment with tamoxifen but not vehicle induced CD44 promoter activity (Figure 4F). Together, these data support a model wherein an oncogenic RAS/MEK1/2/ERK1/2 and AP-1 cascade triggers transcription and surface expression of CD44 in MOC cells contributing to enhanced tumor aggressiveness.
ERK1/2 and CD44 in human OSCC
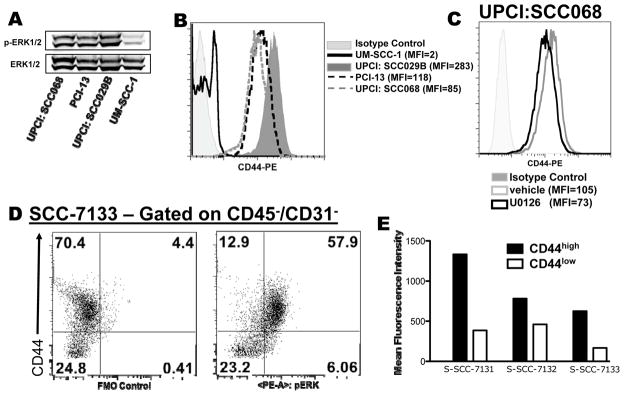
Having established the importance of the ERK1/2 and CD44 pathway in the aggressiveness of this murine model, we next extended these findings to human OSCC. Analysis of human OSCC cell lines revealed a spectrum of ERK1/2 phosphorylation levels. Of the cell lines analyzed, UM-SCC-1 had low levels and, UPCI:SCC029B, UPCI:SCC068, and PCI-13 had high levels of phospho-ERK1/2 (Figure 5A). Interestingly, the primary tumor origin of these cell lines paralleled the MOC cell lines—cell lines with high p-ERK1/2 were derived from advanced tumors (all with nodal metastases) whereas the UM-SCC-1 cell line arose from a T2N0 primary tumor (32, 33). Similar to the MOC lines, FACS analysis of CD44 showed a strict correlation with ERK1/2 phosphorylation with low levels in UM-SCC-1 and high levels in UPCI:SCC029B, UPCI:SCC068, and PCI-13 (Figure 5B). Inhibition of ERK1/2 activity by U0126 led to a modest decrease in cell surface CD44 expression (Figure 5C and Figure S9D). In this particular model, it is possible that ERK1/2 activation is not the only driver of CD44 surface expression, and thus MEK inhibition may not be sufficient in all contexts to decrease CD44 levels. Finally, analysis of freshly resected primary human OSCCs by intracellular phospho-flow cytometry showed that the CD44high tumor cells had approximately 2- to 4-fold higher levels of phospho-ERK1/2 compared to the CD44low tumor cells (Figure 5D,E and S10). Moreover, transplantation of sorted CD44low and CD44high primary human OSCC cells into NOD/SCID/gamma mice replicated published findings showing that CD44high tumor cells engraft better (data not shown) (12). Thus, findings in the MOC lines parallel human OSCC.
Figure 5. Human OSCC also display a p-ERK1/2 and CD44 relationship.
A. Western analysis of pERK1/2 and ERK1/2 of the indicated human OSCC lines. B. FACS analysis of cell surface CD44 expression on human OSCC lines with MFI as indicated. Isotype control (from UPCI: SCC029B) is represented by the gray shaded curve. C. FACS analysis of UPCI:SCC068 cell surface CD44 expression after cells were treated with vehicle control (gray line) or U0126 (10μM, black line) for 48 hours. The gray shaded curve represents isotype control. D. Freshly resected primary human OSCC also display a relationship between p-ERK1/2 and CD44. Primary tumor was digested, hematopoietic cells were excluded, and tumor cells were then stained for CD44 and intracellular p-ERK1/2. Dot plots show control (left) and enhanced p-ERK1/2 staining (right) in the CD44high vs CD44low S-SCC-7133 tumor cells. E. MFI of p-ERK1/2 expression in CD44high and CD44low tumor populations from three separate primary human tumors.
Discussion
Herein, we describe a panel of transplantable syngeneic C57BL/6 cell lines that parallel the human disease in their (1) chemical carcinogenesis based mechanism of formation, (2) intrinsic signaling aberrations and (3) histopathology and in vivo biology including lymphatic metastasis. Salley first described the use of DMBA as an oral carcinogen in the hamster cheek (34) but Crowe and colleagues only recently adapted this protocol to mice (26), which we then used to generate primary OSCCs and the cell lines. Using these resources, we identified that ERK1/2 activation and CD44 expression are linked, paralleling findings in human OSCC, and showed that ERK1/2 transcriptionally targets CD44 to contribute to an aggressive phenotype. These data define one mechanism whereby ERK1/2 mediates tumor aggressiveness and highlight the possibility of therapeutically targeting ERK1/2 in primary human OSCC.
The development of this murine model of OSCC represents a significant advance in the available pre-clinical tools for studying this disease. Currently used reagents include genetically engineered mouse models, such as those with oral mucosa specific inactivation of SMAD4, carcinogen induced primary OSCC and human tumor xenografts in immunodeficient mice (26, 35–37). Of these, the latter remain the most widely used by OSCC investigators. Xenograft models, while recapitulating the intrinsic signaling of the human disease, are not appropriate for in vivo studies of host-tumor interactions because they must be grown in immunocompromised mice. Transplantable syngeneic models for head and neck cancer have been described using oral keratinocytes either transformed in vitro with 4-nitroquinolone or by expression of HPV E6/E7/H-RAS, but neither of these have been generally used and do not display fidelity with human OSCC in terms of lymphatic metastases (38, 39). Others have used the C3H/HeJ derived SCCVII cell line, which is of cutaneous origin, as a surrogate for OSCC (40). Therefore, the panel of cell lines that we have generated helps fill the void of pre-clinical OSCC models by providing a transplantable, orally-derived tumor system that can be grown in the universally used, immunocompetent C57BL/6 mice. Furthermore, and of paramount importance, this tumor system faithfully recapitulates several aspects of the human disease, including lymphatic metastasis.
We identified that the indolent MOC1/22 had an H-RAS mutation whereas the more aggressive MOC2/7/10 cell lines had K-RAS mutations with two-fold higher active K-RAS. This association between specific mutant RAS isoforms and tumor aggressiveness was similar to the finding of Chodosh and colleagues, who identified that in a c-MYC or Wnt1 driven mouse model of breast cancer, tumors bearing K-RAS mutations had decreased oncogene dependence and increased ERK1/2 activation compared to ones with H-RAS mutations (41). K-RAS may induce additional effector pathways enhancing ERK1/2 phosphorylation as illustrated by studies of cell line resistance to the MEK inhibitor AZD6244 (42). Although mutant RAS and B-Raf oncogenic activators have been documented in human melanomas, pancreas, colon and other cancers, the frequency of these mutations is far less in OSCC (43–45). In particular, RAS mutations (both in H- and K-RAS) have been reported at low frequencies in Western populations with OSCC. Interestingly, in India, where a common risk factor is the use of betel nut chew, the rate of RAS mutations in OSCC is as high as 35–50%, which may reflect an underlying genetic susceptibility or sensitivity to specific carcinogens (44–46).
In addition to the mutant RAS differences amongst the tumor cell lines, the major intracellular signaling alteration we identified was the degree of ERK1/2 phosphorylation. The slow growing MOC1/22 cell lines had markedly less ERK1/2 activation compared to the MOC2/7/10 lines. Activation of RAS/MEK1/2/ERK1/2 is commonly seen in malignancies and drives tumor cell proliferation, migration and invasion by inducing anti-apoptotic and proliferative pathways (47). ERK1/2 can also promote metastasis by inducing Slug, Snail, and EMT (30, 48). Recently, Blenis and colleagues showed that ERK2, but not ERK1, activated a Fra-1 mediated induction of ZEB1/2 to induce EMT in breast cancer (49). Extending these data, the connection we highlighted between ERK1/2 and CD44 in mouse and human OSCC cell lines and primary human tumors suggests a generalized connection between these molecules and is consistent with the description by Weinberg and colleagues of parallels between EMT and CSC (13). Interestingly, Basu and colleagues recently identified that tumor heterogeneity, likely due to EMT, establishes chemotherapy resistance in a mesenchymal like subset of cells and that drugs that target this population are ideal candidates for future therapeutics (50). Although MEK inhibitors have shown significant associated toxicities in clinical trials, including blurred vision and neurotoxicity, newer compounds are being developed that may have reduced effects in critical organs and may eventually contribute to oral cancer management (47). We speculate that ERK1/2 can therefore be targeted to prevent not only metastasis, but also to treat chemoresistant CSC that may express high levels of CD44 and ERK1/2 activity.
We found a key role for CD44 as a downstream effector of ERK1/2-mediated tumor aggressiveness. shRNA mediated reduction of CD44 led to delayed tumor growth and the emergence of escape variants with parental levels of CD44, similar to the findings of Weinberg and colleagues (31). However, because these tumors recovered CD44 expression, we cannot make any definitive conclusion about the role of CD44 on metastasis with the knockdown approach. Thus, our data highlights the role of ERK1/2 and its downstream mediator CD44 in promoting OSCC aggressiveness using a new syngeneic model of transplantable OSCC. Our current work is focused on further mechanistic dissection of this link and therapeutically targeting both ERK1/2 and CD44 to alter the biology and outcomes of OSCC.
Supplementary Material
Acknowledgments
We thank Robert Schreiber, Ruby Chan, Charles Rickert, Steven Wang and Greg Longmore for guidance. We thank Jessica Archambault and Michael White for advice on mouse work. Experimental support provided by the RCAVS Histology (Brian Faddis and Pat Keller) and Clinical/Translational Core (Dorina Kallogjeri and Jay Piccirillo) (NIH P30DC04665) and the Speed Congenics Facility of the Rheumatic Diseases Core.
RU was supported by the NCI (K08CA090403) and the Veteran’s Affairs Research Service. NPJ and JHL were supported by NIH-T32DC00022.
Footnotes
Disclosure: No conflict of interest
References
- 1.Kademani D. Oral cancer. Mayo Clin Proc. 2007 Jul;82(7):878–87. doi: 10.4065/82.7.878. [DOI] [PubMed] [Google Scholar]
- 2.Haddad RI, Shin DM. Recent advances in head and neck cancer. N Engl J Med. 2008 Sep 11;359(11):1143–54. doi: 10.1056/NEJMra0707975. [DOI] [PubMed] [Google Scholar]
- 3.Rogers SN, Brown JS, Woolgar JA, Lowe D, Magennis P, Shaw RJ, et al. Survival following primary surgery for oral cancer. Oral Oncol. 2009 Mar;45(3):201–11. doi: 10.1016/j.oraloncology.2008.05.008. [DOI] [PubMed] [Google Scholar]
- 4.Agrawal N, Frederick MJ, Pickering CR, Bettegowda C, Chang K, Li RJ, et al. Exome Sequencing of Head and Neck Squamous Cell Carcinoma Reveals Inactivating Mutations in NOTCH1. Science. 2011 Jul 28; doi: 10.1126/science.1206923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stransky N, Egloff AM, Tward AD, Kostic AD, Cibulskis K, Sivachenko A, et al. The Mutational Landscape of Head and Neck Squamous Cell Carcinoma. Science. 2011 Jul 28; doi: 10.1126/science.1208130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lu SL, Herrington H, Reh D, Weber S, Bornstein S, Wang D, et al. Loss of transforming growth factor-beta type II receptor promotes metastatic head-and-neck squamous cell carcinoma. Genes Dev. 2006 May 15;20(10):1331–42. doi: 10.1101/gad.1413306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rubin Grandis J, Melhem MF, Gooding WE, Day R, Holst VA, Wagener MM, et al. Levels of TGF-alpha and EGFR protein in head and neck squamous cell carcinoma and patient survival. J Natl Cancer Inst. 1998 Jun 3;90(11):824–32. doi: 10.1093/jnci/90.11.824. [DOI] [PubMed] [Google Scholar]
- 8.Molinolo AA, Amornphimoltham P, Squarize CH, Castilho RM, Patel V, Gutkind JS. Dysregulated molecular networks in head and neck carcinogenesis. Oral Oncol. 2009 Apr-May;45(4–5):324–34. doi: 10.1016/j.oraloncology.2008.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hoover AC, Strand GL, Nowicki PN, Anderson ME, Vermeer PD, Klingelhutz AJ, et al. Impaired PTPN13 phosphatase activity in spontaneous or HPV-induced squamous cell carcinomas potentiates oncogene signaling through the MAP kinase pathway. Oncogene. 2009 Nov 12;28(45):3960–70. doi: 10.1038/onc.2009.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bancroft CC, Chen Z, Dong G, Sunwoo JB, Yeh N, Park C, et al. Coexpression of proangiogenic factors IL-8 and VEGF by human head and neck squamous cell carcinoma involves coactivation by MEK-MAPK and IKK-NF-kappaB signal pathways. Clin Cancer Res. 2001 Feb;7(2):435–42. [PubMed] [Google Scholar]
- 11.Wang SJ, Bourguignon LY. Role of hyaluronan-mediated CD44 signaling in head and neck squamous cell carcinoma progression and chemoresistance. Am J Pathol. 2011 Mar;178(3):956–63. doi: 10.1016/j.ajpath.2010.11.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Prince ME, Sivanandan R, Kaczorowski A, Wolf GT, Kaplan MJ, Dalerba P, et al. Identification of a subpopulation of cells with cancer stem cell properties in head and neck squamous cell carcinoma. Proc Natl Acad Sci U S A. 2007 Jan 16;104(3):973–8. doi: 10.1073/pnas.0610117104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mani SA, Guo W, Liao MJ, Eaton EN, Ayyanan A, Zhou AY, et al. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell. 2008 May 16;133(4):704–15. doi: 10.1016/j.cell.2008.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Toole BP. Hyaluronan-CD44 Interactions in Cancer: Paradoxes and Possibilities. Clin Cancer Res. 2009 Dec 15;15(24):7462–8. doi: 10.1158/1078-0432.CCR-09-0479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Orian-Rousseau V, Chen L, Sleeman JP, Herrlich P, Ponta H. CD44 is required for two consecutive steps in HGF/c-Met signaling. Genes Dev. 2002 Dec 1;16(23):3074–86. doi: 10.1101/gad.242602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ponta H, Sherman L, Herrlich PA. CD44: from adhesion molecules to signalling regulators. Nat Rev Mol Cell Biol. 2003 Jan;4(1):33–45. doi: 10.1038/nrm1004. [DOI] [PubMed] [Google Scholar]
- 17.Godar S, Ince TA, Bell GW, Feldser D, Donaher JL, Bergh J, et al. Growth-inhibitory and tumor- suppressive functions of p53 depend on its repression of CD44 expression. Cell. 2008 Jul 11;134(1):62–73. doi: 10.1016/j.cell.2008.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cheng C, Yaffe MB, Sharp PA. A positive feedback loop couples Ras activation and CD44 alternative splicing. Genes Dev. 2006 Jul 1;20(13):1715–20. doi: 10.1101/gad.1430906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brown RL, Reinke LM, Damerow MS, Perez D, Chodosh LA, Yang J, et al. CD44 splice isoform switching in human and mouse epithelium is essential for epithelial-mesenchymal transition and breast cancer progression. J Clin Invest. 2011 Mar 1;121(3):1064–74. doi: 10.1172/JCI44540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu C, Kelnar K, Liu B, Chen X, Calhoun-Davis T, Li H, et al. The microRNA miR-34a inhibits prostate cancer stem cells and metastasis by directly repressing CD44. Nat Med. 2011 Feb;17(2):211–5. doi: 10.1038/nm.2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Joshua B, Kaplan MJ, Doweck I, Pai R, Weissman IL, Prince ME, et al. Frequency of cells expressing CD44, a Head and Neck cancer stem cell marker: Correlation with tumor aggressiveness. Head Neck. 2011 Feb 14; doi: 10.1002/hed.21699. [DOI] [PubMed] [Google Scholar]
- 22.de Jong MC, Pramana J, van der Wal JE, Lacko M, Peutz-Kootstra CJ, de Jong JM, et al. CD44 expression predicts local recurrence after radiotherapy in larynx cancer. Clin Cancer Res. 2010 Nov 1;16(21):5329–38. doi: 10.1158/1078-0432.CCR-10-0799. [DOI] [PubMed] [Google Scholar]
- 23.Hancock WW, Lu B, Gao W, Csizmadia V, Faia K, King JA, et al. Requirement of the chemokine receptor CXCR3 for acute allograft rejection. J Exp Med. 2000 Nov 20;192(10):1515–20. doi: 10.1084/jem.192.10.1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Foster LC, Arkonac BM, Sibinga NE, Shi C, Perrella MA, Haber E. Regulation of CD44 gene expression by the proinflammatory cytokine interleukin-1beta in vascular smooth muscle cells. J Biol Chem. 1998 Aug 7;273(32):20341–6. doi: 10.1074/jbc.273.32.20341. [DOI] [PubMed] [Google Scholar]
- 25.Scholl FA, Dumesic PA, Khavari PA. Mek1 alters epidermal growth and differentiation. Cancer Res. 2004 Sep 1;64(17):6035–40. doi: 10.1158/0008-5472.CAN-04-0017. [DOI] [PubMed] [Google Scholar]
- 26.Ku TK, Nguyen DC, Karaman M, Gill P, Hacia JG, Crowe DL. Loss of p53 expression correlates with metastatic phenotype and transcriptional profile in a new mouse model of head and neck cancer. Mol Cancer Res. 2007 Apr;5(4):351–62. doi: 10.1158/1541-7786.MCR-06-0238. [DOI] [PubMed] [Google Scholar]
- 27.Brenner JC, Graham MP, Kumar B, Saunders LM, Kupfer R, Lyons RH, et al. Genotyping of 73 UM-SCC head and neck squamous cell carcinoma cell lines. Head Neck. 2010 Apr;32(4):417–26. doi: 10.1002/hed.21198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Weaver BK, Bohn E, Judd BA, Gil MP, Schreiber RD. ABIN-3: a molecular basis for species divergence in interleukin-10-induced anti-inflammatory actions. Mol Cell Biol. 2007 Jul;27(13):4603–16. doi: 10.1128/MCB.00223-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Quintanilla M, Brown K, Ramsden M, Balmain A. Carcinogen-specific mutation and amplification of Ha-ras during mouse skin carcinogenesis. Nature. 1986 Jul 3–9;322(6074):78–80. doi: 10.1038/322078a0. [DOI] [PubMed] [Google Scholar]
- 30.Turley EA, Veiseh M, Radisky DC, Bissell MJ. Mechanisms of disease: epithelial-mesenchymal transition--does cellular plasticity fuel neoplastic progression? Nat Clin Pract Oncol. 2008 May;5(5):280–90. doi: 10.1038/ncponc1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hofmann M, Rudy W, Gunthert U, Zimmer SG, Zawadzki V, Zoller M, et al. A link between ras and metastatic behavior of tumor cells: ras induces CD44 promoter activity and leads to low-level expression of metastasis-specific variants of CD44 in CREF cells. Cancer Res. 1993 Apr 1;53(7):1516–21. [PubMed] [Google Scholar]
- 32.Lin CJ, Grandis JR, Carey TE, Gollin SM, Whiteside TL, Koch WM, et al. Head and neck squamous cell carcinoma cell lines: established models and rationale for selection. Head Neck. 2007 Feb;29(2):163–88. doi: 10.1002/hed.20478. [DOI] [PubMed] [Google Scholar]
- 33.White JS, Weissfeld JL, Ragin CC, Rossie KM, Martin CL, Shuster M, et al. The influence of clinical and demographic risk factors on the establishment of head and neck squamous cell carcinoma cell lines. Oral Oncol. 2007 Aug;43(7):701–12. doi: 10.1016/j.oraloncology.2006.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Salley JJ. Experimental carcinogenesis in the cheek pouch of the Syrian hamster. J Dent Res. 1954 Apr;33(2):253–62. doi: 10.1177/00220345540330021201. [DOI] [PubMed] [Google Scholar]
- 35.Bornstein S, White R, Malkoski S, Oka M, Han G, Cleaver T, et al. Smad4 loss in mice causes spontaneous head and neck cancer with increased genomic instability and inflammation. J Clin Invest. 2009 Nov;119(11):3408–19. doi: 10.1172/JCI38854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vitale-Cross L, Czerninski R, Amornphimoltham P, Patel V, Molinolo AA, Gutkind JS. Chemical carcinogenesis models for evaluating molecular-targeted prevention and treatment of oral cancer. Cancer Prev Res (Phila) 2009 May;2(5):419–22. doi: 10.1158/1940-6207.CAPR-09-0058. [DOI] [PubMed] [Google Scholar]
- 37.Sano D, Myers JN. Xenograft models of head and neck cancers. Head Neck Oncol. 2009;1:32. doi: 10.1186/1758-3284-1-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thomas GR, Chen Z, Oechsli MN, Hendler FJ, Van Waes C. Decreased expression of CD80 is a marker for increased tumorigenicity in a new murine model of oral squamous-cell carcinoma. Int J Cancer. 1999 Jul 30;82(3):377–84. doi: 10.1002/(sici)1097-0215(19990730)82:3<377::aid-ijc11>3.0.co;2-9. [DOI] [PubMed] [Google Scholar]
- 39.Spanos WC, Hoover A, Harris GF, Wu S, Strand GL, Anderson ME, et al. The PDZ binding motif of human papillomavirus type 16 E6 induces PTPN13 loss, which allows anchorage-independent growth and synergizes with ras for invasive growth. J Virol. 2008 Mar;82(5):2493–500. doi: 10.1128/JVI.02188-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kim S. Animal models of cancer in the head and neck region. Clin Exp Otorhinolaryngol. 2009 Jun;2(2):55–60. doi: 10.3342/ceo.2009.2.2.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jang JW, Boxer RB, Chodosh LA. Isoform-specific ras activation and oncogene dependence during MYC- and Wnt-induced mammary tumorigenesis. Mol Cell Biol. 2006 Nov;26(21):8109–21. doi: 10.1128/MCB.00404-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Little AS, Balmanno K, Sale MJ, Newman S, Dry JR, Hampson M, et al. Amplification of the Driving Oncogene, KRAS or BRAF, Underpins Acquired Resistance to MEK1/2 Inhibitors in Colorectal Cancer Cells. Sci Signal. 2011;4(166):ra17. doi: 10.1126/scisignal.2001752. [DOI] [PubMed] [Google Scholar]
- 43.Karnoub AE, Weinberg RA. Ras oncogenes: split personalities. Nat Rev Mol Cell Biol. 2008 Jul;9(7):517–31. doi: 10.1038/nrm2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hardisson D. Molecular pathogenesis of head and neck squamous cell carcinoma. Eur Arch Otorhinolaryngol. 2003 Oct;260(9):502–8. doi: 10.1007/s00405-003-0581-3. [DOI] [PubMed] [Google Scholar]
- 45.Weber A, Langhanki L, Sommerer F, Markwarth A, Wittekind C, Tannapfel A. Mutations of the BRAF gene in squamous cell carcinoma of the head and neck. Oncogene. 2003 Jul 24;22(30):4757–9. doi: 10.1038/sj.onc.1206705. [DOI] [PubMed] [Google Scholar]
- 46.Saranath D, Chang SE, Bhoite LT, Panchal RG, Kerr IB, Mehta AR, et al. High frequency mutation in codons 12 and 61 of H-ras oncogene in chewing tobacco-related human oral carcinoma in India. Br J Cancer. 1991 Apr;63(4):573–8. doi: 10.1038/bjc.1991.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fremin C, Meloche S. From basic research to clinical development of MEK1/2 inhibitors for cancer therapy. J Hematol Oncol. 2010;3:8. doi: 10.1186/1756-8722-3-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen H, Zhu G, Li Y, Padia RN, Dong Z, Pan ZK, et al. Extracellular signal-regulated kinase signaling pathway regulates breast cancer cell migration by maintaining slug expression. Cancer Res. 2009 Dec 15;69(24):9228–35. doi: 10.1158/0008-5472.CAN-09-1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shin S, Dimitri CA, Yoon SO, Dowdle W, Blenis J. ERK2 but not ERK1 induces epithelial-to-mesenchymal transformation via DEF motif-dependent signaling events. Mol Cell. 2010 Apr 9;38(1):114–27. doi: 10.1016/j.molcel.2010.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Basu D, Montone KT, Wang LP, Gimotty PA, Hammond R, Diehl JA, et al. Detecting and targeting mesenchymal-like subpopulations within squamous cell carcinomas. Cell Cycle. 2011 Jun 15;10(12) doi: 10.4161/cc.10.12.15883. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.