Abstract
Background
Thyroid cancer diagnosis in the United States has increased by 2.3-folds in the last three decades. Up to 30% of thyroid fine-needle aspiration biopsy (FNAB) results are inconclusive. Several differentially expressed microRNAs (miRNAs) have been identified as candidate diagnostic markers for thyroid nodules. We hypothesized that these differentially expressed miRNAs may improve the accuracy of FNAB in difficult to diagnose thyroid nodules.
Methods
Expression levels of four miRNAs (miR-7, -126, -374a, and let-7g) were analyzed using quantitative real-time reverse transcription-polymerase chain reaction in 95 FNAB samples as the training set. A predictor model was formulated based on the most differentially expressed miRNA (miR-7) ΔCt value and the model was applied on a separate cohort of 59 FNAB samples as the validation set.
Results
miR-7 was the best predictor to distinguish benign from malignant thyroid FNAB samples. The other three miRNAs were co-expressed and did not significantly contribute to the predictor model. miR-7 had a sensitivity of 100%, specificity of 29%, positive predictive value (PPV) of 36%, negative predictive value (NPV) of 100%, and overall accuracy of 76% when applied to the validation set. In subgroup analysis of preoperative nondiagnostic, indeterminate, or suspicious FNAB samples, the predictor model had an overall accuracy of 37% with sensitivity of 100%, specificity of 20%, PPV of 25%, and NPV of 100%.
Conclusions
miR-7 may be a helpful adjunct marker to thyroid FNAB in tumor types which are inconclusive. Given the high NPV of miR-7, a patient with a benign result based on the predictor model may be followed as opposed to performing an immediate diagnostic thyroidectomy. Future prospective clinical trials evaluating its accuracy in a larger cohort are warranted to determine its clinical utility.
Introduction
Thyroid cancer is the most common endocrine malignancy and its incidence has increased by 2.3-fold in last three decades (1). Thyroid cancers are classified into papillary, follicular, Hürthle cell, medullary, and undifferentiated or anaplastic thyroid carcinomas with papillary thyroid carcinoma comprising the most predominant histology (80%) (2,3). While the prevalence of thyroid nodules is relatively common (4), fine-needle aspiration biopsy (FNAB) demonstrating malignancy is less common occurring in 3%–8% of FNAB, and the majority of results (70%) will demonstrate benign cytologic features (5–7). Up to 30% of FNAB samples containing an adequate sample are indeterminate as the cytologic features are inconclusive for distinguishing benign from malignant lesions solely based on cytology. In these cases, a diagnostic thyroidectomy is usually necessary for definitive diagnosis on histology (8).
There have been several factors which could influence the need for diagnostic thyroidectomy, such as repeat FNAB or secondary review of FNAB samples (8–11). There has been a significant effort focused on combining the FNAB result with clinical information or combining the information from other diagnostic modalities such as ultrasonography, immunocytochemical markers, and molecular markers to improve the accuracy of thyroid FNAB (12–19). Within the area of molecular markers there have been several studies evaluating the potential of small noncoding RNA called microRNAs (miRNAs) as diagnostic markers for thyroid cancer.
miRNAs are ∼22 nucleotides long and are responsible for gene regulation at the mRNA level (20). miRNA regulate gene expression by binding to the 3’untranslated region of their target mRNAs and can either lead to repression of translation or mRNA degradation depending on the level of the complementarity to their target mRNA sequence (21). There have been several studies which have investigated the miRNA signatures in thyroid cancer. Some of them have identified dysregulated miRNAs in papillary thyroid cancer compared with normal thyroid tissue or multinodular goiter (22–26) and some studies have demonstrated the feasibility of testing these dysregulated miRNAs in FNAB samples (22,24,26,27). A study by Weber et al. addressed in particular the dysregulated miRNAs in follicular thyroid carcinoma (FTC) compared with follicular adenoma (FA) and found that two miRNAs (miR-197 and -346) were significantly overexpressed in FTC compared with their benign counterparts (28). Another study by Sheu et al. studied the diagnostic utility of miR-146b, -181b, -21, -221, and -222 in follicular variant of papillary thyroid cancer (FPTC) and FTC and they found that these five miRNAs can distinguish common variants of papillary thyroid cancer from FA/multinodular goiter (29). Several miRNA expression levels have also been implicated in thyroid tumorigenesis (24,28,30) and miR-146b in particular has been found to be associated with thyroid cancer with high risk features (31).
Recently our group found downregulation of miR-126 and miR-7 in thyroid cancer (32). We compared the two broad categories of benign and malignant tumor samples, which included the challenging histologic subtypes which are difficult to distinguish by FNAB (i.e., FA/carcinoma, Hürthle cell adenoma/carcinoma, and FPTC). Discovery of these miRNAs in difficult to diagnose histologic subtypes led to our current effort in validating this information in FNAB samples. Given the evidence suggesting suboptimal applicability of tissue expression data to FNAB samples, the goal of this study was to formulate a model that would predict benign from malignant samples based on a group of FNAB samples as the training set and validate this model on a separate cohort of FNAB samples as a validation set.
Materials and Methods
FNAB samples and patient information
FNAB samples, patient demographics, and histopathological information were obtained at the Johns Hopkins University School of Medicine, Baltimore, MD under an institutional review board approved protocol after written informed consent. All the FNA samples were obtained under ultrasound guidance, intraoperatively, before an incision was made. The samples were placed immediately into RNAlater® (Applied biosystems/Ambion, Foster City, CA) and stored at −80°C until they were ready for RNA extraction. The samples were classified based on The Bethesda System for Reporting Thyroid Cytopathology. A total of 95 FNAB samples were used as the training set to derive a predictor model and the model was applied to 59 FNAB samples as the validation set. The 59 FNAB validation samples were initially tested blinded without the cytological or histological diagnosis. Patient demographic and clinical information are summarized in Table 1.
Table 1.
Patient Demographics and Clinical Characteristics of Study Cohort
| Variables | Training set (n=95/93)a | Validation set (n=59) | p-Value |
|---|---|---|---|
| Average age (years)±SD (range) | 48±14 (19–84) | 48±13 (19–80) | 0.934 |
| Sex, n (%) | |||
| Male | 22 (24) | 8 (14) | |
| Female | 71 (76) | 51 (86) | 0.143 |
| Ethnicity, n (%) | |||
| White | 70 (75) | 39 (66) | |
| Asian/Pacific Islander | 2 (2) | 3 (5) | |
| African American | 12 (13) | 13 (22) | |
| Hispanic | 2 (2) | 2 (3) | |
| Other/unknown | 7 (8) | 2 (3) | 0.348 |
| Family history of thyroid cancer, n (%) | 7 (8) | 1 (2) | 0.151 |
| Prior history of radiation, n (%) | 2 (2) | 7 (12) | 0.028 |
| BSRTC, n (%) | |||
| I. Nondiagnostic or unsatisfactory | 16 (17) | 3 (5) | |
| II. Benign | 21 (22) | 20 (34) | |
| III. Atypia of undetermined significance or follicular lesion of undetermined significance | 17 (18) | 8 (14) | |
| IV. Follicular neoplasm or suspicious for a follicular neoplasm | 9 (9) | 7 (12) | |
| V. Suspicious for malignancy | 5 (5) | 6 (10) | |
| VI. Malignant | 20 (21) | 15 (25) | |
| NA | 7 (7) | 0 (0) | 0.052 |
| Average tumor diameter (cm)±SD (range) | 2.56±1.72 (0.2–8.0) | 2.87±1.65 (0.3–7.5) | 0.182 |
| Final pathology | |||
| Benign | 67 (71) | 42 (71) | |
| Hyperplastic nodule | 30 | 24 | |
| Hyperplastic nodule with Hürthle cell changes | 10 | 3 | |
| Follicular adenoma | 5 | 3 | |
| Hürthle cell adenoma | 12 | 4 | |
| Hyperplastic nodule with thyroiditis | 10 | 8 | 1.00 |
| Malignant | 28 (29) | 17 (29) | |
| Follicular variant of papillary thyroid cancer | 5 | 5 | |
| Classic papillary thyroid cancer | 23 | 12 | 1.00 |
| TNM stage of malignant tumors | |||
| Stage I | 17 | 13 | |
| Stage II | 2 | 2 | |
| Stage III | 4 | 1 | |
| Stage IVa | 5 | 1 | 0.483 |
Group 1 consisted of 93 patients with 95 nodules.
BSRTC, Bethesda System for Reporting Thyroid Cytology; TNM, tumor–node–metastasis; SD, standard deviation; NA, not available.
RNA isolation
RNA extraction of FNAB samples was performed using miRCURY™ RNA isolation kit (Exiqon, Woburn, MA) according to the manufacturer's protocol. RNA quantity and quality were determined using NanoDrop (Thermo Scientific, Wilmington, DE) and RNA was stored at −80°C until they were used for quantitative real-time reverse transcription-polymerase chain reaction (qRT-PCR).
miRNA expression analysis using qRT-PCR
qRT-PCR was performed on the 95 FNAB training cohort for miRNA-7 (assay #000268) and miRNA-126 (#002228) based on the model previously proposed by our group (32). Two additional miRNAs (miR-374a [assay #000563] and let-7g [assay #002282]), which were the third and fourth most significantly expressed miRNAs in our prior study, were also included in this analysis. qRT-PCR was performed on the 59 FNAB validation cohort for miRNA-7 (assay #000268) and miRNA-126 (#002228). TaqMan® microRNA assay probes were purchased from Applied Biosystems (Foster City, CA). cDNA was created using 5ng of total RNA and miRNA specific hairpin primers with TaqMan microRNA Reverse Transcription Kit (PN 4366597; Applied Biosystems). Five nanograms of total RNA were used in a 15 μL reaction to create the cDNA template. We used 2.5 μL of cDNA in a 10 μL reaction and all qRT-PCR reactions were performed in triplicates using miRNA specific primers (TaqMan microRNA assay) and TaqMan Universal PCR Master Mix on an ABI 7900HT Fast Real-Time PCR system (Applied Biosystems). miR-625* (assay #002432) was used as the endogenous control based on our previous data, which showed the least variability across thyroid tumor types (32). miRNA expression levels were calculated as the delta cycle threshold (ΔCt), which was calculated from cycle threshold (Ct) of the miRNA of interest subtracted by the Ct value of the endogenous control.
Statistical analysis
An initial screening procedure was performed on the ΔCt values of 95 FNAB training set samples in which the distribution of each of the miRNAs was compared between the two groups of interest (benign vs. malignant) using an exact Wilcoxon rank sum test. Any miRNAs that were found to have possible differences in distribution, as exhibited by having univariate p-values from the screening procedure of <0.05, were further individually evaluated and then jointly in a logistic regression model. All p-values were analyzed as two-tailed tests. The Mann–Whitney U test was used to compare miRNA expression levels between benign and malignant samples.
Results
Patient demographics and clinicopathological information
The clinical and demographical data for FNAB samples are summarized in Table 1. All the clinicopathological features were relatively similar between the training and the validation cohort, except there were slightly more patients with a history of radiation in the validation cohort. Final histology of FNAB samples was primarily papillary thyroid cancer and FPTC. The majority of malignant samples were comprised of American Joint Committee on Cancer tumor–node–metastasis classification system stage I tumors.
miRNA expression levels and predictor model derivation based on 95 FNAB training set
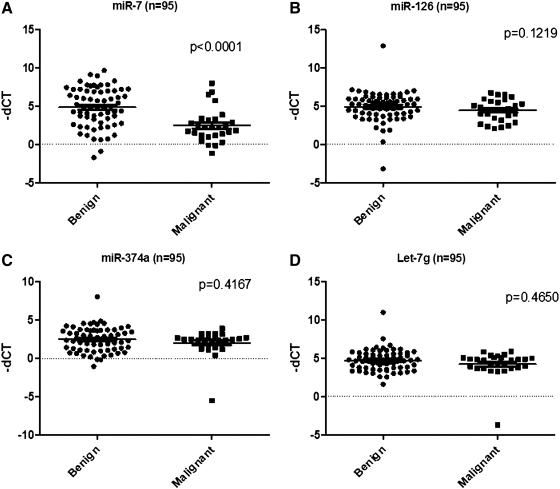
Expression levels of miR-7, -126, -374a, and let-7g were tested using qRT-PCR (Fig. 1). Only miR-7 was significantly differentially expressed between benign and malignant samples. miR-7 was significantly downregulated in malignant samples compared with benign with a p-value of <0.0001.
FIG. 1.
Scatter-plot of miRNAs [miR-7 (A), -126 (B), -374a (C), and let-7g (D)] in 95 FNAB training set samples. Each value represents a sample and the expression levels are expressed as negative (−) ΔCt values. p-values were calculated using the Mann–Whitney U test. All the values were normalized to the internal control (miR-625*). FNAB, fine-needle aspiration biopsy; miRNAs, microRNAs.
Based on the exact Wilcoxon rank sum test and logistic regression model, miR-7 was the only miRNA associated with the ability to classify tumors as benign or malignant. The cutoff value for malignant classification was miR-7 ΔCt ≥−3.40456; classify benign if miR-7 ΔCt <−3.40456. This model had a sensitivity of 82%, specificity of 73%, positive predictive value (PPV) of 56%, negative predictive value (NPV) of 91%, and overall accuracy of 76%.
miRNA expression levels and result of the predictor model in the FNAB validation set
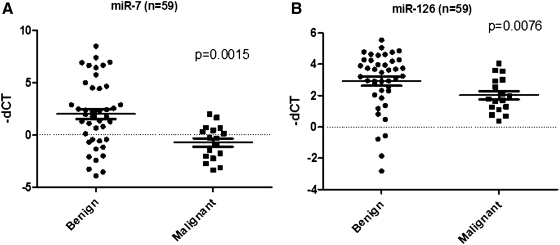
Expression levels of miR-7 and -126 were tested using qRT-PCR. Both miR-7 and miR-126 were significantly downregulated in malignant samples compared with benign ones with p-values of 0.0015 and 0.0076, respectively (Fig. 2).
FIG. 2.
Scatter-plot of the 2 most differentially expressed miRNAs [miR-7 (A) and -126 (B)] on the 59 FNAB validation set samples. Each dot represents a sample and the expression levels are expressed as −ΔCt values. p-values were calculated using the Mann–Whitney U test. All the values were normalized to the internal control (miR-625*).
When the predictor model derived from the FNAB training set was applied to the 59 samples, it had a sensitivity of 100%, specificity of 29%, PPV of 36%, NPV of 100%, and overall accuracy of 76%. When the model was specifically applied to the subgroup preoperatively designated as nondiagnostic, indeterminate, or suspicious it had a sensitivity of 100%, specificity of 20%, PPV of 25%, and NPV of 100%.
Discussion
In this study, we formulated a predictor model that would distinguish benign from malignant samples based on miRNA expression levels. We tested the expression levels of four reported miRNAs (miR-7, -126, -374a, and let-7g) that were most significantly differentially expressed based on tissue microarray and qRT-PCR (32) analysis in 95 FNAB training set samples. We then applied the model on a separate cohort of 59 FNAB samples as the validation set. The application of the model based on the 95 FNAB training set on the 59 FNAB validation set demonstrated a sensitivity of 100% with NPV of 100% but with suboptimal specificity and PPV. When the model was applied to the subgroup of patients with inconclusive FNAB findings, it retained high sensitivity and NPV of 100% but with lower specificity and PPV.
The introduction of routine use of FNAB has significantly reduced the number of unnecessary thyroidectomies for benign disease while increasing the yield of malignancy in thyroidectomy specimen (33–35). However, indeterminate lesions which may comprise up to 30% of FNAB result still pose significant challenge to clinicians because many of these patients require a thyroidectomy for definitive histologic diagnosis. There have been numerous studies trying to improve the accuracy of FNAB to obviate the need for unnecessary thyroid surgeries and to allow for more complete initial thyroid operations. Among these efforts, several studies have focused on miRNA expression profiling of thyroid cancer. Within these studies, miR-222, -221, and -146b are consistently demonstrated to be upregulated in papillary thyroid cancer (22–24). However, in our initial microarray analysis of difficult-to-diagnose thyroid nodules on FNA cytology, we did not find these miRs to be differentially expressed on quantitative RT-PCR validation of the microarray data (32). Numerous studies having identified differentially expressed miRNAs in thyroid cancer, specific miRNA(s) which would distinguish benign from malignant thyroid tumor in indeterminate FNAB samples have not yet been validated in clinical FNAB samples.
We found miR-7 to be the most accurate miRNA to distinguish benign from malignant thyroid FNAB samples. There currently is no available data on miR-7 in thyroid cancer or thyroid tumorigenesis. miR-7 has been characterized as a tumor suppressor in some malignancies [e.g., tongue squamous cell carcinoma (36) and glioblastoma (37)] but also has been characterized as an oncogenic miRNA “oncomiR” in lung cancer (38). It has also been associated with aggressiveness of lymph-node negative, estrogen-receptor positive human breast cancers (39). Further, miR-7 has been found to target several different proto-oncogenes such as epidermal growth factor receptor (EGFR) (40,41), insulin-like growth factor 1 receptor (IGF1R) (36), and p2-activated kinase 1 (Pak1) (42). There are some studies that suggest that miR-7 acts upstream of EGFR as its regulator (40,41). However, other investigators suggest that miR-7 is downstream of EGFR, suggesting that EGFR regulates miR-7 expression (38). Nonetheless, there appears to be an intricate relationship between miR-7 expression levels with EGFR and with other proto-oncogenes that will require further functional studies to understand its role in thyroid cancer initiation and/or progression.
Our previous study is the first to demonstrate the downregulation of miR-7 in thyroid cancer and this was demonstrated in both thyroid tissue and FNAB samples (32). To our knowledge, our study is also the first to evaluate the miRNA expression profile of the specific difficult to diagnose histologic subgroups on FNAB and validate these candidate diagnostic miRNAs in FNAB samples. Many patients undergo a diagnostic thyroidectomy for inconclusive thyroid FNAB results. Although the accuracy of our predictor model was suboptimal, it had a sensitivity and NPV of 100% in both the validation cohort and the subgroup of indeterminate FNAB samples. This implies that when the predictor model predicts negative for malignancy based on miR-7 measurement such patients may be closely followed rather than undergoing immediate diagnostic thyroidectomy. However, due to its low specificity and PPV, a positive result would not be accurate enough to make a decision on the appropriate extent of the initial thyroidectomy. We recognized additional studies with even larger sample size, showing similarly high sensitivity and NPV of 100%, will be necessary for clinical implementation of miR-7 expression level analysis as an adjunct to FNAB.
In summary, miR-7 was identified to be the most significantly differentially expressed miRNAs between benign and malignant thyroid FNAB samples and was the best predictor for distinguishing benign from malignant FNAB samples. Our predictor model for miR-7 has excellent NPV. Future prospective clinical trials should be considered and conducted in a larger cohort of FNAB samples to validate its clinical utility.
Acknowledgment
This research was supported by the Intramural Research Program, Center for Cancer Research, National Cancer Institute, National Institutes of Health.
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Sipos JA. Mazzaferri EL. Thyroid cancer epidemiology and prognostic variables. Clin Oncol (R Coll Radiol) 2010;22:395–404. doi: 10.1016/j.clon.2010.05.004. [DOI] [PubMed] [Google Scholar]
- 2.Hundahl SA. Fleming ID. Fremgen AM. Menck HR. A National Cancer Data Base report on 53,856 cases of thyroid carcinoma treated in the U.S., 1985–1995 [see comments] Cancer. 1998;83:2638–2648. doi: 10.1002/(sici)1097-0142(19981215)83:12<2638::aid-cncr31>3.0.co;2-1. [DOI] [PubMed] [Google Scholar]
- 3.Clark OH. Predictors of thyroid tumor aggressiveness. West J Med. 1996;165:131–138. [PMC free article] [PubMed] [Google Scholar]
- 4.Dean DS. Gharib H. Epidemiology of thyroid nodules. Best Pract Res Clin Endocrinol Metab. 2008;22:901–911. doi: 10.1016/j.beem.2008.09.019. [DOI] [PubMed] [Google Scholar]
- 5.Amrikachi M. Ramzy I. Rubenfeld S. Wheeler TM. Accuracy of fine-needle aspiration of thyroid. Arch Pathol Lab Med. 2001;125:484–488. doi: 10.5858/2001-125-0484-AOFNAO. [DOI] [PubMed] [Google Scholar]
- 6.Yang J. Schnadig V. Logrono R. Wasserman PG. Fine-needle aspiration of thyroid nodules: a study of 4703 patients with histologic and clinical correlations. Cancer. 2007;111:306–315. doi: 10.1002/cncr.22955. [DOI] [PubMed] [Google Scholar]
- 7.Wang CC. Friedman L. Kennedy GC. Wang H. Kebebew E. Steward DL. Zeiger MA. Westra WH. Wang Y. Khanafshar E. Fellegara G. Rosai J. Livolsi V. Lanman RB. A large multicenter correlation study of thyroid nodule cytopathology and histopathology. Thyroid. 2010;21:243–251. doi: 10.1089/thy.2010.0243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Faquin WC. Baloch ZW. Fine-needle aspiration of follicular patterned lesions of the thyroid: diagnosis, management, and follow-up according to National Cancer Institute (NCI) recommendations. Diagn Cytopathol. 2010;38:731–739. doi: 10.1002/dc.21292. [DOI] [PubMed] [Google Scholar]
- 9.Davidov T. Trooskin SZ. Shanker BA. Yip D. Eng O. Crystal J. Hu J. Chernyavsky VS. Deen MF. May M. Artymyshyn RL. Routine second-opinion cytopathology review of thyroid fine needle aspiration biopsies reduces diagnostic thyroidectomy. Surgery. 2010;148:1294–1299. doi: 10.1016/j.surg.2010.09.029. discussion 9–301. [DOI] [PubMed] [Google Scholar]
- 10.Oertel YC. Miyahara-Felipe L. Mendoza MG. Yu K. Value of repeated fine needle aspirations of the thyroid: an analysis of over ten thousand FNAs. Thyroid. 2007;17:1061–1066. doi: 10.1089/thy.2007.0159. [DOI] [PubMed] [Google Scholar]
- 11.Tan YY. Kebebew E. Reiff E. Caron NR. Ogilvie JB. Duh QY. Clark OH. Ljung BM. Miller T. Does routine consultation of thyroid fine-needle aspiration cytology change surgical management? J Am Coll Surg. 2007;205:8–12. doi: 10.1016/j.jamcollsurg.2007.02.075. [DOI] [PubMed] [Google Scholar]
- 12.Shibru D. Chung KW. Kebebew E. Recent developments in the clinical application of thyroid cancer biomarkers. Curr Opin Oncol. 2008;20:13–18. doi: 10.1097/CCO.0b013e3282f27e49. [DOI] [PubMed] [Google Scholar]
- 13.Banks ND. Kowalski J. Tsai HL. Somervell H. Tufano R. Dackiw AP. Marohn MR. Clark DP. Umbricht CB. Zeiger MA. A diagnostic predictor model for indeterminate or suspicious thyroid FNA samples. Thyroid. 2008;18:933–941. doi: 10.1089/thy.2008.0108. [DOI] [PubMed] [Google Scholar]
- 14.Chudova D. Wilde JI. Wang ET. Wang H. Rabbee N. Egidio CM. Reynolds J. Tom E. Pagan M. Rigl CT. Friedman L. Wang CC. Lanman RB. Zeiger M. Kebebew E. Rosai J. Fellegara G. Livolsi VA. Kennedy GC. Molecular classification of thyroid nodules using high-dimensionality genomic data. J Clin Endocrinol Metab. 2010;95:5296–5304. doi: 10.1210/jc.2010-1087. [DOI] [PubMed] [Google Scholar]
- 15.Kouniavsky G. Zeiger MA. Thyroid tumorigenesis and molecular markers in thyroid cancer. Curr Opin Oncol. 2010;22:23–29. doi: 10.1097/CCO.0b013e328333846f. [DOI] [PubMed] [Google Scholar]
- 16.Vriens MR. Schreinemakers JM. Suh I. Guerrero MA. Clark OH. Diagnostic markers and prognostic factors in thyroid cancer. Future Oncol. 2009;5:1283–1293. doi: 10.2217/fon.09.85. [DOI] [PubMed] [Google Scholar]
- 17.Stang MT. Carty SE. Recent developments in predicting thyroid malignancy. Curr Opin Oncol. 2009;21:11–17. doi: 10.1097/CCO.0b013e32831db2af. [DOI] [PubMed] [Google Scholar]
- 18.Chiu CG. Strugnell SS. Griffith OL. Jones SJ. Gown AM. Walker B. Nabi IR. Wiseman SM. Diagnostic utility of galectin-3 in thyroid cancer. Am J Pathol. 2010;176:2067–2081. doi: 10.2353/ajpath.2010.090353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Adeniran AJ. Theoharis C. Hui P. Prasad ML. Hammers L. Carling T. Udelsman R. Chhieng DC. Reflex BRAF testing in thyroid fine-needle aspiration biopsy with equivocal and positive interpretation: a prospective study. Thyroid. 2011;21:717–723. doi: 10.1089/thy.2011.0021. [DOI] [PubMed] [Google Scholar]
- 20.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 21.Esquela-Kerscher A. Slack FJ. Oncomirs—microRNAs with a role in cancer. Nat Rev Cancer. 2006;6:259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 22.Nikiforova MN. Tseng GC. Steward D. Diorio D. Nikiforov YE. MicroRNA expression profiling of thyroid tumors: biological significance and diagnostic utility. J Clin Endocrinol Metab. 2008;93:1600–1608. doi: 10.1210/jc.2007-2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.He H. Jazdzewski K. Li W. Liyanarachchi S. Nagy R. Volinia S. Calin GA. Liu CG. Franssila K. Suster S. Kloos RT. Croce CM. de la Chapelle A. The role of microRNA genes in papillary thyroid carcinoma. Proc Natl Acad Sci USA. 2005;102:19075–19080. doi: 10.1073/pnas.0509603102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pallante P. Visone R. Ferracin M. Ferraro A. Berlingieri MT. Troncone G. Chiappetta G. Liu CG. Santoro M. Negrini M. Croce CM. Fusco A. MicroRNA deregulation in human thyroid papillary carcinomas. Endocr Relat Cancer. 2006;13:497–508. doi: 10.1677/erc.1.01209. [DOI] [PubMed] [Google Scholar]
- 25.Tetzlaff MT. Liu A. Xu X. Master SR. Baldwin DA. Tobias JW. Livolsi VA. Baloch ZW. Differential expression of miRNAs in papillary thyroid carcinoma compared to multinodular goiter using formalin fixed paraffin embedded tissues. Endocr Pathol. 2007;18:163–173. doi: 10.1007/s12022-007-0023-7. [DOI] [PubMed] [Google Scholar]
- 26.Chen YT. Kitabayashi N. Zhou XK. Fahey TJ., 3rd Scognamiglio T. MicroRNA analysis as a potential diagnostic tool for papillary thyroid carcinoma. Mod Pathol. 2008;21:1139–1146. doi: 10.1038/modpathol.2008.105. [DOI] [PubMed] [Google Scholar]
- 27.Mazeh H. Mizrahi I. Halle D. Ilyayev N. Stojadinovic A. Trink B. Mitrani-Rosenbaum S. Roistacher M. Ariel I. Eid A. Freund HR. Nissan A. Development of a microRNA-based molecular assay for the detection of papillary thyroid carcinoma in aspiration biopsy samples. Thyroid. 2011;21:111–118. doi: 10.1089/thy.2010.0356. [DOI] [PubMed] [Google Scholar]
- 28.Weber F. Teresi RE. Broelsch CE. Frilling A. Eng C. A limited set of human MicroRNA is deregulated in follicular thyroid carcinoma. J Clin Endocrinol Metab. 2006;91:3584–3591. doi: 10.1210/jc.2006-0693. [DOI] [PubMed] [Google Scholar]
- 29.Sheu SY. Grabellus F. Schwertheim S. Worm K. Broecker-Preuss M. Schmid KW. Differential miRNA expression profiles in variants of papillary thyroid carcinoma and encapsulated follicular thyroid tumours. Br J Cancer. 2010;102:376–382. doi: 10.1038/sj.bjc.6605493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ricarte-Filho JC. Fuziwara CS. Yamashita AS. Rezende E. da-Silva MJ. Kimura ET. Effects of let-7 microRNA on cell growth and differentiation of papillary thyroid cancer. Transl Oncol. 2009;2:236–241. doi: 10.1593/tlo.09151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chou CK. Chen RF. Chou FF. Chang HW. Chen YJ. Lee YF. Yang KD. Cheng JT. Huang CC. Liu RT. miR-146b is highly expressed in adult papillary thyroid carcinomas with high risk features including extrathyroidal invasion and the BRAF(V600E) mutation. Thyroid. 2010;20:489–494. doi: 10.1089/thy.2009.0027. [DOI] [PubMed] [Google Scholar]
- 32.Kitano M. Rahbari R. Patterson EE. Xiong Y. Prasad NB. Wang Y. Zeiger MA. Kebebew E. Expression profiling of difficult-to-diagnose thyroid histologic subtypes shows distinct expression profiles and identify candidate diagnostic microRNAs. Ann Surg Oncol. 2011;18:3443–3452. doi: 10.1245/s10434-011-1766-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hamberger B. Gharib H. Melton LJ., 3rd Goellner JR. Zinsmeister AR. Fine-needle aspiration biopsy of thyroid nodules. Impact on thyroid practice and cost of care. Am J Med. 1982;73:381–384. [PubMed] [Google Scholar]
- 34.Castro MR. Gharib H. Thyroid fine-needle aspiration biopsy: progress, practice, and pitfalls. Endocr Pract. 2003;9:128–136. doi: 10.4158/EP.9.2.128. [DOI] [PubMed] [Google Scholar]
- 35.Hadi M. Gharib H. Goellner JR. Heerden JA. Has fine-needle aspiration biopsy changed thyroid practice? Endocr Pract. 1997;3:9–13. doi: 10.4158/EP.3.1.9. [DOI] [PubMed] [Google Scholar]
- 36.Jiang L. Liu X. Chen Z. Jin Y. Heidbreder CE. Kolokythas A. Wang A. Dai Y. Zhou X. MicroRNA-7 targets IGF1R (insulin-like growth factor 1 receptor) in tongue squamous cell carcinoma cells. Biochem J. 2010;432:199–205. doi: 10.1042/BJ20100859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kefas B. Godlewski J. Comeau L. Li Y. Abounader R. Hawkinson M. Lee J. Fine H. Chiocca EA. Lawler S. Purow B. microRNA-7 inhibits the epidermal growth factor receptor and the Akt pathway and is down-regulated in glioblastoma. Cancer Res. 2008;68:3566–3572. doi: 10.1158/0008-5472.CAN-07-6639. [DOI] [PubMed] [Google Scholar]
- 38.Chou YT. Lin HH. Lien YC. Wang YH. Hong CF. Kao YR. Lin SC. Chang YC. Lin SY. Chen SJ. Chen HC. Yeh SD. Wu CW. EGFR promotes lung tumorigenesis by activating miR-7 through a Ras/ERK/Myc pathway that targets the Ets2 transcriptional repressor ERF. Cancer Res. 2010;70:8822–8831. doi: 10.1158/0008-5472.CAN-10-0638. [DOI] [PubMed] [Google Scholar]
- 39.Foekens JA. Sieuwerts AM. Smid M. Look MP. de Weerd V. Boersma AW. Klijn JG. Wiemer EA. Martens JW. Four miRNAs associated with aggressiveness of lymph node-negative, estrogen receptor-positive human breast cancer. Proc Natl Acad Sci USA. 2008;105:13021–13026. doi: 10.1073/pnas.0803304105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Duex JE. Comeau L. Sorkin A. Purow B. Kefas B. USP18 regulates EGF receptor expression and cancer cell survival via microRNA-7. J Biol Chem. 2011;286:25377–25386. doi: 10.1074/jbc.M111.222760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Webster RJ. Giles KM. Price KJ. Zhang PM. Mattick JS. Leedman PJ. Regulation of epidermal growth factor receptor signaling in human cancer cells by microRNA-7. J Biol Chem. 2009;284:5731–5741. doi: 10.1074/jbc.M804280200. [DOI] [PubMed] [Google Scholar]
- 42.Reddy SD. Ohshiro K. Rayala SK. Kumar R. MicroRNA-7, a homeobox D10 target, inhibits p21-activated kinase 1 and regulates its functions. Cancer Res. 2008;68:8195–8200. doi: 10.1158/0008-5472.CAN-08-2103. [DOI] [PMC free article] [PubMed] [Google Scholar]