SUMMARY
The identity of higher-order neurons and circuits playing an associative role to control feeding is unknown. We injected pseudorabies virus, a retrograde tracer, into masseter muscle, salivary gland and tongue of BAC-transgenic mice expressing GFP in specific neural populations and identified several CNS regions that project multi-synaptically to the periphery. MCH and orexin neurons were identified in the lateral hypothalamus, and Nurr1 and Cnr1 in the amygdala and insular/rhinal cortices. Cholera Toxin-β tracing showed that insular Nurr1+ and Cnr1+ neurons project to the amygdala or lateral hypothalamus, respectively. Finally we show that cortical Cnr1+ neurons show increased Cnr1 mRNA and c-Fos expression after fasting, consistent with a possible role for Cnr1+ neurons in feeding. Overall, these studies define a general approach for identifying specific molecular markers for neurons in complex neural circuits. These markers now provide a means for functional studies of relevant neurons in feeding or other complex behaviors.
INTRODUCTION
The homeostatic control of energy balance is essential for survival. Caloric intake studies in humans reveal a precise regulation of adipose tissue mass over the span of decades (Weigle, 1994). In rodents, following forced overfeeding, food intake is temporarily reduced until body weight returns to baseline at which point food intake returns to normal (Wilson et al., 1990). The identification of leptin and other hormones with potent CNS effects that modulate food intake provides a mechanism by which homeostatic control of energy is maintained (Friedman and Halaas, 1998). These effects are mediated by direct effects on neurons in the hypothalamus and elsewhere in the brain (Schwartz et al., 2000). However, many other sensory, metabolic and emotional factors also influence the likelihood of initiating feeding suggesting that the hypothalamus and specific CNS centers interact with other brain regions that in aggregate comprise a complex neural circuit that integrates multiple signals relevant for this behavior.
Feeding is thus a complex motivated behavior (Geiselman, 1996) that requires the integration of hunger and satiety cues conveyed by internal signals, information relayed by the physical and chemical senses, the overall emotional state and, in humans, the conscious motivation of an individual. In contrast to a reflex, no single stimulus can be predicted to trigger this complex behavior with certainty, with each of these inputs influencing the likelihood that the coordinated effector events required for feeding will be initiated. However, neither the precise neuroanatomical location nor the identities of the neurons in the higher brain centers that integrate the aforementioned inputs are known. The delineation of these neural pathways will be necessary in order to develop a better understanding of the neural basis of appetitive behavior.
Once the decision to eat has been made, proper temporal coordination of a series of outputs is required to ensure that the motor process of eating will be effectively completed. These events include the production and secretion of saliva and the proper function of buccal/orofacial muscles required for chewing and swallowing (Thexton, 1992). The CNS loci responsible for the coordination of these motor and autonomic efferents are largely unknown and it is likely that such an associative motor center(s) would communicate with other brain regions that convey relevant sensory information.
The hypothalamus plays an important role in the maintenance of energy homeostasis and also regulates efferent autonomic pathways that regulate metabolism (Williams et al., 2001). Animals with specific lesions in the lateral hypothalamus (LH) become anorectic and lose weight, while animals with lesions of the ventromedial hypothalamus overeat and become obese, (King, 2006; Vettor et al., 2002). However it is not well understood how the hypothalamus interacts with higher motor centers controlling feeding, nor other regions that alter feeding after lesions, such as the amygdala (Rollins and King, 2000) or cortical and limbic areas whose function is altered in response to feeding cues as shown in imaging studies of humans (Tataranni et al., 1999).
In this report we sought to delineate the hierarchy of the anatomical connections among diverse regions involved in motor and autonomic elements of feeding, and begin to establish the molecular identity of the neurons that compose these circuits. We identified these neural circuits using the Bartha strain of pseudorabies virus (PRV), a well validated tool for neural tracing that is propagated retrogradely over chains of synaptically connected neurons (Ekstrand et al., 2008; Pomeranz et al., 2005). Light (Card et al., 1990) and transmission electron (Card et al., 1993) microscopy studies show that PRV replicates within synaptically linked populations of neurons and the course of its infection recapitulates connections for circuits that have been characterized using other methods. In these studies it was shown that attenuated PRV “moves through CNS in a circuit-specific fashion” without producing “widespread necrotizing patterns of infection” expected from indiscriminate PRV spreading via extracellular space or cell fusion. In addition, glial PRV infection is consistent with a proposed role of restricting the spreading of virus from extremely infected neurons which further ensures trans-neuronal transfer of PRV virions (Rinaman et al., 1993). Glial function would prevent PRV escaping from neurons through their dendritic arbor, acting as a de facto “PRV sink”.
In this report, we used PRV strains encoding different reporters (i.e. green fluorescence protein, EGFP; red fluorescence protein, mRFP and beta galactosidase, LacZ) to map brain regions that project via descending pathways to innervate the submandibular salivary gland, masseter muscle and the intrinsic muscles of the tongue. We then made use of a series of BAC transgenic mice expressing EGFP, available through the GENSAT project (Gong et al., 2003), to identity markers for subclasses of neurons in these regions. Finally we confirmed whether neurons bearing those markers projected directly to areas belonging to the feeding neural circuit and whether those neurons were sensitive to changes in feeding status, specifically fasting.
These methods represent a general strategy for studying the neuroanatomical and molecular components of complex neural circuits. By identifying markers for neurons in complex circuits, this approach now enables functional analyses of those circuits using recent developments in optogenetics (Miller, 2006) and methods for cell-specific gene expression analysis or neural tracing (Cahoy et al., 2008; Emery and Barres, 2008).
RESULTS
The execution of a complex behavior such as feeding requires the coordinated motor and autonomic activity of several organs and tissues. For example, the secretion of saliva is critical for mastication, bolus formation and swallowing. Saliva secretion is controlled mainly by a reflex arch induced by mechanical and gustatory stimuli that operates via the salivary center in the brainstem (Anderson and Hector, 1987; Pedersen et al., 2002). As first suggested by Pavlov, saliva production is also regulated via inputs from higher order brain centers projecting to the salivary center (Hubschle et al., 1998).
Mastication and swallowing are also critical to ensure proper ingestion of food and are the result of the intricate interaction between a number of orofacial muscles in the jaw, face and tongue (Fay and Norgren, 1997a, b, c). Importantly, mastication and swallowing are also modulated by inputs from higher order cortical centers (Thexton, 1992; Mosier and Bereznaya, 2001).
In this report, we set out to identify higher-order brain regions and neurons that are integrated into circuits regulating end organs which are critical motor and autonomic elements of ingestion: the submandibular salivary gland (SAL), the masseter muscle (MAS) and the intrinsic muscles of the anterior, lateral and posterior regions of the tongue (TA, TL and TP). These anatomic loci were selected both because 1) they are key elements of feeding and 2) they are controlled by distinct efferent cranial nerves whose nuclei are in proximity and at similar coordinates in the brainstem making it possible to trace CNS outputs to these regions simultaneously.
We began these studies by injecting PRV Bartha, a retrograde neuronal tracer, individually into the salivary gland, the tongue and the masseter muscle, as a means for identifying common brain areas that innervate all these tissues via descending neural pathways.
Innervation of Feeding-Related Organs
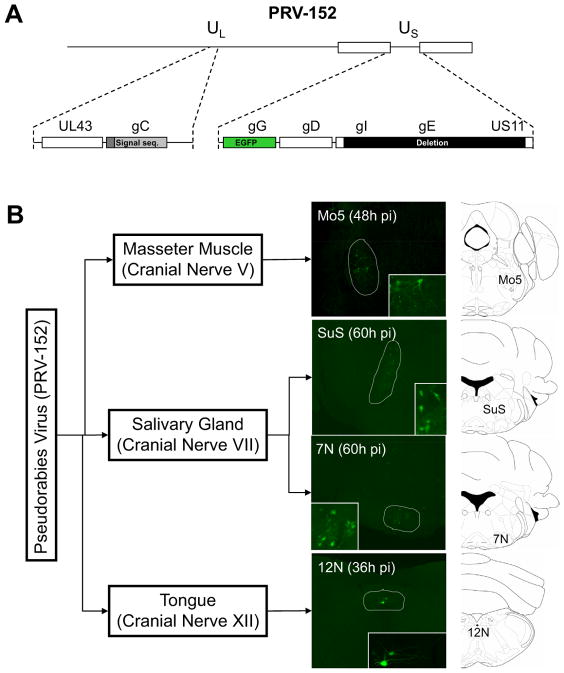
In these initial studies, we used PRV-152 (Fig. 1A), a Bartha PRV strain which expresses EGFP constitutively (Smith et al., 2000). PRV-152 was injected bilaterally into the submandibular salivary gland (SAL), the masseter muscle (MAS), or unilaterally into the midline of the posterior part of the tongue (TP). Mice were sacrificed 36, 48 or 60 hrs post-infection. PRV has a 12–24h cycle during which the virus infects a neuron’s axonal terminals, moves to the soma, replicates and moves to a pre-synaptic target. Thus at these early times only a limited number of neurons is infected. Immunofluorescence for EGFP in the brains of infected animals was assayed to catalogue infected CNS sites after tracing from each of the peripheral sites. In each case, the motor nucleus for the expected cranial nerve was the first region where PRV infection was detected, (Fig. 1B).
Figure 1. Brainstem Cranial Nerve Motor Nuclei are the First CNS Infected Targets Following PRV Injection Into Peripheral Tissues.
A) PRV-152, a Bartha PRV strain genetically modified to express EGFP constitutively (Smith et al., 2000) was used in the present studies. B) PRV-152 was injected bilaterally into the submandibular salivary gland (SAL), the masseter muscle (MAS), or unilaterally into the posterior part of the tongue (TP). 36, 48 or 60 hrs after the injections, EGFP immunofluorescence was analyzed in the brains of infected animals. The motor nucleus for the indicated cranial nerves was the first region where PRV infection was detected, following PRV injection into the indicated tissues. The motor nucleus of the cranial nerve V (Mo5) was the first brainstem area infected after PRV-152 injection into the masseter muscle (MAS), the motor nucleus of the cranial nerve VII (7N) and the superior salivatory nucleus (SuS) were identified after PRV injection into the salivary gland (SAL). Finally, the motor nucleus of the cranial nerve XII (12N) was the only place where PRV was readily observed after PRV injection into the posterior region of the tongue (TP).
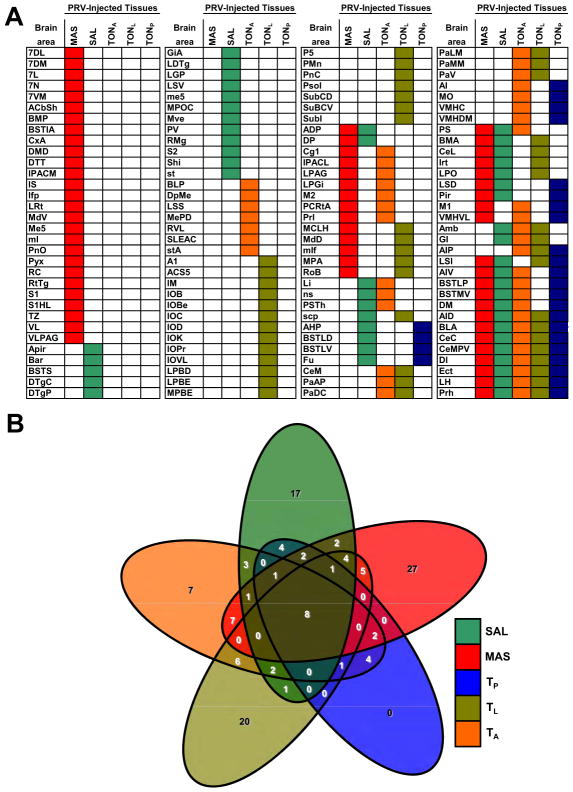
We next set out to identify higher order brain regions infected by PRV at a later time point after PRV injection by compiling a list of forebrain regions infected 96–120 hours after PRV injection in the three peripheral sites (SAL, MAS and tongue ) (Fig. 2A). In this analysis we sought to identify common brain regions projecting (indirectly) to more than one peripheral site. With the goal of improving the ability of our approach to identify a limited number of brain regions projecting to these peripheral tissues, we reasoned that inclusion in our analysis of an additional 4th and 5th peripheral site would allow us to refine the number of brain regions projecting to multiple sites. Since we were limited to sites at a similar anatomic level to the masseter and salivary gland, and because of the complex organization of the intrinsic muscles of the tongue, we decided to add data from animals in which PRV injections into the lateral (TL) and anterior (TA) subdivisions of this organ were performed. This analysis revealed that while many brain areas were infected by PRV 96 hours after viral injection into each individual site (see color-coded columns in Fig. 2A), there were only a small number of brain regions that were common to the descending neural circuits projecting to the salivary gland, the masseter muscle and all three regions of the tongue.
Figure 2. Brain Areas Infected by PRV After Injection Into Multiple Peripheral Tissues.
A) Distribution of forebrain areas infected by PRV-152 96hr after five distinct PRV injections (SAL, MAS, TP, TL, TA) were made into peripheral tissues. This analysis revealed that several brain areas were infected by PRV in each circuit studied (color- coded columns). Very few brain regions were common to the descending neural circuits projecting to all peripheral tissues injected. These regions included the lateral hypothalamus, basolateral and central amygdala and insular, ectorhinal and perirhinal cortices (see multiple color-coded rows in the lower-right corner). B) Symmetrical, non-proportional, 5-way Venn diagram illustrating the number of PRV-infected brain regions after injection in 1 to 5 peripheral sites. It can be appreciated the vast number of subsets (32) to be considered when analyzing multiple circuits and how our proposed approach can identify brain regions belonging to only one such subset. n=3 per injection site.
These regions included the lateral hypothalamus (LH), basolateral and central amygdala (Amy) and insular (Ins), and ectorhinal and perirhinal (Rhi) cortices (see multiple color-coded rows at the bottom right of Fig. 2A). Thus while pair-wise comparisons between masseter, salivary gland and/or specific tongue circuits did show several different subsets of brain regions, only the aforementioned regions were labeled after infection into all of the peripheral sites. These data are quantitatively represented in a symmetrical, non-proportional, 5-way Venn diagram illustrating the number of PRV-infected brain regions after injection into one or more of these peripheral sites (Fig. 2B). This analysis reveals the number of CNS sites with PRV infection after injection into one or more peripheral sites, and shows that only the five sites listed above show GFP immunofluorescence after PRV injection into all of the peripheral sites. It is noteworthy that despite the fact that the approach we employed was unbiased, each of these regions of overlap (LH, Amy, Ins and Rhi cortices) has been previously suggested to play a role to regulate feeding, based on data from lesioning or imaging studies (Rollins and King, 2000; Tataranni and DelParigi, 2003).
We next sought to determine whether individual neurons in these regions were integrated into more than one circuit. This possibility was tested by injecting isogenic viruses with different reporters simultaneously into more than one peripheral site (Fig. S1). The use of isogenic PRV strains allows for the co-infection of a neuron by more than one virus strain so long as the superinfection (infection of a specific neuron by 2 or more different viral strains) of a given neuron takes place within a specific time-window.
PRV-152 and PRV-614 (Banfield et al., 2003; encodes mRFP) were injected simultaneously in pair-wise combinations of SAL, MAS or TP tissues. . Double labeled neurons were evident in the insular cortex, lateral hypothalamus, amygdala and rhinal cortex (see arrowheads in Fig. S1) of all pair-wise tissue combinations: SAL-MAS, SAL-TP and MAS-TP. The distribution of EGFP and mRFP signals 96–120h post injection was analyzed by immunohistochemistry of EGFP (green) and direct mRFP fluorescence (red), respectively. As expected there were also subsets of neurons that showed only EGFP signal or the mRFP signal alone and still other neurons that were unlabeled. We conclude that some neurons in the insular and rhinal cortices, the amygdala and the lateral hypothalamus project to more than one peripheral site, others project to individual peripheral sites and still others to none of these sites. However, because viral infection eventually renders cells resistant to a super-infection after 12–24 hours, both viruses need to infect neurons within a narrow time window for co-infection to be evident. While co-infection can be used to identify neurons projecting to multiple sites, for a variety of reasons, it is not possible to precisely quantify the precise number of double labeled cells, since in some cases a neuron that projects to multiple sites would not be labeled by a second virus if the superinfection fell outside this time window. Indeed, reports indicate that there is not complete overlap of PRV infection even when isogenic PRV virus strains are injected into the same organ (Cano et al., 2004). In addition, there is a strong dose dependence for PRV infection, as the number of neurons that are superinfected depends on the initial effective dose administered at the injection point and on how many virion particles cross trans-synaptically within a given time window (Card et al., 1999). Finally, we had to assess RFP expression by direct fluorescence because of the lack of an antibody to detect mRFP so neurons with low levels of mRFP may go undetected (while they could be detected if an antibody were available). Nonetheless, data from Fig. S1 unequivocally show that a subset of neurons in several different higher brain regions is integrated into circuits projecting to at least two different end organs relevant for feeding.
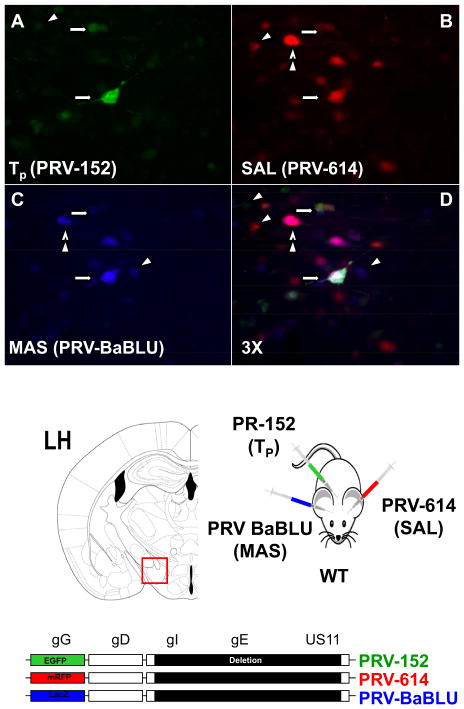
We next set out to determine whether there were individual neurons projecting to all three peripheral tissues, using EGFP and mRFP expressing viruses and also PRV-BaBLU (Standish et al., 1995), another PRV strain isogenic to PRV-152 and PRV-614 (see bottom of Fig. 3). We injected animals with all three viruses (PRV-152 in TP, PRV-614 in SAL and PRV-BaBLU in MAS) and identified triple labeled neurons within the LH (Fig. 3, see arrows), as well as other neurons displaying single (see arrowheads) or double (see double arrowheads) PRV infection. This is a technically difficult study because as mentioned, all three PRV strains need to infect a specific neuron within a narrow time window that allows all three reporters to be expressed at detectable levels. Here again it is likely that we underestimated the actual number of neurons projecting to the three peripheral tissues originally injected. Nonetheless, the fact that specific neurons can be simultaneously infected by different PRV strains injected into three different peripheral sites provides strong evidence that those neurons can project to multiple peripheral sites. Indeed the same line of evidence was used previously to show dual projections of specific neurons to peripheral sites involved in the fight or flight response (Jansen et al, 1995). Moreover, the current observation that some LH neurons are wired into circuits that project to three peripheral sites is consistent with its role as an important element of the neural circuit regulating feeding. An important next step would be to establish the role of specific neurons in LH (and elsewhere) in controlling motor outputs but a critical evaluation of the function of LH neurons and those in other regions, would require identifying neuron-specific promoters to thus making possible subsequent functional studies.
Figure 3. Triple PRV Labeling in the LH Identifies a Neuronal Population Projecting to Masseter Muscle, Salivary Gland and Tongue.
A triple PRV injection paradigm (PRV-152 in TP, PRV-614 in SAL and PRV-BaBLU in MAS) was used to identify triple labeled neurons. PRV-BaBLU, is isogenic to PRV-152 and PRV-614, and encodes LacZ (see scheme at the bottom). Triple labeled neurons were observed (arrows) in the LH, as well as neurons displaying single (arrowheads) or double (double arrowheads) PRV infection.
Toward that end, we used immunohistochemistry in wild type and GFP expressing transgenic mice to identify molecular markers for the neuronal populations in the LH and the other regions that were labeled after injections into the SAL and MAS.
Melanocortin concentrating hormone (MCH) (Saito and Nagasaki, 2008) and orexin (Sakurai, 2003) are neuropeptides that define two important neuronal populations in the LH that regulate feeding. PRV-152 infections into SAL and MAS were repeated in wild type mice and immunohistochemistry against PRV-encoded EGFP and either MCH or orexin was performed. These studies revealed that both MCH+ and orexin+ neurons were infected following PRV-injection into both the salivary gland and masseter muscle (see arrowheads in Fig. S2) suggesting that both neural populations can project via descending pathways to these two peripheral tissues. Because these neurons are known to control feeding, these data validate the approach we developed for identifying cell types that play a role in feeding.
Molecular Markers for Feeding-Related Neurons
Based on the previous result, we reasoned that neurons in other anatomic sites which project to multiple peripheral cell types (Amy, Ins and Rhi) could also play a functional role and thus set out to identify markers for specific neurons in these regions. With such markers, one could use specific promoters to enable conditional PRV tracing (DeFalco et al., 2001; Wintermantel et al., 2006) or to express cassettes that induce neuron-specific ablation (Alon and Friedman, 2006; Hara et al., 2001), or other gain and/or loss of function systems to modulate neural activity, including optogenic approaches (Lerchner et al., 2007; Redfern et al., 1999; Wehr et al., 2009). The difficulty in the case of other brain regions was that in contrast to the LH where defined neural populations have already been identified, there is a paucity of molecular information for neurons in the Amy, Ins and Rhi. We thus set out to develop an approach for identifying such markers in these other less well characterized brain regions.
As an alternative to using immunohistochemistry which was used to detect orexin and MCH, we took an empiric approach by injecting PRV-614 (mRFP) into BAC transgenic mice from the GENSAT project in which EGFP was already known to be expressed in subpopulations of neurons in the amygdala or the cortex (see scheme in Fig. 4). In those cases where both mRFP (from PRV) and EGFP (from the transgenic mouse) were detected in the same neuron, we conclude that the gene whose promoter was directing EGFP expression would provide a marker for neurons integrated into the circuit projecting to the tissue infected by PRV-614. Toward this end, we scanned the GENSAT data base (Gong et al., 2003) to identify BAC transgenic lines that show expression of EGFP in the insular and rhinal cortices and the amygdala and injected PRV-614 into the MAS or SAL of these transgenic mice (Table S1). Because our primary objective was to identify markers, we simply made use of available lines known to express GFP in the regions of interest and did not make distinctions based on the function of the genes driving BAC expression.
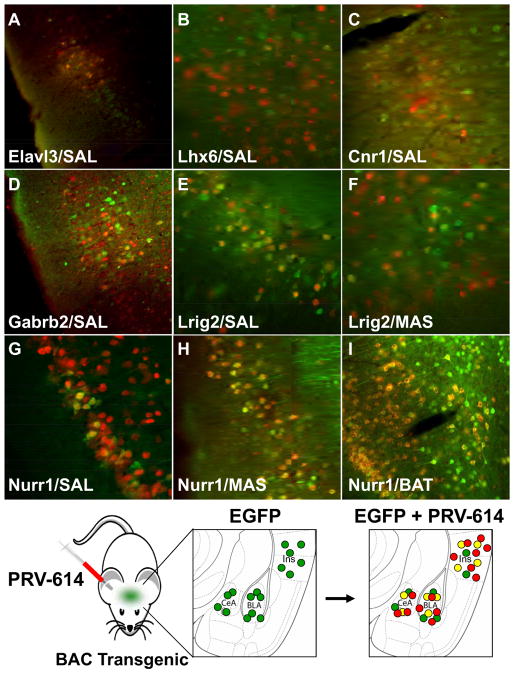
Figure 4. Colocalization of PRV-Encoded mRFP and Transgenic EGFP Reveals Markers for Neurons in Feeding Neuronal Circuits.
Elavl3+/PRV+ neurons (A) were detected in the rhinal cortex following a SAL PRV injection. Lhx6+/PRV+ neurons (B) were detected in the insular cortex after a MAS PRV injection. Cnr1+/PRV+ neurons (C) were detected in the insular cortex following a PRV injection into SAL. ; Gabrb2+/PRV+ neurons (D) were detected in the insular cortex following a PRV injection into SAL. Lrig2+/PRV+ neurons were detected in the insular cortex following PRV injections into SAL (E) and MAS (F). Nurr1+/PRV+ neurons were detected in the insular cortex following SAL (G), MAS (H) and Brown Adipose Tissue (I) PRV injections. (J) Diagram showing our approach to identify markers for neuronal populations from a specific neural circuit (as indicated by their susceptibility to PRV infection). Mice in which EGFP is known to be expressed in a specific brain region are infected in the periphery with PRV-614 (encodes mRFP) and in those cases where both mRFP (from PRV) and EGFP (from the transgenic mouse) are detected in the same neuron, it can be asserted that the gene whose promoter directs EGFP expression is a marker for neurons integrated into the circuit projecting to the tissue infected by PRV-614.
11 out of 17 transgenic mouse lines studied in this manner (~65%) failed to show a “yellow” signal (i.e. red mRFP from the virus plus green EGFP from the transgenic mouse’s neurons) indicating that there was no co-localization of EGFP (potential marker) and mRFP (PRV infection). However in 6 transgenic mouse lines (~35%), “yellow” neurons were readily identifiable (Table S1 and Fig. 4) in amygdala, insula and/or rhinal cortices following PRV infection in MAS and/or SAL. In some cases there was colocalization only after PRV injection into the MAS (Lhx6) or the SAL (Cnr1, Gabrb2, Elav3) and in the case of Nurr1 and Lrig2 colocalization was detected after injection in both tissues. Furthermore, in most cases only one region was positive for the marker (Insular cortex: Lhx6, Lrig2, Gabrb2, Cnr1; Rhinal cortex: Elav3), but in the case of Nurr1 two distinct anatomic regions displayed “yellow” neurons (Insular cortex and Amygdala). The distribution of these markers in PRV-infected neurons in both the insula and rhinal cortices spans cortical layers II through VI with neurons displaying a morphology characteristic of spiny (layer IV) or pyramidal neurons (layers V and VI) (Thomson and Lamy, 2007).
Overall, this analysis identified a set of molecular markers for neurons that project to one or more of the peripheral sites that were studied. These markers include ElavL3 (Fig. 4A), a transcription factor that belongs to Elav family of RNA-binding proteins, which is a highly conserved gene family that plays a crucial role in neurogenesis (Akamatsu et al., 1999). Lhx6 (Fig. 4B), is a LIM homeodomain transcription factor that has been implicated in a pathway mediating innate behaviors involving the amygdala and the hypothalamus (Choi et al., 2005). Cnr1 (Fig. 4C), is a cannabinoid receptor and is known to play a role in appetite regulation and consumption of palatable foods (Kirkham, 2009). It has also been reported that ligand binding to Cnr1 expressed in the insular cortex is enhanced in mice exposed to a high fat diet (South and Huang, 2008). Gabrb2 (Fig. 4D), is a Subunit of the GABA A receptor (Halonen et al., 2009) and Lrig2 (Fig. 4E-F), is a member of the leucine-rich repeats and immunoglobulin-like domains (LRIG) family (Guo et al., 2004). Nurr1 is of particular interest because it was the only instance where we observed marked gene expression in more than one brain region i.e; the amygdala and insula (Table S1, Fig. 4G-H). Nurr1 is a nuclear receptor that has been associated to food aversion in the amygdala (Ge et al., 2003) as well as the development of dopaminergic neurons (Perlmann and Wallen-Mackenzie, 2004). Nurr1 is present throughout the cortex and is found mostly in cortical layer V and VI (Hirokawa et al., 2008; Watakabe et al., 2007) which suggests that Nurr1+ neurons may be elements of cortico-cortical and cortico-thalamic projections (Thomson and Lamy, 2007).
To further identify circuits in which Nurr1+ neurons are integrated, we tested whether Nurr1+ neurons and PRV colocalized after injecting PRV into the tongue (TP) and the brown adipose tissue, an organ involved in energy homeostasis (Schulz, 1987). After PRV injections into the tongue, we observed “yellow” neurons only in the amygdala (data not shown), while after brown adipose tissue PRV injections we observed colocalization on the insular cortex (Fig. 4I). Also, to determine whether Nurr1+ neurons were able to project to two different organs simultaneously, we injected PRV-BaBLU and PRV-614 into the masseter muscle and salivary glands of Nurr1::EGFP transgenic mice (see bottom of Fig. 5) and identified Nurr1+ neurons in the insular cortex and the amygdala that project to both neural circuits (see Fig. 5A-B.) These data show that Nurr1+ neurons in the amygdala and insula are integrated into multiple efferent neural circuits. It is important to note that while Nurr1 and the other markers identified reveal neurons that are “infectable” by PRV, the gene products themselves may or may not play a direct role in modulating the feeding-related properties of those neurons.
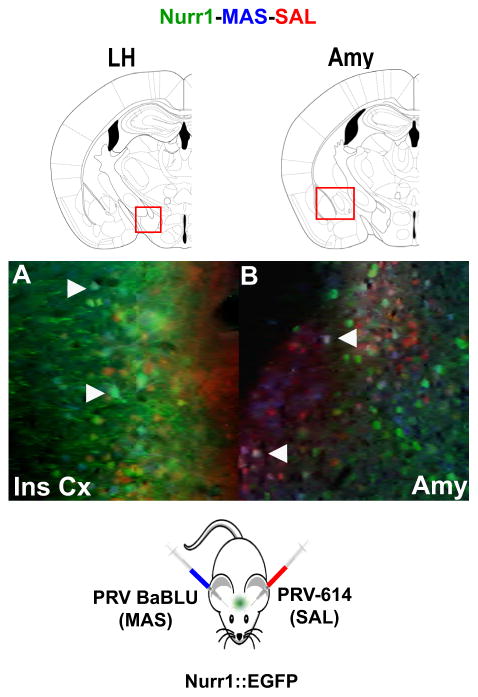
Figure 5. Nurr1 is a Marker of Neurons in Neural Circuits Projecting to Both Salivary Gland and Masseter Muscle.
PRV-BaBLU and PRV-614 were injected in MAS and SAL of Nurr1::EGFP transgenic mice (see scheme at the bottom). Nurr1+ neurons contributing to both neural circuits (positive for both viral reporters) were identified in the insular cortex (A) and the amygdala (B).
To further delineate the connections within the feeding neural circuit elements identified by PRV tracing and confirm the prediction that Cnr1+ and Nurr1+ neurons should project to the LH and/or the amygdala, we injected Cholera Toxin β (ChToxβ), a non-PRV tracer, into the LH and amygdala of Cnr1 and Nurr1 GFP-BAC transgenic mice. ChToxβ is a retrograde tracer that is taken by axonal terminals and then travels to the soma of the respective neurons. Following injections of ChToxβ into the Amy, we were able to detect tracer in Nurr1+ neurons in the insular cortex (Fig. 6A). After ChToxβ injections into the LH, tracer was also found in Cnr1+ neurons in the insula (Fig. 6B ). Further studies combining PRV and ChToxβ tracing will provide additional details about the specific wiring of Nurr1+ and Cnr1+ neurons in feeding neural circuits and possible heterogeneity with respect to the sites of projections of these neurons.
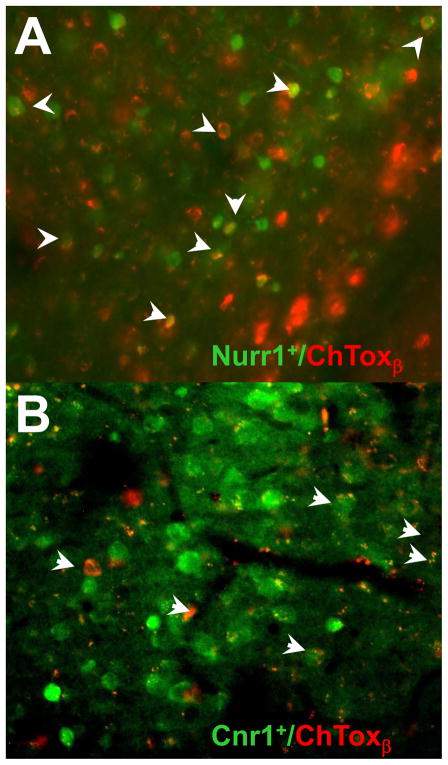
Figure 6. Cholera Toxin β Tracing Confirms Connectivity of Insular Nurr1+ And Cnr1+ Neurons Predicted by PRV Tracing.
Cholera toxin β conjugated to Alexa Fluor 594 was injected into the LH or amygdala of Cnr1::EGFP and Nurr1::EGFP mice (n=3). One week after the stereotaxic injection of the tracer, it was detected in Nurr1+ neurons after injections in the amygdala (A) and also in Cnr1+ neurons following injections into the LH (B).
We next tested whether these neuronal populations are sensitive to nutritional perturbations. First, we determined whether mRNA levels of Cnr1 increased in the insular cortex of mice that had experienced a 36 hour fasting. Taqman assays showed that insular cortex levels of Cnr1 mRNA increased significantly in fasted mice compared with mice that had received ad libitum access to food (Fig. 7A). We next assayed c-Fos expression in Cnr1+ neurons in the insular cortex in control and 36 hour fasted mice. The expression of c-Fos is an indicator of the activation of a given neural population following the onset of a specific stimulus. Immunohistochemical analysis of brain sections from Cnr1::EGFP mice that underwent a 36 hours fasting indicated that the number of double labeled neurons doubles in the posterior insular cortex (statistically significant changes in the posterior agranular insula and visceral insula) (Fig. 7B). In these studies, we analyzed the response to 36 hours of food restriction and can now study c-Fos and insular Cnr1+ gene expression over shorter (and longer) intervals. Nonetheless, these results suggest that Cnr1+ neurons of the insular cortex are sensitive to perturbations in an animal’s nutritional status.
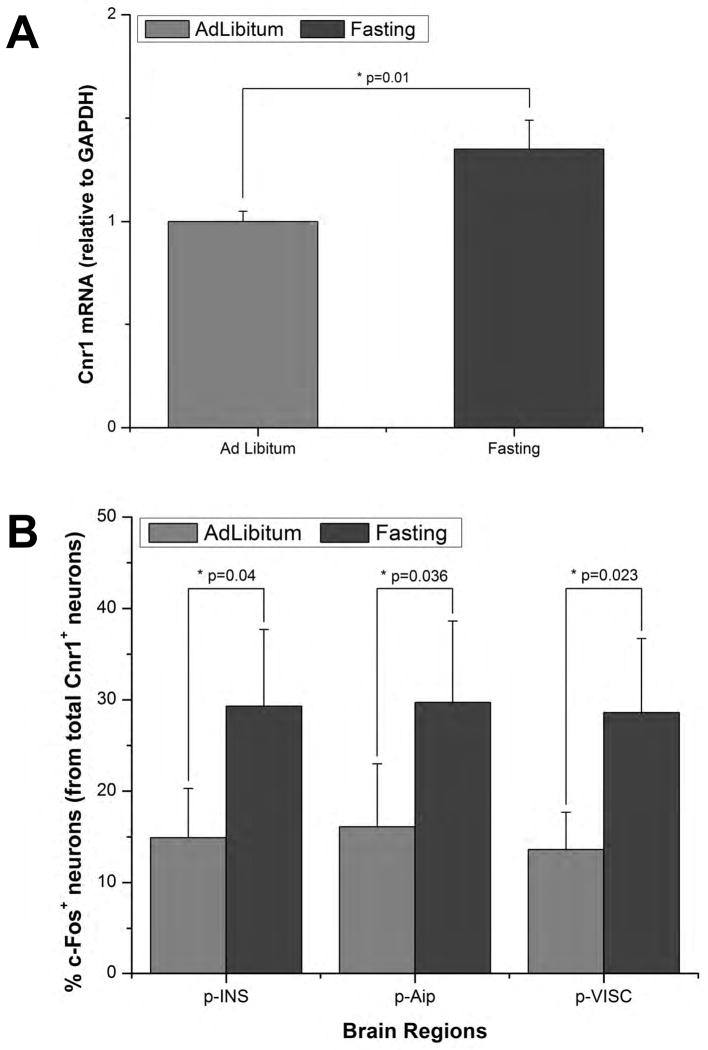
Figure 7. Insular Cnr1+ Neurons are Sensitive to Fasting.
(A) Insular cortex Cnr1 mRNA levels were measured in wild type mice fed ad libitum vs. mice that had fasted for 36 hrs. Using a Taqman assay it was determined that there was a significant increase in Cnr1 mRNA following a 36h fasting period (p=0.01, n=5). (B) We also quantified the number of c-Fos+/Cnr1+ neurons in the insular cortex of Cnr1::EGFP mice in mice with unrestricted access to food vs. mice that had fasted for 36hrs (n=3 per group). It was observed that the number of c-Fos+/Cnr1+ neurons in the posterior insular cortex (p-INS) doubled following fasting (p=0.040); this was also valid for its agranular (p-Aip; p=0.036) and visceral (p-VISC; p=0.023) subdivisions. All data are represented as mean +/- SEM; Student's t test was used for statistical analysis. Significance was assumed for p values < 0.05.
DISCUSSION
In this report, we set out to identify entry points for studying higher order neural circuits for feeding, a complex behavior. We used this approach to identify distinct neural populations expressing Nurr1 and Cnr1 in the amygdala and insular cortex and orexin and MCH neurons in the LH as being integrated into neural circuits that project to multiple peripheral sites that play a motor and/or autonomic role in feeding. We confirmed predictions made from these PRV studies using another neuronal tracer, Cholera Toxin-β. Functional data also suggest that the Cnr1+ neurons in the insula are sensitive to the animal’s nutritional state. The availability of BACs for the markers described in this communication now allows carrying further studies on the effect on feeding of increasing or decreasing the activity of these neurons using channel or halorhodopsin (Gradinaru et al., 2007; Zhang et al., 2007) or BAC-Trap analysis of cell-type-specific gene expression profiles (Doyle et al., 2008; Heiman et al., 2008).
Towards a Molecular Annotation of Neuronal Circuits
The delineation of the cellular elements of complex neural circuits that control behavior is of critical importance for understanding how the brain is organized and how it functions to control diverse behaviors. This level of understanding requires the identification of specific neural populations innervating the peripheral organs necessary for specific behavioral responses. In order to approach this systematically, a means for identifying specific markers for neurons that comprise those circuits is necessary.
Our study establishes PRV tracing combined with GFP expressing transgenic mice in GENSAT as a valid platform to identify unique neuronal populations within specific brain regions innervating multiple organs that potentially play a higher order associative role to control behavior. By applying this approach we were able to identify molecular markers for discrete populations of neurons that project via descending pathways to specific peripheral organs (salivary gland, masseter muscle and tongue) and that could control their coordinated function.
The ability of this unbiased approach to identify key elements of the neural circuit that regulates feeding was validated by the identification of MCH and orexin neurons in the LH, both of which have been previously shown to modulate feeding behavior. Furthermore, ablation studies for those two neuronal classes (Alon and Friedman, 2006; Hara et al., 2001) highlight the usefulness of molecular approaches to establish the role of a neuronal population for a specific behavior. In addition to the LH, imaging studies and lesion experiments have shown that the amygdala, insula, and rhinal cortex may also play a role in the control of feeding (Rollins and King, 2000; Tataranni and DelParigi, 2003; Tataranni et al., 1999). Because the brain areas identified in our studies also correspond to regions classically associated to higher cognitive functions (emotion, interoception and other complex behaviors) it is thus possible that neurons in these brain regions play a key integrative function. The neuroanatomical approach for neuron and marker identification developed in this report now provides multiple molecular entry points to further asses the role of those brain regions and specific neuronal subtypes in complex behaviors, thus providing a framework for future studies aimed at understanding in detail the cellular organization of feeding centers and the molecular repertoire used by their neurons.
The data in this report also indicate that while many brains regions are components of individual neural circuits projecting to single peripheral sites such as the salivary glands or orofacial/buccal muscles, a limited number of brain regions contribute simultaneously to multiple circuits, suggesting that these regions may play an important role in coordinating the function of those organs. The double and triple PRV labeling studies further indicate that specific and discrete neuronal populations project indirectly to multiple sites. The connectivity of these neurons thus suggests that they could function as de facto “command” neurons as suggested previously. (Jansen et al., 1995). Whether these neurons ultimately show the properties and characteristics of command neurons seen in invertebrates is now testable with the identification of molecular markers for these putative command neurons.
The availability of promoters marking these neurons makes now possible gene-profiling studies based on a RNA harvesting approaches (Doyle et al., 2008; Heiman et al., 2008). This should shed light on the molecular organization of feeding neural circuits and will enable us to identify “druggable” targets (Hopkins and Groom, 2002) and functionally validate them. The availability of conditional PRV strains allows the mapping of efferent connections into specific neuronal classes (DeFalco et al., 2001; Wintermantel et al., 2006) and the transgenic expression of constructs using varied gain and/or loss of function strategies (Lerchner et al., 2007; Redfern et al., 1999; Wehr et al., 2009) should provide a good starting point in deciphering the contribution of specific neuronal classes to a particular behavior.
In summary, the delineation of circuits involved in the generation, modulation and/or cessation of complex behaviors will be essential for our understanding of brain function. The experimental approach outlined in this report provides a platform for identifying and testing the functional role of neurons in brain regions that project to specific peripheral sites that are critical for feeding and potentially other complex, motivational behaviors.
EXPERIMENTAL PROCEDURES
Animals
Wild type FVB/NJ mice were obtained from Jackson Laboratories (Ban-Harbor, ME). Transgenic strains used were available from the GENSAT project (Gong et al., 2003). Animal care and experimental procedures were performed with approval from the Animal Care and Use Committee of Rockefeller University under established guidelines.
Pseudorabies Virus
Stocks of PRV152 and PRV-BaBlu (generous gifts of Prof L Enquist, Dept. of Molecular Biology, Princeton University) and PRV-614 (a generous gift of Prof Banfield, Dept. of Microbiology and Immunology, Queen’s University, Ontario, Canada) were prepared as previously described (DeFalco et al., 2001) to concentrations of 3–9×108 pfu/ml.
Mice were anaesthetized with ketamine (60 mg/kg, i.p., Fort Dodge Animal Health, Fort Dodge, IA) and xylazine (7 mg/kg, i.p.); all chemicals were obtained from Sigma-Aldrich, St. Louis, MO, unless otherwise noted. An injection of PRV-152, PRV-614 or PRV-BaBlu was performed over 30 seconds via a Hamilton syringe fitted with a 30G1/2 needle. The quantity of virus injected was based on pilot studies that indicated that 5–10 ul of the original PRV stocks would allow a robust visualization of the neuronal circuits innervating each organ. Topical application to control for virus leakage showed no evidence of CNS virus infection.
Dual injections were performed using PRV-152 and PRV-614 and triple injections included the use of PRV-BaBlu.
Immunofluorescence
Immunofluorescence and double labeling were performed as previously described (DeFalco et al., 2001). Briefly, anesthetized animals were perfused transcardially with saline solution (0.9% NaCl) followed by 4% paraformaldehyde (PFA) in PBS. Brains were post-fixed overnight and serial 50 μm coronal sections were cut on a vibratome (Leica, Bannockburn, IL). Free-floating brain sections were washed then incubated in blocking solution (0.1% Triton X-100, 3% bovine serum albumin, 2% goat serum in PBS) for 2 hrs at RT, followed by an overnight incubation with primary antibody at 4°C. Sections were washed and incubated with the secondary antibody at RT for 1 hr followed by a final wash. Sections were mounted with Fluoromount (Southernbiotech, Birmingham, AL). Primary antibodies and their final concentrations were as follows: rabbit anti-GFP antibody (1:1000, Molecular Probes, Invitrogen, Carlsbad, CA), chicken anti-GFP antibody (1:1000, Abcam, Cambridge, MA), rabbit anti-PRV antibody (1:250, Affinity Bioreagents, Rockford, IL), rabbit anti-β Gal antibody (1:500, MP Biomedicals, Solon, OH), rabbit anti-MCH (1:500, Phoenix Pharmaceuticals, Burlingame, CA) and rabbit anti-Orexin (1:500, Abcam). Secondary antibodies were as follows: goat anti-rabbit Alexa 488, goat anti-rabbit Alexa 594, goat anti-rabbit Alexa 360, goat anti-chicken Alexa 488 (1:1000, Molecular Probes). Sections were visualized with a Zeiss Axioplane microscope (Zeiss, Peabody, MA). Anatomical analysis and nomenclature were based on a mouse brain atlas (Paxinos and Franklin, 2001).
Cholera Toxin β Tracing
Mice were placed in a Kopf stereotaxic frame and 200nl of ChToxβ conjugated to Alexa Fluor 594 (Invitrogen) were injected into the LH or amygdala of Cnr1::EGFP mice or Nurr1::EGFP mice, following coordinates determined from the mouse brain atlas (Paxinos and Franklin, 2001). One week post ChToxβ injection mice were perfused with 4% PFA and brains were processed for immunohistochemistry as described above. GFP was detected with a chicken antibody anti-GFP and ChToxβ was detected by determining the intrinsic fluorescence of the Alexa Fluor 594 conjugate.
Quantitative Real Time PCR
Male FvBN mice were euthanized by decapitation. Brains were carefully removed and total RNA was isolated from the insular cortex following dissection based on information from the mouse brain atlas (Paxinos and Franklin, 2001), using RNeasy (Qiagen, Valencia, CA). RNA concentrations were determined using a nanodrop (Thermo Scientific, Wilmington, DE) and RNA quality verified with Agilent Bioanalyzer (Agilent Technologies, Santa Clara, CA). Reverse transcription was performed on 1 μg of RNA using Superscript II reverse transcriptase (Invitrogen).
The relative abundance of Cnr1 transcripts in the insular cortex of mice subjected to a normal diet (ad libitum) or 36h fasting was assessed by TaqMan qRT-PCR. TaqMan gene expression assays were obtained from Applied Biosystems (Foster City, CA, USA). qRT-PCR reactions were performed in triplicates using TaqMan Universal PCR Master Mix (Applied Biosystems, Carlsbad, CA). Measurements were performed on three independent RNA preparations from the ad libitum or fasted groups. Data were normalized based on the control gene glyceraldehyde-3-phosphate dehydrogenase. Assays ID are Cnr1: Mm00432621_s1 and GAPD: Mm99999915_g1
c-Fos Studies
Colocalization of c-Fos and GFP in Cnr1::EGFP mice was determined by double immunohistochemistry as described above, using the chicken anti-GFP antibody and a rabbit polyclonal antibody anti-c-Fos (Santa Cruz Biotechnology, Santa Cruz, CA). The secondary antibody for c-Fos detection was a goat anti-rabbit antibody conjugated to Alexa Fluor 555. Double labeled neurons were identified using an Upright LSM 510 laser scanning confocal microscope Zeiss available at the Rockefeller University Bio-Imaging Resource Center. Two groups of mice (n=3) were used, one with unrestricted access to food and other that was fasted for 36 hrs.
Supplementary Material
Acknowledgments
We would like to thank Prof. Allyn Mark for his helpful comments on the manuscript, R. Bellani and X. Lapointe-Gagner for technical assistance, and S. Korres for administrative support. This work was supported by NIH RO1 DA018799–03 (JMF), a Sjögren’s Syndrome Foundation Research Fellowship (CAP) and a Clinician Scientist Fellowship from the Medical Research Council (SAS). JFM is a Howard Hughes Medical Institute Investigator.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Akamatsu W, Okano HJ, Osumi N, Inoue T, Nakamura S, Sakakibara S, Miura M, Matsuo N, Darnell RB, Okano H. Mammalian ELAV-like neuronal RNA-binding proteins HuB and HuC promote neuronal development in both the central and the peripheral nervous systems. Proc Natl Acad Sci USA. 1999;96:9885–9890. doi: 10.1073/pnas.96.17.9885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alon T, Friedman JM. Late-onset leanness in mice with targeted ablation of melanin concentrating hormone neurons. J Neurosci. 2006;26:389–397. doi: 10.1523/JNEUROSCI.1203-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson DJ, Hector MP. Periodontal mechanoreceptors and parotid secretion in animals and man. J Dent Res. 1987;66:518–523. doi: 10.1177/00220345870660022201. [DOI] [PubMed] [Google Scholar]
- Banfield BW, Kaufman JD, Randall JA, Pickard GE. Development of pseudorabies virus strains expressing red fluorescent proteins: new tools for multisynaptic labeling applications. J Virol. 2003;77:10106–10112. doi: 10.1128/JVI.77.18.10106-10112.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cahoy JD, Emery B, Kaushal A, Foo LC, Zamanian JL, Christopherson KS, Xing Y, Lubischer JL, Krieg PA, Krupenko SA, et al. A transcriptome database for astrocytes, neurons, and oligodendrocytes: a new resource for understanding brain development and function. J Neurosci. 2008;28:264–278. doi: 10.1523/JNEUROSCI.4178-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cano G, Card JP, Sved AF. Dual viral transneuronal tracing of central autonomic circuits involved in the innervation of the two kidneys in rat. J Comp Neurol. 2004;471:462–481. doi: 10.1002/cne.20040. [DOI] [PubMed] [Google Scholar]
- Card JP, Enquist LW, Moore RY. Neuroinvasiveness of pseudorabies virus injected intracerebrally is dependent on viral concentration and terminal field density. J Comp Neurol. 1999;407:438–452. doi: 10.1002/(sici)1096-9861(19990510)407:3<438::aid-cne11>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- Card JP, Rinaman L, Lynn RB, Lee BH, Meade RP, Miselis RR, Enquist LW. Pseudorabies virus infection of the rat central nervous system: ultrastructural characterization of viral replication, transport, and pathogenesis. J Neurosci. 1993;13:2515–2539. doi: 10.1523/JNEUROSCI.13-06-02515.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Card JP, Rinaman L, Schwaber JS, Miselis RR, Whealy ME, Robbins AK, Enquist LW. Neurotropic properties of pseudorabies virus: uptake and transneuronal passage in the rat central nervous system. J Neurosci. 1990;10:1974–1994. doi: 10.1523/JNEUROSCI.10-06-01974.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi GB, Dong HW, Murphy AJ, Valenzuela DM, Yancopoulos GD, Swanson LW, Anderson DJ. Lhx6 delineates a pathway mediating innate reproductive behaviors from the amygdala to the hypothalamus. Neuron. 2005;46:647–660. doi: 10.1016/j.neuron.2005.04.011. [DOI] [PubMed] [Google Scholar]
- DeFalco J, Tomishima M, Liu H, Zhao C, Cai X, Marth JD, Enquist L, Friedman JM. Virus-assisted mapping of neural inputs to a feeding center in the hypothalamus. Science. 2001;291:2608–2613. doi: 10.1126/science.1056602. [DOI] [PubMed] [Google Scholar]
- Doyle JP, Dougherty JD, Heiman M, Schmidt EF, Stevens TR, Ma G, Bupp S, Shrestha P, Shah RD, Doughty ML, et al. Application of a translational profiling approach for the comparative analysis of CNS cell types. Cell. 2008;135:749–762. doi: 10.1016/j.cell.2008.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekstrand MI, Enquist LW, Pomeranz LE. The alpha-herpesviruses: molecular pathfinders in nervous system circuits. Trends Mol Med. 2008;14:134–140. doi: 10.1016/j.molmed.2007.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emery B, Barres BA. Unlocking CNS cell type heterogeneity. Cell. 2008;135:596–598. doi: 10.1016/j.cell.2008.10.031. [DOI] [PubMed] [Google Scholar]
- Fay RA, Norgren R. Identification of rat brainstem multisynaptic connections to the oral motor nuclei in the rat using pseudorabies virus. II Facial muscle motor systems. Brain Res Rev. 1997a;25:276–290. doi: 10.1016/s0165-0173(97)00027-1. [DOI] [PubMed] [Google Scholar]
- Fay RA, Norgren R. Identification of rat brainstem multisynaptic connections to the oral motor nuclei using pseudorabies virus. I Masticatory muscle motor systems. Brain Res Rev. 1997b;25:255–275. doi: 10.1016/s0165-0173(97)00026-x. [DOI] [PubMed] [Google Scholar]
- Fay RA, Norgren R. Identification of rat brainstem multisynaptic connections to the oral motor nuclei using pseudorabies virus. III Lingual muscle motor systems. Brain Res Rev. 1997c;25:291–311. doi: 10.1016/s0165-0173(97)00028-3. [DOI] [PubMed] [Google Scholar]
- Friedman JM, Halaas JL. Leptin and the regulation of body weight in mammals. Nature. 1998;395:763–770. doi: 10.1038/27376. [DOI] [PubMed] [Google Scholar]
- Ge H, Chiesa R, Pena de Ortiz S. Hzf-3 expression in the amygdala after establishment of conditioned taste aversion. Neuroscience. 2003;120:1–4. doi: 10.1016/s0306-4522(03)00265-3. [DOI] [PubMed] [Google Scholar]
- Geiselman PJ. Control of food intake. A physiologically complex, motivated behavioral system. Endocrinol Metab Clin North Am. 1996;25:815–829. doi: 10.1016/s0889-8529(05)70356-x. [DOI] [PubMed] [Google Scholar]
- Gong S, Zheng C, Doughty ML, Losos K, Didkovsky N, Schambra UB, Nowak NJ, Joyner A, Leblanc G, Hatten ME, et al. A gene expression atlas of the central nervous system based on bacterial artificial chromosomes. Nature. 2003;425:917–925. doi: 10.1038/nature02033. [DOI] [PubMed] [Google Scholar]
- Gradinaru V, Thompson KR, Zhang F, Mogri M, Kay K, Schneider MB, Deisseroth K. Targeting and readout strategies for fast optical neural control in vitro and in vivo. J Neurosci. 2007;27:14231–14238. doi: 10.1523/JNEUROSCI.3578-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo D, Holmlund C, Henriksson R, Hedman H. The LRIG gene family has three vertebrate paralogs widely expressed in human and mouse tissues and a homolog in Ascidiacea. Genomics. 2004;84:157–165. doi: 10.1016/j.ygeno.2004.01.013. [DOI] [PubMed] [Google Scholar]
- Halonen LM, Sinkkonen ST, Chandra D, Homanics GE, Korpi ER. Brain regional distribution of GABA(A) receptors exhibiting atypical GABA agonism: roles of receptor subunits. Neurochem Int. 2009;55:389–396. doi: 10.1016/j.neuint.2009.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara J, Beuckmann CT, Nambu T, Willie JT, Chemelli RM, Sinton CM, Sugiyama F, Yagami K, Goto K, Yanagisawa M, et al. Genetic ablation of orexin neurons in mice results in narcolepsy, hypophagia, and obesity. Neuron. 2001;30:345–354. doi: 10.1016/s0896-6273(01)00293-8. [DOI] [PubMed] [Google Scholar]
- Heiman M, Schaefer A, Gong S, Peterson JD, Day M, Ramsey KE, Suarez-Farinas M, Schwarz C, Stephan DA, Surmeier DJ, et al. A translational profiling approach for the molecular characterization of CNS cell types. Cell. 2008;135:738–748. doi: 10.1016/j.cell.2008.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopkins AL, Groom CR. The druggable genome. Nat Rev Drug Discov. 2002;1:727–730. doi: 10.1038/nrd892. [DOI] [PubMed] [Google Scholar]
- Hubschle T, McKinley MJ, Oldfield BJ. Efferent connections of the lamina terminalis, the preoptic area and the insular cortex to submandibular and sublingual gland of the rat traced with pseudorabies virus. Brain Res. 1998;806:219–231. doi: 10.1016/s0006-8993(98)00765-3. [DOI] [PubMed] [Google Scholar]
- Jansen AS, Nguyen XV, Karpitskiy V, Mettenleiter TC, Loewy AD. Central command neurons of the sympathetic nervous system: basis of the fight-or-flight response. Science. 1995;270:644–646. doi: 10.1126/science.270.5236.644. [DOI] [PubMed] [Google Scholar]
- King BM. The rise, fall, and resurrection of the ventromedial hypothalamus in the regulation of feeding behavior and body weight. Physiol Behav. 2006;87:221–244. doi: 10.1016/j.physbeh.2005.10.007. [DOI] [PubMed] [Google Scholar]
- Kirkham TC. Cannabinoids and appetite: food craving and food pleasure. Int Rev Psychiatry. 2009;21:163–171. doi: 10.1080/09540260902782810. [DOI] [PubMed] [Google Scholar]
- Lerchner W, Xiao C, Nashmi R, Slimko EM, van Trigt L, Lester HA, Anderson DJ. Reversible silencing of neuronal excitability in behaving mice by a genetically targeted, ivermectin-gated Cl- channel. Neuron. 2007;54:35–49. doi: 10.1016/j.neuron.2007.02.030. [DOI] [PubMed] [Google Scholar]
- Miller G. Optogenetics. Shining new light on neural circuits. Science. 2006;314:1674–1676. doi: 10.1126/science.314.5806.1674. [DOI] [PubMed] [Google Scholar]
- Mosier K, Bereznaya I. Parallel cortical networks for volitional control of swallowing in humans. Exp Brain Res. 2001;140:280–289. doi: 10.1007/s002210100813. [DOI] [PubMed] [Google Scholar]
- Paxinos G, Franklin KBJ. The Mouse Brain in Stereotaxic Coordinates. 2. San Diego: Academic Press; 2001. [Google Scholar]
- Pedersen AM, Bardow A, Jensen SB, Nauntofte B. Saliva and gastrointestinal functions of taste, mastication, swallowing and digestion. Oral Dis. 2002;8:117–129. doi: 10.1034/j.1601-0825.2002.02851.x. [DOI] [PubMed] [Google Scholar]
- Perlmann T, Wallen-Mackenzie A. Nurr1, an orphan nuclear receptor with essential functions in developing dopamine cells. Cell Tissue Res. 2004;318:45–52. doi: 10.1007/s00441-004-0974-7. [DOI] [PubMed] [Google Scholar]
- Pomeranz LE, Reynolds AE, Hengartner CJ. Molecular biology of pseudorabies virus: impact on neurovirology and veterinary medicine. Microbiol Mol Biol Rev. 2005;69:462–500. doi: 10.1128/MMBR.69.3.462-500.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redfern CH, Coward P, Degtyarev MY, Lee EK, Kwa AT, Hennighausen L, Bujard H, Fishman GI, Conklin BR. Conditional expression and signaling of a specifically designed Gi-coupled receptor in transgenic mice. Nat Biotechnol. 1999;17:165–169. doi: 10.1038/6165. [DOI] [PubMed] [Google Scholar]
- Rinaman L, Card JP, Enquist LW. Spatiotemporal responses of astrocytes, ramified microglia, and brain macrophages to central neuronal infection with pseudorabies virus. J Neurosci. 1993;13:685–702. doi: 10.1523/JNEUROSCI.13-02-00685.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rollins BL, King BM. Amygdala-lesion obesity: what is the role of the various amygdaloid nuclei? Am J Physiol Regul Integr Comp Physiol. 2000;279:R1348–1356. doi: 10.1152/ajpregu.2000.279.4.R1348. [DOI] [PubMed] [Google Scholar]
- Saito Y, Nagasaki H. The melanin-concentrating hormone system and its physiological functions. Results Probl Cell Differ. 2008;46:159–179. doi: 10.1007/400_2007_052. [DOI] [PubMed] [Google Scholar]
- Sakurai T. Orexin: a link between energy homeostasis and adaptive behaviour. Curr Opin Clin Nutr Metab Care. 2003;6:353–360. doi: 10.1097/01.mco.0000078995.96795.91. [DOI] [PubMed] [Google Scholar]
- Schulz LO. Brown adipose tissue: regulation of thermogenesis and implications for obesity. J Am Diet Assoc. 1987;87:761–764. [PubMed] [Google Scholar]
- Schwartz MW, Woods SC, Porte D, Jr, Seeley RJ, Baskin DG. Central nervous system control of food intake. Nature. 2000;404:661–671. doi: 10.1038/35007534. [DOI] [PubMed] [Google Scholar]
- Smith BN, Banfield BW, Smeraski CA, Wilcox CL, Dudek FE, Enquist LW, Pickard GE. Pseudorabies virus expressing enhanced green fluorescent protein: A tool for in vitro electrophysiological analysis of transsynaptically labeled neurons in identified central nervous system circuits. Proc Natl Aca Sci USA. 2000;97:9264–9269. doi: 10.1073/pnas.97.16.9264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- South T, Huang XF. Temporal and site-specific brain alterations in CB1 receptor binding in high fat diet-induced obesity in C57Bl/6 mice. J Neuroendocrinol. 2008;20:1288–1294. doi: 10.1111/j.1365-2826.2008.01785.x. [DOI] [PubMed] [Google Scholar]
- Standish A, Enquist LW, Miselis RR, Schwaber JS. Dendritic morphology of cardiac related medullary neurons defined by circuit-specific infection by a recombinant pseudorabies virus expressing beta-galactosidase. J Neurovirol. 1995;1:359–368. doi: 10.3109/13550289509111025. [DOI] [PubMed] [Google Scholar]
- Tataranni PA, DelParigi A. Functional neuroimaging: a new generation of human brain studies in obesity research. Obes Rev. 2003;4:229–238. doi: 10.1046/j.1467-789x.2003.00111.x. [DOI] [PubMed] [Google Scholar]
- Tataranni PA, Gautier JF, Chen K, Uecker A, Bandy D, Salbe AD, Pratley RE, Lawson M, Reiman EM, Ravussin E. Neuroanatomical correlates of hunger and satiation in humans using positron emission tomography. Proc Natl Acad Sci USA. 1999;96:4569–4574. doi: 10.1073/pnas.96.8.4569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thexton AJ. Mastication and swallowing: an overview. Br Dent J. 1992;173:197–206. doi: 10.1038/sj.bdj.4808002. [DOI] [PubMed] [Google Scholar]
- Thomson AM, Lamy C. Functional maps of neocortical local circuitry. Front Neurosci. 2007;1:19–42. doi: 10.3389/neuro.01.1.1.002.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vettor R, Fabris R, Pagano C, Federspil G. Neuroendocrine regulation of eating behavior. J Endocrinol Invest. 2002;25:836–854. doi: 10.1007/BF03344047. [DOI] [PubMed] [Google Scholar]
- Wehr M, Hostick U, Kyweriga M, Tan A, Weible AP, Wu H, Wu W, Callaway EM, Kentros C. Transgenic silencing of neurons in the mammalian brain by expression of the allatostatin receptor (AlstR) J Neurophysiol. 2009;102:2554–2562. doi: 10.1152/jn.00480.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigle DS. Appetite and the regulation of body composition. FASEB J. 1994;8:302–310. doi: 10.1096/fasebj.8.3.8143936. [DOI] [PubMed] [Google Scholar]
- Williams G, Bing C, Cai XJ, Arrold JA, King PJ, Liu XH. The hypothalamus and the control of energy homeostasis: different circuits, different purposes. Physiol Behav. 2001;74:683–701. doi: 10.1016/s0031-9384(01)00612-6. [DOI] [PubMed] [Google Scholar]
- Wilson BE, Meyer GE, Cleveland JC, Jr, Weigle DS. Identification of candidate genes for a factHor regulating body weight in primates. Am J Physiol. 1990;259:R1148–1155. doi: 10.1152/ajpregu.1990.259.6.R1148. [DOI] [PubMed] [Google Scholar]
- Wintermantel TM, Campbell RE, Porteous R, Bock D, Grone HJ, Todman MG, Korach KS, Greiner E, Perez CA, Schutz G, et al. Definition of estrogen receptor pathway critical for estrogen positive feedback to gonadotropin-releasing hormone neurons and fertility. Neuron. 2006;52:271–280. doi: 10.1016/j.neuron.2006.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Aravanis AM, Adamantidis A, de Lecea L, Deisseroth K. Circuit-breakers: optical technologies for probing neural signals and systems. Nat Rev Neurosci. 2007;8:577–581. doi: 10.1038/nrn2192. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.