Abstract
Armenia, situated between the Black and Caspian Seas, lies at the junction of Turkey, Iran, Georgia, Azerbaijan and former Mesopotamia. This geographic position made it a potential contact zone between Eastern and Western civilizations. In this investigation, we assess Y-chromosomal diversity in four geographically distinct populations that represent the extent of historical Armenia. We find a striking prominence of haplogroups previously implicated with the Agricultural Revolution in the Near East, including the J2a-M410-, R1b1b1*-L23-, G2a-P15- and J1-M267-derived lineages. Given that the Last Glacial Maximum event in the Armenian plateau occured a few millennia before the Neolithic era, we envision a scenario in which its repopulation was achieved mainly by the arrival of farmers from the Fertile Crescent temporally coincident with the initial inception of farming in Greece. However, we detect very restricted genetic affinities with Europe that suggest any later cultural diffusions from Armenia to Europe were not associated with substantial amounts of paternal gene flow, despite the presence of closely related Indo-European languages in both Armenia and Southeast Europe.
Keywords: Armenia, phylogenetics, Y-chromosome, SNPs, neolithic
Introduction
Located at the crossroads of Europe and the Middle East, the Armenian plateau has long served as both a recipient of and potential conduit for gene flow between the two regions. Although archaeological evidence for human as well as Neanderthal activity in Armenia during the Paleolithic era exists,1, 2 the Last Glacial Maximum (LGM) likely made permanent settlements of the region infeasible3 until the glacial recessions between 16 and 18 kya.4 While Mesolithic sites in Sasun are known,5 the improving climatic conditions during this period allowed the Armenian plateau to gradually transform into a region characterized by bountiful water supply and wealth of fertile plains.6 The transition from the LGM to a hospitable landscape that would support stable and detectable human occupation is expected to have progressed over millennia. These conditions as well as its proximity to the Fertile Crescent catalyzed the region's emergence as one of the earliest recipients of agriculture (∼8 kya) during the Neolithic Revolution.7, 8 In addition to the relatively early appearance of agriculture in Armenia, the Armenian highlands seem to have been instrumental in the dispersal of obsidian,9 leather footwear10 and viticulture;11 technologies that would later acculturate across the Near East and eventually enter Europe. The extent to which the Armenian gene pool has been structured and influenced by events during and since the Neolithic, however, remains unknown.
Presently, Armenians are characterized as a distinct ethnotype, speaking a single Indo-European language, Armenian. Linguistic analyses have found that Armenian represents one of the oldest living Indo-European languages and exhibits its greatest affinities with Greek and Balkan languages.12 With some linguists placing the origins of the Proto-Indo-European and Indo-European languages in either Anatolia or Transcaucasia,12 it has been proposed that Armenians represent close descendants of the ancestral Indo-European population and that subsequent migrations from Armenia into Greece were responsible for the language group's dispersal into Europe.13, 14 However, a lack of archaeological support for this notion has led to the alternative supposition that invasions from Balkan or Anatolian tribes15 introduced an Indo-European language into Armenia, resulting in the observed similarities between Armenian and the Southeastern European languages.16, 17
In the efforts to gain a comprehensive understanding of the impact that complex historical migrations and events have had upon the genetic structure of populations, the human Y-chromosome has emerged as a highly effective tool.18 Prior examinations of the paternal lineages within Armenia have revealed population-expansion times corresponding to the Neolithic emergence of agriculture,19 as well as genetic affinities toward both Near Eastern and European populations;20, 21 results that are largely corroborated by mitochondrial DNA22 and Alu insertion (PAI) studies.23
It should be noted, however, that the above-mentioned patrilineal studies are hindered by their utilization of a limited set of Y-chromosomal markers that severely restricted their ability to define phylogenetic relationships. In particular, none of the previous paternal investigations of Armenia resolved the predominantly European haplogroup R beyond the level of R1*(xR1a), which leaves the precise relationship of Armenians to Europeans ambiguous. In the current study, we perform, for the first time, a high resolution analysis of the Y-chromosome structure in four distinct Armenian populations that encompass the breadth of historical Armenia to investigate the role the region had in the Neolithic and Indo-European expansions, as well as finely assess the effects of Armenia's complex history upon its patrilineal genetic architecture.
Materials and methods
Sample collection and DNA isolation
Buccal swabs were acquired with informed consent from 413 unrelated individuals under the ethical guidelines asserted by the Florida International University Institutional Review Board. These individuals, collected from four distinct regions of the historical expanse of Armenia (Figure 1), the Ararat Valley (n=110), Gardman (n=96), Lake Van (n=103) and Sasun (n=104), were assessed to be ethnic Armenians for at least two generations. DNA was isolated with the Gentra Buccal Cell Kit using the protocol described in the manufacturer's specifications (Puregene, Gentra Systems, Minneapolis, MN, USA).
Figure 1.
Geographic locations of the Armenian populations examined. The dashed line indicates the boundary of the greatest extent of historical Armenia.
Y-SNP and Y-STR genotyping
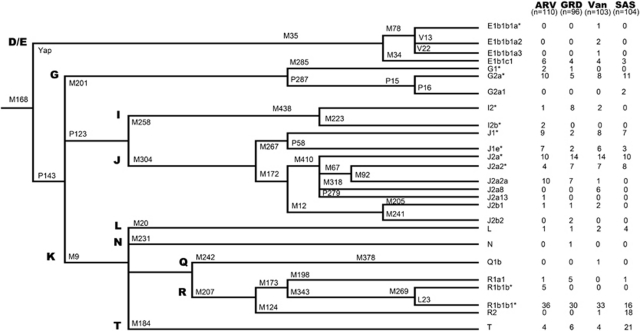
A total of 70 Y-chromosomal binary markers (Figure 2) were hierarchally examined for the purpose of categorizing each sample into particular Y-haplogroups that can be used to trace phylogenetic relationships. Genotypes were identified using standard PCR–RFLP, allele-specific PCR and the YAP PAI24, 25 procedures. Y-haplogroup nomenclature was assigned as in Karafet et al26 and Myres et al.27 To determine Y-STR haplotypes for individuals belonging to haplogroups R1b-M343, J2-M172 and T-M184, 17 loci (DYS19, DYS385 a/b, DYS389I/II, DYS390, DYS391, DYS392, DYS393, DYS437, DYS438, DYS439, DYS448, DYS456, DYS458, DYS635 and Y-GATA H4) were amplified using the AmpFSTR Yfiler PCR amplification kit (Applied Biosystems). The number of repeats at each locus was determined by first separating the PCR products in an ABI Prism 3100 × 1 Genetic Analyzer (Applied Biosystems, Foster City, CA, USA) and then analyzing the fragments with the Genemapper v3.2 program. The size of the DYS389I locus was subtracted from that of the DYS389II locus for all analyses.
Figure 2.
Y-haplogroup phylogeography within Ararat Valley, Gardman, Lake Van and Sasun. The following markers were typed, but were found to be non-polymorphic: M84, M290, M286, P18, P37, M62, M365, M390, P56, M367, M369, M322, M68, M138, M158, M289, M318, M339, M340, M419, P81, M76, M357, M175, M458, M412, M320 and P77.
Statistical and phylogenetic analyses
A list of the 25 previously published populations employed in all the phylogenetic analyses, except the MDS based on haplogroup frequencies (see Supplementary Table 1 for the populations utilized in this analysis), is provided in Table 1. Pairwise genetic distances (Fst) as well as Nei's genetic diversity estimates were calculated with the Arlequin v3.5 program (Genetics and Biometry Laboratory, University of Geneva, Geneva, Switzerland).28 The frequencies of the complete set of detected haplogroups were used to determine pairwise Fst distances among the four Armenian populations. MICROSAT29 (Stanford University Medical Center, Stanford, CA, USA) was used to generate Rst distances from STR allelic frequencies in haplogroups Rlb and J2 in Armenia, as well as populations containing appreciable frequencies of haplogroups R1b and J2. These distances were utilized to generate Multidimensional Scaling Analysis (MDS) plots using SPSS v18.0 software (SPSS, 2001, SPSS Inc., Chicogo, IL, USA).
Table 1. Populations analyzed.
| Geographic region and population names | Abbreviation | n | Reference |
|---|---|---|---|
| Caucasus and Armenia | |||
| Armenia | |||
| Ararat Valley | ARV | 110 | Present study |
| Gardman | GRD | 96 | Present study |
| Lake Van | VAN | 103 | Present study |
| Sasun | SAS | 104 | Present study |
| Caucasus | |||
| Caucasusa | CAU | 1103 | Myres et al27 |
| Georgia | GEO | 66 | Battaglia et al39 |
| Near East | |||
| Levant | |||
| Lebanon | LEB | 950 | Zalloua et al49 |
| Syria | SYR | 356 | El-Sibai et al;50 Zalloua et al51 |
| Jordan | JOR | 273 | El-Sibai et al50 |
| Palestine | PAL | 302 | Zalloua et al51 |
| Iran | |||
| Iran | IRN | 410 | El-Sibai et al;50 Haber et al52 |
| Anatolia | |||
| West Turkeyb | TUW | 161 | Cinnioğlu et al53 |
| Central Turkeyc | TUC | 154 | Cinnioğlu et al53 |
| East Turkeyd | TUE | 208 | Cinnioğlu et al53 |
| Europe | |||
| Balkans | |||
| Greece | GRE | 185 | Myres et al;27 King et al40 |
| Crete | CRE | 193 | King et al40 |
| Macedonia | MAC | 149e; 57 | Spiroski et al;54e Battaglia et al39 |
| Romania | ROM | 330 | Myres et al27 |
| East Europe | |||
| Hungary | HUN | 113 | Myres et al27 |
| Poland | POL | 202 | Myres et al27 |
| Slovakia | SLO | 276 | Myres et al27 |
| Slovenia | SVE | 102 | Myres et al27 |
| Russia | RUS | 545 | Roewer et al55 |
| Czech Republic | CZE | 87 | Myres et al27 |
| West Europe | |||
| England | ENG | 102 | Myres et al27 |
| France | FRA | 460 | Myres et al27 |
| Germany | GER | 321 | Myres et al27 |
| Italy | ITA | 282 | Myres et al27 |
| Denmark | DEN | 110 | Myres et al27 |
Includes the following populations: Avars, Bagvalals, Andis, Lezgis, Darginians, Tabasarans, Kumyks, Megrels, Abkhazes, Balkars, Cherkessians, North Osetins and Kabardians.
Includes Turkey sections 1, 8 and 9.
Includes Turkey sections 2, 6 and 7.
Includes Turkey sections 3, 4 and 5.
Whitley's haplogroup predictor was used to determine Macedonian haplotypes belonging to R1b with >90% confidence.
Network diagrams and time estimations
The phylogenetic relationships of the eight loci haplotypes of all individuals within haplogroups R1b, J2 and T were ascertained by creation of median-joining (MJ) network diagrams30 using the NETWORK 4.5. 1.6 software (Fluxus Engineering, Clare, UK). An additional network projection that included only individuals who were found to belong in either haplogroup R1b1b*-M269 or R1b1b1*-L23 was generated to attempt to discern the genetic relationship between Armenians and Europeans with the ancestral M412 allele. Haplotypes were created from the aforementioned set of eight loci available for each reference population and locus weights were assigned as described previously.31
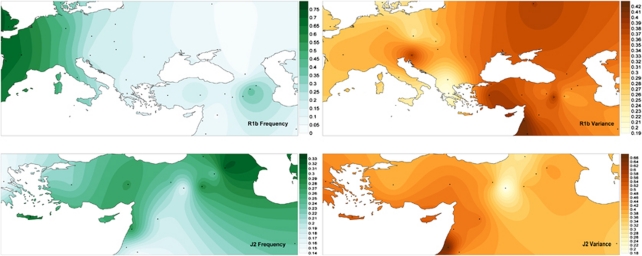
Expansion times for haplogroups R1b, J2 and T were estimated from the eight STR loci dataset using the method described in Zhivotovsky et al32 and modified by Sengupta and collaborators.33 Genealogical mutation rates calculated from family based studies defined for each locus in Ravid-Amir and Rosset34 and Goedbloed et al,35 with an intergeneration time of 25 years were utilized. Owing to the marginal differences in the time estimates generated using both family studies, we provide, throughout the narrative, the expansion values based on Ravid-Amir and Rosset34 mutation rates. We have recalculated the time estimates for the reference populations with the genealogical mutation rate and the Zhivotovsky et al32 method to allow direct comparions of dates. Because of the limited availability of haplotypes within haplogroup T for several of the reference populations, time estimates were calculated using the Levant and Turkey as single groups. Furthermore, because of the elevated levels of haplogroup T in Sasun relative to the other three collections two dates were calculated from Armenian populations, one for Ararat Valley, Gardman and Lake Van, collectively, and a second for Sasun. In addition, spatial gradient maps were generated to illustrate haplogroup frequency and STR allelic variance clines in both haplogroups R1b and J2 using the program Surfer v.9.0.343 (http://www.goldensoftware.com).
Results
Y-haplogroup distribution
For all four Armenian populations analyzed in this study, the most prevalent major haplogroups (Figure 2) are R-M207 (38%, 36%, 33% and 34% in Ararat Valley, Gardman, Lake Van and Sasun, respectively) and J-M304 (38%, 36%, 43% and 27%, respectively). Of the lineages within haplogroup R, the largely Near Eastern27 R1b1b1*-L23 predominates in Ararat Valley, Gardman and Lake Van (33%, 31% and 32%, respectively). Furthermore, in Ararat Valley we find five individuals belonging to the paraphyletic haplogroup R1b1b*-M269. The Sasun collection, meanwhile, contains comparable distributions of haplogroups R1b1b1*-L23 (15%) and R2-M124 (17%). It should be noted that only low frequencies of haplogroup R1a1*-M198, which has been associated with the Indo-Aryan expansions,36 were observed in Ararat Valley (0.9%), Gardman (5.2%) and Sasun (0.9%).
Within haplogroup J-M304, the majority of the collected Armenians belong to the J2a-M410 clade, with the most common lineage being J2a*-M410 (9%, 15%, 14% and 10% in Ararat Valley, Gardman, Lake Van and Sasun, respectively), followed by J2a2*-M67 (4%, 7%, 7% and 8%) and J2a2a-M92 (9%, 7%, 1% and 0%), each of which are largely restricted to Near and Middle Eastern populations.37 Of the remaining J-M304 lineages, appreciable frequencies of haplogroup J1-M267 are found across all four sampled Armenian populations (14.5%, 4.2%, 13.5% and 9.6% in Ararat Valley, Gardman, Lake Van and Sasun, respectively), whereas minimal levels of the South European38, 39 haplogroup J2b-M12 are found only in Ararat Valley (0.9%), Gardman (3.1%) and Lake Van (1.9%).
In each of the examined Armenian populations, we witness moderate levels of haplogroups T-M184, G-M201 and E1b1b1-M35. Interestingly, haplogroup T-M184, which is relatively rare in other Near Eastern populations, as well as in three of the Armenian collections tested here, represents the most prominent descent in Sasun, comprising 20.1% of the samples. The presence of this haplogroup in Ararat Valley, Gardman and Lake Van, by contrast, is more limited, composing only 3.6%, 6.3% and 3.9%, respectively, of the individuals from those collections. Haplogroup G-M201 (10.9%, 6.3%, 7.8% and 12.5% in Ararat Valley, Gardman, Lake Van and Sasun, respectively), on the other hand, is fairly evenly distributed across the four Armenian populations, with sublineage G2a-P15 comprising the vast majority of these chromosomes. Similarly, the E1b1b1-M35 clade (5.5%, 4.2%, 7.8% and 2.9% in Ararat Valley, Gardman, Lake Van and Sasun, respectively) in the four Armenian populations consists almost entirely of haplogroup E1b1b1c1*-M34. However, a small number of individuals from Lake Van belong to the E1b1b1a*-M78 (0.9%), E1b1b1a2-V13 (1.9%) and E1b1b1a3-V22 (0.9%) branches that have been implicated as signals of Greek influence.40
Pairwise Fst distances between the four Armenian populations based on haplogroup frequencies, as well as their corresponding P-values, are provided in Table 2. These results indicate that the genetic distances separating Ararat Valley, Gardman and Lake Van are not statistically significant (P≤0.001). Sasun, however, exhibits statistically significant divergence from the remaining Armenian populations, most likely as the result of the prominence in Sasun of lineages (T-M184 and R2a-M124) found at substantially lower frequencies in Ararat Valley, Gardman and Lake Van. An MDS plot based on Fst distances generated from Y-haplogroup frequencies is provided in Supplementary Figure 1. However, conclusions yielded from this plot must be tempered, as the low haplogroup resolution generates artificial associations that may not exist with an increased number of markers (ie, the observed affinities between Armenia and Europe because of their high levels of R, despite the lack, in Armenia, of the M412-derived allele that predominates in European populations).
Table 2. Fst distances based on haplogroup frequencies.
| ARV | GRD | SAS | VAN | |
|---|---|---|---|---|
| Ararat Valley | — | 0.1441 | 0.0000 | 0.2793 |
| Gardman | 0.0044 | — | 0.0000 | 0.1982 |
| Sasun | 0.04809* | 0.04488* | — | 0.0000 |
| Lake Van | 0.0009 | 0.0032 | 0.04173* | — |
Fst distances and P–values are provided in the lower and upper triangles, respectively.
*P<0.001.
Haplogroup R1b
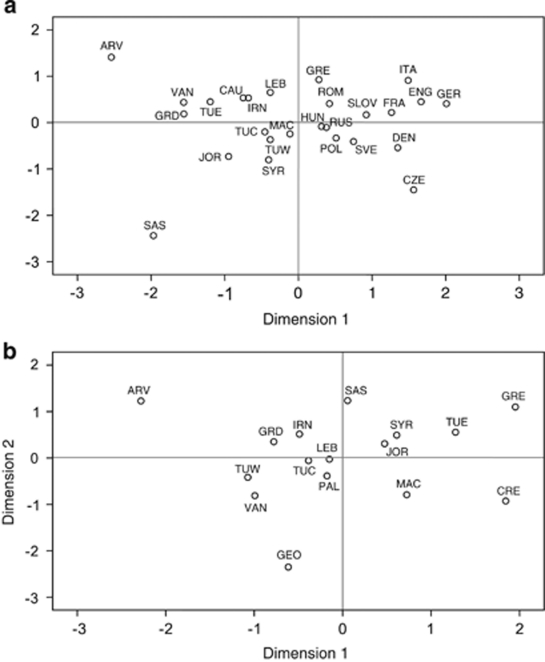
Haplotypes for the 120 Armenians belonging to haplogroup R1b-M343 are available in Supplementary Table 2. In addition, the number of observed haplotypes, gene diversity and allelic variance within haplogroup R1b are provided in Table 3. An MDS analysis of Rst distances calculated from individuals possessing the derived R1b-M343 allele are illustrated in Figure 3a. Despite the elevated frequencies of haplogroup R1*(xR1a) in Armenia, likely driving the European influences seen in the MDS plot based on haplogroup frequencies (Supplementary Figure 1), Armenian populations group further from the European cluster than do most of the Near Eastern collections (Figure 3a). Instead, Gardman and Lake Van segregate closest to the Eastern Turkey population, whereas Sasun and Ararat Valley appear to act as outliers, exhibiting substantial separation from the other two Armenian groups, as well as the reference populations.
Table 3. Haplotype diversity, variance and time estimations for Haplogroup R1b.
| Population | %a | n | Nei's diversity | Haplotype variance | Time estimatesb (kya) | Time estimatesc (kya) |
|---|---|---|---|---|---|---|
| Ararat Valley (8 loci) | 37.3 | 14/41 | 0.8512±0.0421 | 0.3181 | 5.6±1.4 | 6.2±1.7 |
| (17 loci) | — | 22/41 | 0.9390±0.0248 | 0.5038d | — | — |
| Gardman (8 loci) | 31.3 | 23/30 | 0.9770±0.0163 | 0.3213 | 5.0±1.4 | 5.7±2.8 |
| (17 loci) | — | 26/30 | 0.9908±0.0110 | 0.3493d | — | — |
| Lake Van (8 loci) | 32.0 | 23/33 | 0.9697±0.0157 | 0.2912 | 5.3±2.1 | 6.6±4.2 |
| (17 loci) | — | 25/33 | 0.9773±0.0146 | 0.2875d | — | — |
| Sasun (8 loci) | 15.4 | 7/16 | 0.8000±0.0898 | 0.3948 | 4.7±0.8 | 5.5±1.7 |
| (17 loci) | — | 9/16 | 0.8917±0.0603 | 0.3331d | — | — |
| West Turkey | 13.5 | 20/22 | 0.9913±0.0165 | 0.3872 | 6.0±2.0 | 8.9±4.2 |
| Central Turkey | 19.1 | 26/29 | 0.9901±0.0130 | 0.3544 | 5.3±1.2 | 6.1±1.9 |
| East Turkey | 12.0 | 15/25 | 0.9569±0.0202 | 0.3215 | 4.9±1.5 | 6.0±2.9 |
| Syria | 3.7 | 12/13 | 0.9872±0.0354 | 0.3429 | 5.9±1.7 | 6.4±1.9 |
| Jordan | 9.2 | 16/20 | 0.9737±0.0250 | 0.4138 | 6.6±2.0 | 8.1±3.5 |
| Lebanon | 7.6 | 37/50 | 0.9771±0.0113 | 0.4194 | 5.6±1.0 | 6.5±1.8 |
| Iran | 8.6 | 36/44 | 0.9873±0.089 | 0.3062 | 4.1±1.1 | 4.4±1.4 |
| Caucasus | 3.0 | 25/33 | 0.9678±0.0195 | 0.3499 | 4.6±0.8 | 5.8±1.8 |
| Greece | 13.0 | 16/24 | 0.9565±0.0250 | 0.1936 | 3.2±2.0 | 4.0±1.9 |
| Macedonia | 12.8 | 14/19 | 0.9357±0.0472 | 0.2142 | 3.3±1.1 | 3.8±1.7 |
Figure 3.
MDS analyses based on Rst distances calculated from STR allelic frequencies from eight loci of individuals within haplogroups (a) R1b (Stress=0.14116) and (b) J2 (Stress=0.20418). Stress values <0.23 were considered acceptable as determined by Sturrock and Rocha.56
An MJ network diagram generated from the Y-STR haplotypes of all R1b-M343 individuals is provided in Supplementary Figure 2a. Haplotypes from each of the four Armenian populations are found across a wide distribution of clusters and are commonly associated with haplotypes from Near Eastern populations. However, we note very limited haplotype sharing among Armenian and European populations, an observation congruent with the MDS plot (Figure 3a). Similarly, the network based only on individuals belonging to the R1b1b*-M269 and R1b1b1*-L23 lineages (Supplementary Figure 2b) exhibits a wide distribution of Armenian haplotypes and does not identify a segregation between individuals of Armenian and East European descent, which is not inconsistent with older gene flow between the two regions. Expansion time estimates for haplogroup R1b-M343 are provided in Table 3. Overall, the dates calculated using the genealogical mutation rates from Ravid-Amir and Rosset34 and Goedbloed et al35 are comparable, with the latter yielding slightly older dates and greater standard errors. These estimates seem to indicate that the Armenian expansion times are intermediate to those of the Levant and Europe; a cline illustrated in a spatial gradient map of mean variance within R1b-M343 (Figure 4). In addition, the dates generated from this analysis appear to agree with late to early Neolithic expansions for R1b-M343 across the four Armenian collections and most reference populations. However, owing to the contentions associated with the current calibrations of the Y-STR mutation rates,32, 34, 35, 41 as well as the limitations of the assumptions utilized by the methodologies for time estimations, the absolute dates generated in this study should only be taken as rough estimates of upper bounds.
Figure 4.
Spatial gradient maps illustrating clines in frequency and mean STR allelic variance (eight loci) within haplogroups R1b and J2.
Haplogroup J2
Supplementary Table 3 provides the haplotypes for the 105 J2-M172-derived individuals. An MDS plot based on Rst distances from individuals under the J2 lineages (Figure 3b) illustrates Gardman, Sasun and Lake Van plotting among the Turkish, Iranian and Levantine populations, while demonstrating segregation away from Crete and Greece. The phylogenetic relationships of haplotypes within haplogroup J2 are displayed in an MJ network in Supplementary Figure 3. As in the R1b-M343 network, the low genetic diversity of Ararat Valley and Sasun is clearly observed in the partitioning of samples from these populations into only a few haplotypes. The relative expansion times for haplogroup J2-M172 (Table 4) generally correspond with those yielded for R1b-M343, with the exception of Greece and Crete, which, unlike haplogroup R1b-M343, are slightly older than the dates yielded for several of the Near Eastern groups as well as the four Armenian populations.
Table 4. Haplotype diversity, variance and time estimations for Haplogroup J2.
| Population | %a | n | Nei's diversity | Haplotype variance | Time estimatesb (kya) | Time estimatesc (kya) |
|---|---|---|---|---|---|---|
| Ararat Valley (8 loci) | 23.6 | 15/26 | 0.9292±0.0317 | 0.3567 | 4.8±0.8 | 5.1±1.2 |
| (17 loci) | — | 18/26 | 0.9538±0.0327 | 0.4948d | — | — |
| Gardman (8 loci) | 32.3 | 22/31 | 0.9699±0.0169 | 0.4081 | 4.8±0.9 | 5.7±1.6 |
| (17 loci) | — | 26/31 | 0.9892±0.0108 | 0.5907d | — | — |
| Lake Van (8 loci) | 29.1 | 21/30 | 0.9609±0.0218 | 0.3819 | 4.3±1.0 | 3.9±0.8 |
| (17 loci) | — | 25/30 | 0.9816±0.0209 | 0.6566d | — | — |
| Sasun (8 loci) | 17.3 | 7/18 | 0.7908±0.0711 | 0.1614 | 2.2±0.6 | 2.8±1.2 |
| (17 loci) | — | 12/18 | 0.9216±0.0510 | 0.5662d | — | — |
| West Turkey | 21.9 | 31/35 | 0.9916±0.0097 | 0.3794 | 4.3±0.9 | 4.3±1.1 |
| Central Turkey | 27.3 | 39/42 | 0.9954±0.0069 | 0.4598 | 5.6±1.0 | 6.1±1.4 |
| East Turkey | 23.6 | 35/49 | 0.9804±0.0093 | 0.3928 | 4.7±0.7 | 5.1±0.9 |
| Syria | 19.1 | 46/63 | 0.9811±0.0081 | 0.4740 | 6.3±1.4 | 8.1±3.2 |
| Jordan | 14.7 | 28/38 | 0.9758±0.0140 | 0.4127 | 4.3±0.8 | 4.5±1.0 |
| Lebanon | 27.6 | 117/248 | 0.9828±0.0029 | 0.4619 | 5.3±0.8 | 5.3±0.9 |
| Palestine | 19.3 | 29/40 | 0.9808±0.0101 | 0.6740 | 7.8±1.5 | 9.9±3.2 |
| Iran | 24.1 | 65/97 | 0.9875±0.0039 | 0.3517 | 4.4±0.3 | 4.9±0.7 |
| Greece | 14.6 | 22/25 | 0.9900±0.0142 | 0.4629 | 5.7±0.9 | 6.7±1.2 |
| Crete | 30.6 | 37/56 | 0.9708±0.0125 | 0.5218 | 6.4±3.0 | 7.7±4.5 |
| Georgia | 27.3 | 17/18 | 0.9935±0.0210 | 0.2904 | 2.9±0.9 | 2.8±1.0 |
Haplogroup T
The haplotypes of the 35 individuals belonging to Haplogroup T are provided in Supplementary Table 4. The distribution of haplotypes within Haplogroup T-M184 illustrated in an MJ network (Supplementary Figure 4) depicts a positioning of clusters composed almost entirely of Levantine individuals in the center of the diagram, with all other groups emanating from this area. This potentially points toward an origin of these haplotypes in the Levant, a supposition that is further supported by the observation of high genetic diversity in the network among levantine individuals within haplogroup T-M184 in the Levant. Time estimates for T-M184 individuals (Supplementary Table 5) corroborate the supposition that the lineage originated in the Levant and entered the Armenian Plateau at a later time, as the dates generated from Levantine (6.2±1.8 kya using Ravid-Amir and Rosset34) and Iranian (5.9±1.3 kya) individuals are older than those yielded from Turkey (5.7±1.2 kya), Sasun (5.0±1.2 kya) and the remaining three Armenian populations (average 3.9±1.0 kya).
Discussion
Genetic support for Neolithic origins of the Armenian populations
The results of this study suggest that the majority of Armenian Y-chromosomes belong to lineages believed to have originated and expanded during or following the Neolithic, including E1b1b1c-M123, G-M201, J1-M267, J2-M172 and R1b1b1-L23.27, 38, 42, 43, 44, 45, 39 Previous investigations have found some of these haplogroups, including J1-M267 and J2-M172, to illustrate patterns of distributions that reflect the spread of agriculture and domestication from the Fertile Crescent,37, 45 and recently, the distribution of R1b1b-M269 haplotypes across the Near East and Europe has been proposed to be best explained by a Neolithic dispersal.44 Of particular prominence in Armenia are haplogroups R1b-M343 and J2-M172, which are detected in Ararat Valley, Gardman and Lake Van at frequencies higher than those observed in any of the Near Eastern populations analyzed in this study. STR haplotypes within these two lineages exhibit clear genetic affinities with individuals from the Near East, particularly the Levantine populations of Jordan and Lebanon as demonstrated in the MDS plots (Figures 3a and b) and network diagrams (Supplementary Figures 1 and 2). These affiliations suggest gene flow between the Levant and Armenia, which, given the time estimates calculated, likely coincided with the expansion of agriculturalists. Examination of mean haplotype variance and expansion times for R1b-M343 and J2-M172 revealed less variance and slightly younger dates in Armenia relative to the surrounding regions. Therefore, our data likely reflects a directionality of these migrations from an origin in the Levant before reaching the Armenian highlands. However, more recent gene flow, possibly during the numerous imperial expansions from Assyria, Persia or even Europe, cannot be fully excluded as potential causes for the observed distribution patterns of genetic diversity. Yet, it is difficult to imagine such events leading to the genetic homogeny observed among Ararat Valley, Gardman and Lake Van.
Given the inhospitable climatic conditions4 and paucity of archaeological remains3 in Armenia from the millennia preceding the Neolithic, the predominance of Neolithic Y-chromosomes in Armenia suggests that the region was sparsely settled before the arrival of early farmers.5 Settlement during the Mesolithic, such as those observed near Sasun,4 were likely only fleeting. We envision a hypothetical, yet compelling, scenario in which Neolithic agriculturalists from the Levant occupied a vacant Armenian Plateau. This is reflected in the contour plots in Figure 4, where variance estimates indicate a demic decreasing gradient from the Levant toward Armenia, while frequencies illustrate an inverse cline, supporting the idea that the migrants from the Levant constitute a great proportion of the Armenian population. According to this contention, most Paleolithic genetic signals currently detected in the Armenian highlands represent influx of chromosomes from continuously settled areas and not vestiges from older occupations. This is supported by time estimates for Haplogroup T-M184, a clade believed to have originated in the Near East during the Paleolithic,46 as we observe dates in Armenia (∼12–13 kya) lower than those in other regions of the Near East, including Iran and the Levant (∼20 kya). Such a disparity suggests the Haplogroup T lineages presently observed in Armenia were introduced by migrations that are more recent than those that carried Haplogroup T to the Levant and Iran. Given the difficulties associated with absolute dating, it is also possible that these markers were carried by migrants entering Armenia during an even later era, such as the Bronze Age if the T lineage within genetic contributors has been lost to genetic drift.
Limited evidence for Indo-European genetic associations
Shortly after the arrival of early farmers in Armenia and Anatolia (8 kya), agriculture spread to Greece and the Balkans, before rapidly expanding across Europe.47.Furthermore, the classification of Armenian as an old Indo-European language with similarities to the ancestral Proto-Indo-European languages has led to the supposition that agriculturalists migrating from Armenia into Europe were responsible for the establishment of Indo-European languages in the continent.13, 14 However, despite the close linguistic relationship between Armenians and the Indo-European speaking populations of Europe,12 we see little genetic support for this claim. The derived M412 allele, which is found in nearly all haplogroup R1b1b1*-L23 chromosomes in Europe,27 is absent in the sampled Armenians, which also exhibit a scarcity of haplotype sharing with Europeans, suggesting a limited role for Armenians in the introduction of R1b into Europe.
Several authors have proposed that the Indo-European language presently spoken by Armenians arose during the Bronze Age, when Indo-European speaking tribes from the Balkans and Greece invaded Anatolia and Transcaucasia, leading to the subsequent spread of their culture and language.16, 17 In this study, we have detected a number of lineages that are prominent in the Balkans (I2*, I2b*, J2b1 and J2b2) at low levels throughout Ararat Valley, Gardman and Lake Van, the latter of which also contains haplogroups commonly associated with Bronze Age Greece (ie, J2a8-M319 (4.9%), and E1b1b1-M78 and its sublineages (3.9%)). While this may suggest genetic input from early Greek or Phrygian tribes, it is also possible that these low levels of Balkan lineages arrived in Armenia at a later time, such as during one of the many incursions into the area during the reign of the Macedonian, Roman and Byzantine empires. It should be noted that these results only reflect the paternal history of Armenia and studies on a maternal or gender-neutral system may reveal distinct conclusions.
Although the Armenian paternal gene pool exhibits limited genetic affinities with modern European Y-chromosomes, a paleogenetic study examining the gene pool of Neolithic farmers in Europe (∼6 kya) found that these fossils displayed greater genetic similarity with individuals from the modern Near East than to modern Europe.48 These ancient individuals were characterized by haplogroups G2a and F*(xGHIJK), which, in the present day gene pool, are mostly restricted to the Near East and are not prevalent in modern Europeans. These results suggest that the genetic profile currently observed throughout Europe potentially originated from migrations that took place subsequent to the Neolithic era. Such a scenario may explain the lack of segregation separating Armenians and Europeans that was observed in the network containing only the older lineages, R1b1b*-M269 and R1b1b1*-L23, as these haplogroups may represent remnants of an older European population. As a result, comparisons of the paternal component of Armenians and a larger dataset of ancient European samples may illuminate the nature of the migrations into Europe with greater fidelity.
Conclusion
Our results indicate a prevalence of Neolithic paternal chromosomes that are associated with the Agricultural Revolution, namely E1b1b1c-M123, G-M201, J1-M267, J2a-M410 and R1b1b1*-L23, which collectively comprise 77% (58% in Sasun and an average of 84% in Ararat Valley, Gardman and Lake Van) of the observed paternal lineages in the Armenian Plateau. Furthermore, Y-STR variance and haplotype distributions suggest that these lineages were likely introduced into Armenia from the Levant. However, later migrations, such as from Armenia to Europe, do not appear to have been associated with any paternal gene flow.
Acknowledgments
The authors thank Marisil Wright, Priscilla Torres, Luis Rivera, Michelle Hernandez, Evelyn Ramirez, Emmanuel Martinez, Tanya Simms and Maria Cristina Terreros for their technical assistance. In addition, we also gratefully acknowledge Tenzin Gayden and Shilpa Chennakrishnaiah for their constructive criticisms of the manuscript.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on European Journal of Human Genetics website (http://www.nature.com/ejhg)
Supplementary Material
References
- Arslanov KA, Dolukhanov PM, Gei NA. Climate, Black Sea levels and human settlements in Caucasus Littoral 50,000-9000 BP. Quatern Int. 2007;168:121–127. [Google Scholar]
- Pinhasi R, Gasparian B, Wilkinson K, et al. Hovk 1 and the Middle and Upper Paleolithic of Armenia: a preliminary framework. J Hum Evol. 2008;55:803–816. doi: 10.1016/j.jhevol.2008.04.005. [DOI] [PubMed] [Google Scholar]
- Dolukhanov P, Aslanyan S, Kolpakov E, Belyayeva E.Prehistoric sites in northern Armenia Antiquity 2004 , http://antiquity.ac.uk/projgall/dolukhanov301 .
- Akçar N, Yavuz V, Ivy-Ochs S, Kubik PW, Vardar M, Schlüchter C. Paleoglacial records from Kavron Valley, NE Turkey: Field and cosmogenic exposure dating evidence. Quatern Int. 2007;165:170–183. [Google Scholar]
- Kartal M. Anatolian epi-paleolithic period assemblages: problems, suggestions, evaluations and various approaches. Anatolia. 2003;24:45–62. [Google Scholar]
- Lang DM. Armenia, Cradle of Civilization. London and Boston: Allen and Unwin; 1980. [Google Scholar]
- Kushnareva KK. The Southern Caucasus in Prehistory: Stages of Cultural and Socioeconomic Development from the Eighth to the Second Millennium B.C. Ephrata, PA: Science Press; 1997. [Google Scholar]
- Hovsepyan R, Willcox G. The earliest finds of cultivated plants in Armenia: evidence from charred remains and crop processing residues in pisé from the Neolithic settlements of Aratashen and Aknashen. Veget Hist Archaeobot. 2008;17:S63–S71. [Google Scholar]
- Glascock MD. Provenance studies on obsidian artifacts from early Neolithic Iran. Geol Soc Am Abstr Programs. 2009;41:553. [Google Scholar]
- Pinhasi R, Gasparian B, Areshian G, et al. First direct evidence of Chalcolithic footwear from the Near Eastern highlands PLoS One 20105e10984doi: 10.1371/journal.pone.0010984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnard H, Dooley AN, Areshian G, Gasparyan B, Faull KF.Chemical evidence for wine production around 4000 BCE in the Late Chalcolithic Near Eastern highlands J Archaeol Sci 2011. doi: 10.1016/j.jas.2010.11.012 [DOI]
- Gray RD, Atkinson QD. Language-tree divergence times support the Anatolian theory of Indo-European origin. Nature. 2003;426:435–438. doi: 10.1038/nature02029. [DOI] [PubMed] [Google Scholar]
- Renfrew C. Archeology and Language: The Puzzle of Indo European Origins. London: Jonathon Cape; 1987. [Google Scholar]
- Gamkrelidze TV, Ivanov VV. Indo-European and the Indo-Europeans: A Reconstruction and Historical Analysis of a Proto-Language and a Proto-Culture. Berlin: de Gruyter Mouton; 1995. [Google Scholar]
- Alinei M.Towards and invasionless model of Indoeuropean origins: the continuity theoryin Pearce M, Tosi M (eds):Papers from the EEA Third Annual Meeting at Ravenna 1997 – Vol. I. Pre-and ProtohistoryBAR International Series 717199831–33.
- Diakonoff IM.The formation of the Armenian peoplein Jennings L (ed): The Pre-History of the Armenian People Delmar, NY: Caravan Books; 1984101–130. [Google Scholar]
- Greppin JA. Some effects of the Hurro-Urartian people and their languages upon the earliest Armenians. J Am Orient Soc. 1991;111:720–730. [Google Scholar]
- Underhill PA, Kivisild T. Use of Y Chromosome and Mitochondrial DNA population structure in tracing human migrations. Genetics. 2007;41:539–564. doi: 10.1146/annurev.genet.41.110306.130407. [DOI] [PubMed] [Google Scholar]
- Weale ME, Yepiskoposyan L, Jager RF, et al. Armenian Y chromosome haplotypes reveal strong regional structure within a single ethno-national group. Hum Genet. 2001;109:659–674. doi: 10.1007/s00439-001-0627-9. [DOI] [PubMed] [Google Scholar]
- Nasidze I, Sarkisian T, Kerimov A, Stoneking M. Testing hypotheses of language replacement in the Caucasus: evidence from the Y-chromosome. Hum Genet. 2003;112:255–261. doi: 10.1007/s00439-002-0874-4. [DOI] [PubMed] [Google Scholar]
- Nasidze I, Ling EYS, Quinque D, et al. Mitochondrial DNA and Y-chromosome variation in the Caucasus. Ann Hum Genet. 2004;68:205–221. doi: 10.1046/j.1529-8817.2004.00092.x. [DOI] [PubMed] [Google Scholar]
- Nasidze I, Stoneking M. Mitochodnrial DNA variation and language replacements in the Caucasus. Proc Biol Sci. 2001;268:1197–1206. doi: 10.1098/rspb.2001.1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasidze I, Risch GM, Robichaux M, Sherry ST, Batzer MA, Stoneking M. Alu insertion polymorphisms and the genetics structure of human populations from the Caucasus. Eur J Hum Genet. 2001;9:267–272. doi: 10.1038/sj.ejhg.5200615. [DOI] [PubMed] [Google Scholar]
- Gayden T, Regueiro M, Martinez L, Cadenas AM, Herrera RJ. Human Y-chromosome haplotyping by allele-specific polymerase chain reaction. Electrophoresis. 2008;29:2419–2423. doi: 10.1002/elps.200700702. [DOI] [PubMed] [Google Scholar]
- Hammer MF, Horai S. Y chromosomal DNA variation and the peopling of Japan. Am J Hum Genet. 1995;56:951–962. [PMC free article] [PubMed] [Google Scholar]
- Karafet TM, Mendez FL, Meilerman MB, Underhill PA, Zegura SL, Hammer MF. New binary polymorphisms reshape and increase resolution of the human Y chromosomal haplogroup tree. Genome Res. 2008;18:830–838. doi: 10.1101/gr.7172008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myres NM, Rootsi S, Lin AA, et al. A major Y-chromosome founder effect in central and western Europe. Eur J Hum Genet. 2011;19:95–101. doi: 10.1038/ejhg.2010.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Excoffier L, Lischer HEL. Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol Ecol Resour. 2010;10:564–567. doi: 10.1111/j.1755-0998.2010.02847.x. [DOI] [PubMed] [Google Scholar]
- Goodman SJ. Rst Calc: a collection of computer programs for calculating estimates of genetic differentiation from microsatellite data and a determining their significance. Mol. Ecol. 1997;6:881–885. [Google Scholar]
- Bandelt H-J, Forster P, Röhl A. Median-joining networks for inferring intraspecific phylogenies. Mol Biol Evol. 1999;16:37–48. doi: 10.1093/oxfordjournals.molbev.a026036. [DOI] [PubMed] [Google Scholar]
- Martinez L, Underhill PA, Zhivotovsky LA, et al. Paleolithic Y-haplogroup heritage predominates in a Cretan highland plateau. Eur J Hum Genet. 2007;15:485–493. doi: 10.1038/sj.ejhg.5201769. [DOI] [PubMed] [Google Scholar]
- Zhivotovsky LA, Underhill PA, nio∂lu C, et al. The effective mutation rate at Y chromosome short tandem repeats, with application to human population-divergence time. Am J Hum Genet. 2004;74:50–61. doi: 10.1086/380911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sengupta S, Zhivotovsky LA, King R, et al. Polarity and temporality of high-resolution Y-chromosome distribution in India identify both indigenous and exogenous expansions and reveal minor genetic influence of Central Asian pastoralists. Am J Hum Genet. 2006;78:202–221. doi: 10.1086/499411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravid-Amir O, Rosset S. Maximum likelihood estimation of locus-specific mutation rates in Y-chromosome short tandem repeats. Bioinformatics. 2010;26:i440–i445. doi: 10.1093/bioinformatics/btq367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goedbloed M, Vermeulen M, Fang RN, et al. Comprehensive mutation analysis of 17 Y-chromosomal short tandem repeat polymorphisms included in the AmpFlSTR® PCR amplification kit. Intl J Leg Med. 2009;123:471–482. doi: 10.1007/s00414-009-0342-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Underhill PA, Myres NM, Rootsi S, et al. Separating the post-glacial coancestry of European and Asian Y chromosomes within haplogroup R1a. Eur J Hum Genet. 2010;18:479–484. doi: 10.1038/ejhg.2009.194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiaroni J, King RJ, Underhill PA. Correlation of annual precipitation with human Y-chromosome diversity and the emergence of Neolithic agricultural and pastoral economies in the Fertile Crescent. Antiquity. 2008;82:281–289. [Google Scholar]
- Giacomo FD, Luca F, Popa LO, et al. Y chromosomal haplogroup J as a signature of the post-neolithic colonization of Europe. Hum Genet. 2004;115:357–371. doi: 10.1007/s00439-004-1168-9. [DOI] [PubMed] [Google Scholar]
- Battaglia V, Fornarino S, Al-Zahery N, et al. Y-chromosomal evidence of the cultural diffusion of agriculture in southeast Europe. Eur J Hum Genet. 2009;17:820–830. doi: 10.1038/ejhg.2008.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King RJ, Özcan SS, Carter T, et al. Differential Y-chromosome Anatolian influences on the Greek and Cretan Neolithic. Ann Hum Genet. 2008;72:205–214. doi: 10.1111/j.1469-1809.2007.00414.x. [DOI] [PubMed] [Google Scholar]
- Burgarella C, Navascués M. Mutation rate estimates for 110 Y-chromosome STRs combining population and father-son pair data. Eur J Hum Genet. 2011;19:70–75. doi: 10.1038/ejhg.2010.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arredi B, Poloni ES, Paracchini S, et al. A predominantly Neolithic origin for Y-chromosomal DNA variation in North Africa. Am J Hum Genet. 2004;75:338–345. doi: 10.1086/423147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semino O, Magri C, Benuzzi G, et al. Origin, diffusion, and differentiation of Y-chromosome haplogroups E and J: inferences on the Neolithization of Europe and later migratory events in the Mediterranean area. Am J Hum Genet. 2004;74:1023–1034. doi: 10.1086/386295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balaresque P, Bowden GR, Adams SM, et al. A predominantly Neolithic origin for European paternal lineages. PLOS Biol. 2010;8:e1000285. doi: 10.1371/journal.pbio.1000285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiaroni J, King RJ, Myres NM, et al. The emergence of Y-chromosome haplogroup J1e among Arabic-speaking populations. Eur J Hum Genet. 2010;18:348–353. doi: 10.1038/ejhg.2009.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendez FL, Karafet TM, Krahn T, Ostrer H, Soodyall H, Hammer MF. Increased resolution of Y chromosome haplogroup T defines relationships among populations of the Near East, Europe, and Africa. Hum Biol. 2011;83:39–53. doi: 10.3378/027.083.0103. [DOI] [PubMed] [Google Scholar]
- Pinhasi R, Fort J, Ammerman AJ. Tracing the origin and spread of agriculture in Europe. PLOS Biol. 2005;3:2220–2228. doi: 10.1371/journal.pbio.0030410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haak W, Balanovsky O, Sanchez JJ, et al. Ancient DNA from European early Neolithic farmers reveals their near eastern affinities. PLOS Biol. 2010;8:e1000536. doi: 10.1371/journal.pbio.1000536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zalloua PA, Xue Y, Khalife J, et al. Y-chromosomal diversity in Lebanon is structured by recent historical events. Am J Hum Genet. 2008;82:873–882. doi: 10.1016/j.ajhg.2008.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Sibai M, Platt DE, Haber M, et al. Geographical structure of the Y-chromosomal genetic landscape of the Levant: a coastal-inland contrast. Ann Hum Genet. 2009;73:568–581. doi: 10.1111/j.1469-1809.2009.00538.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zalloua PA, Platt DE, El-Sibai M, et al. Identifying genetic traces of historical expansions: Phoenician footprints in the Mediterranean. Am J Hum Genet. 2008;83:633–642. doi: 10.1016/j.ajhg.2008.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haber M, Platt DE, Badro DA, et al. Influences of history, geography, and religion on genetic structure: the Maronites in Lebanon. Eur J Hum Genet. 2011;19:334–340. doi: 10.1038/ejhg.2010.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cinnioǧlu C, King R, Kivisild T, et al. Excavating Y-chromosome haplotype strata in Anatolia. Hum Genet. 2004;114:127–148. doi: 10.1007/s00439-003-1031-4. [DOI] [PubMed] [Google Scholar]
- Spiroski M, Arsov T, Krüger C, Willuweit S, Roewer L. Y-chromosomal STR haplotypes in Macedonian population samples. Forensic Sci Int. 2005;148:69–73. doi: 10.1016/j.forsciint.2004.04.067. [DOI] [PubMed] [Google Scholar]
- Roewer L, Willuweit S, Krüger C. Analysis of Y chromosome STR haplotypes in the European part of Russia reveals high diversities but non-significant genetic distances between populations. Int J Legal Med. 2008;122:219–223. doi: 10.1007/s00414-007-0222-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturrock K, Rocha J. A multidimensional scaling stress evaluation table. Field Methods. 2000;12:49–60. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.