Abstract
Tendon injuries are common clinical problems and are difficult to treat. In particular, the tendon-to-bone insertion site, once damaged, does not regenerate its complex zonal arrangement. A potential treatment for tendon injuries is to replace injured tendons with bioengineered tendons. However, the bioengineering of tendon will require a detailed understanding of the normal development of tendon, which is currently lacking. Here, we use the mouse patellar tendon as a model to describe the spatial and temporal pattern of expression of molecular markers for tendon differentiation from late fetal life to 2 weeks after birth. We found that collagen I, fibromodulin, and tenomodulin were expressed throughout the tendon, whereas tenascin-C, biglycan, and cartilage oligomeric protein were concentrated in the insertion site during this period. We also identified signaling pathways that are activated both throughout the developing tendon, for example, transforming growth factor beta and bone morphogenetic protein, and specifically in the insertion site, for example, hedgehog pathway. Using a mouse line expressing green fluorescent protein in all tenocytes, we also found that tenocyte cell proliferation occurs at highest levels during late fetal life, and declines to very low levels by 2 weeks after birth. These data will allow both the functional analysis of specific signaling pathways in tenocyte development and their application to tissue-engineering studies in vitro.
Introduction
Tendons are a unique component of the musculoskeletal system. They contain differentiated cells, the tenocytes, and a high content of dense, highly polarized extracellular matrix material. Tendons connect and transfer force between the muscles and bones of the body. Tendon injuries are common, and hard to repair.1–4 Treatments such as steroids reduce the pain and inflammation of tendon injury, but do not repair the tendon.5 Surgical repair often leads to chronic pain,6,7 and tendon grafts often lead to donor site morbidity. Recently, attention has focused on the use of tissue-engineering approaches to generate replacement tendon tissue that will improve healing.8–10 However, the generation of correctly differentiated tendon tissue in culture from stem cell populations will require a detailed understanding of the normal gene expression pattern of differentiating tenocytes, and how this is controlled by intercellular signaling during normal tendon development in vivo. Molecular markers of tendon differentiation are needed to assay the success of tissue-engineering approaches, and the normal spatial and temporal pattern of their appearance. This information is lacking for most tendons in the body.
Many cell signaling pathways, such as fibroblast growth factor (FGF), bone morphogenetic protein (BMP), and transforming growth factor beta (TGFβ) signaling, have been shown to be involved in the development of tendons. FGF signaling is required for the specification of axial tendons11–13 as defined by the presence of Scleraxis (SCX)-expressing cells.14 In the limb tendons, the surface ectoderm is required for the appearance of SCX-expressing cells.14,15 It also has been shown that BMP signaling inhibits the formation of SCX-expressing cells, while at the same time stimulating the formation of cartilage. In addition, TGFβ signaling is essential for maintenance and recruitment of tendon progenitors cells during embryonic stages.16
Many details of these processes are not clear. For example, the signals that initiate the formation of limb tendons are unknown. Once specified to form tenocytes, as defined by the expression of SCX, it is not known what precise temporal and spatial pattern of signals controls tenocyte proliferation and differentiation. In fact, the precise temporal and spatial pattern of appearance of tenocyte markers, and the degree to which marker expression differs in the different specialized regions of the tendon (e.g., mid-substance vs. insertion site), have not been documented for most tendons. To create tendons from stem cells in culture, which is one major goal of tissue engineering, it will be necessary to understand the spatial and temporal pattern of tenocyte differentiation in vivo and the signaling pathways that control this process.
Although other animal models, such as rabbits and rats, have been used to study tendon repair, these are not genetic models and lack the reporter lines and targeted mutagenesis lines that are readily available in the mouse. However, only limited descriptions of tendon morphogenesis have been published in the mouse.14,17–19 Therefore, to take advantage of genetic manipulations possible in the mouse, we have set out to develop the murine patellar tendon as a model. The goal of the present study is to establish a spatial and temporal map of the appearance of some major tenocyte markers in the patellar tendon in vivo, and to map the changes in activity of major signaling pathways that accompany these changes in gene expression. This will allow both the mechanistic analysis of normal tendon development and the application of this knowledge to the generation of tendon tissue in vitro.
Materials and Methods
Animals
Knee joints from CD-1® (Charles River), Gli1-LacZ (The Jackson Lab Laboratory, Strain: Gli1tm2Alj), and scleraxis-green fluorescent protein (ScxGFP) mice20 were collected at four specific stages of patellar tendon development; embryonic day seventeen and half (E17.5), and postnatal days 1, 7, and 14 (P1, P7, and P14). Three to five CD-1 and ScxGFP samples from each time point were used for histological and immunostaining studies, and three Gli1-LacZ mice from each time point were used for X-gal staining. Detailed experimental procedures are described in the specific sections next. All procedures described were reviewed and approved by the Institutional Animal Care and Use Committee at CCHMC and were performed in accordance with the Guiding Principles for the Care and Use of Laboratory Animals.
Histological analysis
Knee joints were freshly embedded in OCT (Tissue-Tek) and then frozen immediately in liquid nitrogen. The embedded tissues were cryostat sectioned at 10 μm thickness, then mounted onto slides. Slides were fixed in 2% paraformaldehyde (PFA), washed in phosphate-buffered saline (PBS), and then stained using hematoxylin and eosin (H&E) according to standard procedures.21 Slides were dehydrated and then mounted using Mounting Medium Xylene (Fishier Scientific).
Immunohistochemistry
Knee joints were isolated and embedded in OCT and then frozen immediately in liquid nitrogen. For ScxGFP samples, tissues were fixed in 4% PFA for 2–6 h and then washed with PBS. The samples were cryostat sectioned at 10 μm thickness. For unfixed samples, sections were fixed with 2%–4% PFA for 10 min or with 2% Trichloroacetic acid for 6 min and washed in PBS and permeabilized with PBST (0.1% Trion X-100 in PBS). Slides were then incubated in blocking solution (10% donkey or goat serum, 0.1% Triton X-100, and 4% bovine serum albumin in PBS) for 1 h at room temperature, then incubated in primary antibodies (see below) at 4°C overnight. The next day, sections were washed thrice for 10 min each with PBST and PBS followed by incubation in the corresponding secondary antibodies. After secondary antibody treatment, sections were washed with PBST and PBS thrice for 10 min, counterstained with 4′,6-diamidino-2-phenylindole, and mounted in mounting medium. For color development, sections were treated with diamino benzidine (DAB) substrate using Metal Enhanced DAB Substrate Kit (Thermo Scientific, Cat No.34065). After washing with PBS, slides were counterstained with nuclear fast red (VECTASHIELD®), dehydrated through a graded series of ethanols, and mounted in Mounting Medium Xylene (Fishier Scientific).
The dilutions and sources of primary antibodies used were as follows: anti-collagen I (1:200; Novus Biological Research), anti-Tenomodulin (TNMD) (1:50; Santa Cruz), anti-Bigylcan (1:100; AbCam), anti-cartilage oligomeric protein (COMP) (1:50; AbCam), anti-p-FGFR1 (1:50, antibodies onine.com), anti- Tenascin C (1:1000; AbCam), anti-fibromodulin (FMOD) (1:200; AbCam), anti-Ki67 (1:500; Abcam), and anti-p-Smad1/5/8 (1:100; Cell Signaling). All the secondary antibodies were purchased from Jackson ImmunoResearch Laboratories, Inc. and used according to the manufacturer's instructions.
Image capture and semi-quantitative analysis of immunostaining
A ZEISS AxioPlan 2 microscope (Carl Zeiss MicroImaging LLC) was used for imaging. Images were acquired at 20× using the same exposure settings for all samples within each experimental group. The mean intensity of fluorescence was measured using the outline tools and measurement in the AxioVision Rel 4.8.2.0 software (Carl Zeiss MicroImaging LLC). A total of 12 slides from 3 animals at 4 stages were analyzed per immunostaining within each experiment group. Data were analyzed using one-way analysis of variance (ANOVA) followed by an LSD test for pair-wise comparisons using SPSS (IBM, SPSS Statistics 19).
X-Gal staining
Sections were fixed with 2% PFA for 2 min at 4°C, washed in PBS, X-Gal washing buffer (2 mM MgCl2, 0.02% NP-40 in PBS), and placed in X-gal reaction substrate containing 2 mM MgCl2, 0.02% NP-40, 3.5 mM K ferrocyanide, and 3.5 mM K ferricyanide overnight at 37°C in the dark. After washing with X-Gal washing buffer, slides were counterstained with nuclear fast red (VECTASHIELD), dehydrated through ethanol gradient, and mounted with Mounting Medium Xylene (Fishier Scientific).
Cell proliferation counting
Knee joints were obtained from three CD-1 animals for each time point (E17.5, P1, P7, and P14). Samples were processed according to the immunohistochemistry procedure just described. Ten random sections were selected from a total of 60 sections of each knee joint for imaging. The pictures were taken with a ZEISS AxioPlan 2 microscope (Carl Zeiss MicroImaging LLC). The number of cells in the cell cycle in each section was obtained by counting Ki67-positive cells. Data were analyzed using one-way ANOVA followed by a Tukey test for pair-wise comparisons using SPSS (IBM, SPSS Statistics 19).
Results
The histology of the developing patellar tendon
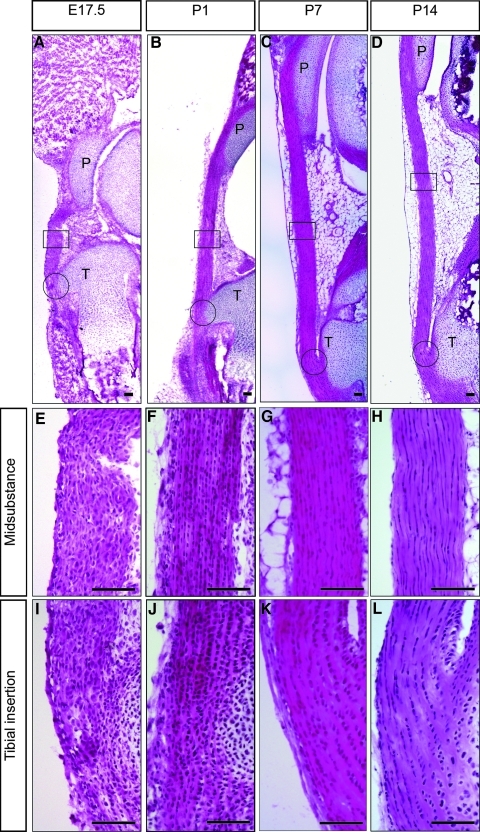
Four time points were used in this study: starting at E17.5, after the patellar tendon has become clearly visible as a discrete structure in the embryo but before birth, and ending at P14 when the animal is fully motile, and, therefore, the tendon is functional. Figure 1 shows representative sagittal sections of patellar tendons from these four stages in their differentiation (E17.5, P1, P7, and P14), all stained with H&E. Higher magnification images from the midsubstance and tibial insertion site at each stage are shown in Figure 1E–L. At E17.5 (Fig. 1A, E, I), the cells in both regions of the tendon are rounded, and separated by relatively little extracellular matrix. There are no obvious differences in cell morphology or intensity of staining between the midsubstance and insertion site at this stage. At P1 (Fig. 1B, F, J), there are clear differences in cell morphology between the midsubstance and insertion site. In the insertion, site cells are becoming aligned into rows of approximately square cell profiles, and their nuclei are rounded. In the midsubstance, cell boundaries are indistinct, but the nuclei are becoming elongated. The cells in both regions are becoming clearly more separated from each other by the increasing extracellular matrix. At P7 (Fig. 1C, G, K) and P14 (Fig. 1D, H, L), this differentiation process continues. In the midsubstance, cells become progressively more elongated, and the extracellular matrix/cell ratio increases. In the tibial insertion site, there is also a continued increase in matrix. However, the cells remain rounded in shape and arranged in linear arrays. We conclude from H and E staining that histological differentiation of the patellar tendon, both in terms of the laying down of extracellular matrix and its spatial differentiation into midsubstance and insertion site, starts in the neonatal period in the mouse.
FIG. 1.
Shows hematoxylin and eosin stained mid-sagittal sections of the mouse patellar tendon at E17.5 (A, E, I), P1 (B, F, J), P7 (C, G, K), and P14 (D, H, L). Higher magnification images of the midsubstance are shown in E–H and of the tibial insertion sites in I–L. P, patella; T, tibial. Scale bar=100 μm. E17.5, embryonic day seventeen and half; P1, P7, P14, postnatal days 1, 7, 14.
Molecular tendon markers expressed throughout the tendon
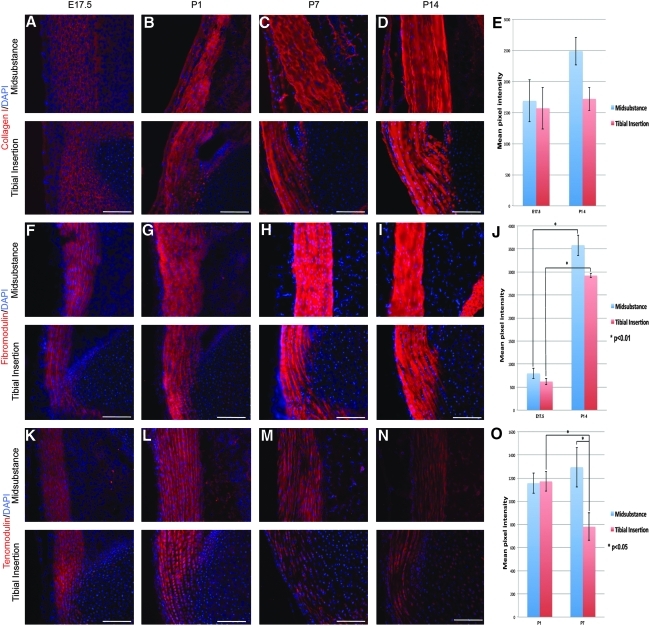
Figure 2 shows the expression pattern of several tendon markers that start to accumulate before birth, and are synthesized approximately equally in both the mid-substance and insertion site. These included type I collagen (COL1) (Fig. 2A–E), the proteoglycan FMOD (Fig. 2F–J), and the glycoprotein TNMD (Fig. 2K–O). All these markers accumulated throughout early postnatal life. In fact, COL1 and FMOD did not show any obvious quantitative differences in expression along the tendon length, whereas the expression of TMOD was reduced significantly in the insertion site starting at P7 (Fig. 2O). The expression of these should, therefore, be controlled by cell signals occurring along the whole length of the tendon.
FIG. 2.
Shows tenocyte markers expressed throughout the patellar tendon. A–D show type I collagen, F–I show fibromodulin, and K–N show tenomodulin expression using specific antibody staining. Specific staining in each case is red, and blue shows nuclear staining using DAPI. E, J, and O show semi-quantitation of the antibody staining by measurement of pixel intensity. Scale bar=100 μm. Error bar represents standard error, and the asterisk represents statistical significance (p<0.05). DAPI, 4′,6-diamidino-2-phenylindole.
Molecular markers expressed differentially in the tendon
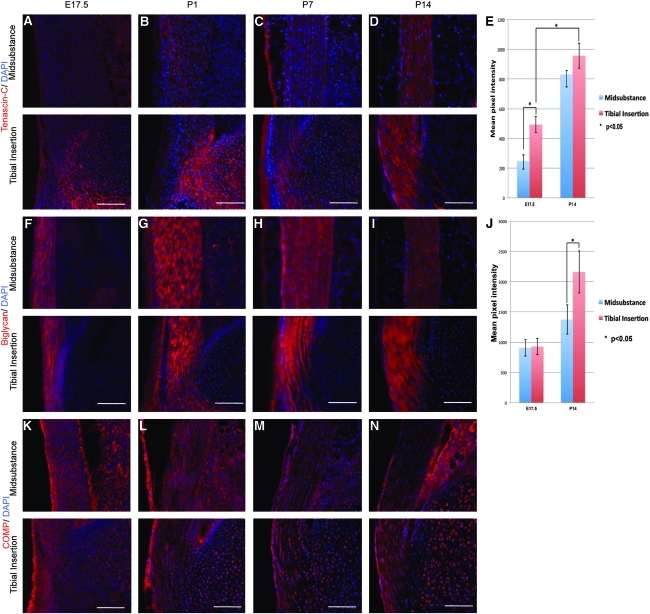
The expression of three tendon markers that are asymmetrically expressed in the tendon is shown in Figure 3. Interestingly, they are also expressed in different temporal patterns. The extracellular matrix glycoprotein tenascin-C (TNC) was found to be expressed only in the insertion site at E17.5 (Fig. 3A, E). After birth, an increasing expression was seen in the mid-substance. However, the protein was concentrated at all stages in the insertion site (Fig. 3B–E). The proteoglycan biglycan (BGN) was synthesized in all regions of the tendon at all stages examined (Fig. 3F–J). However, its expression became more concentrated in the insertion site at P7 and P14 (Fig. 3H–J). A third pattern was seen for COMP, which was expressed at low levels throughout the tendon until P7, when it became concentrated at the insertion site. These data show that the initial differentiation of the tendon into mid-substance and insertion site occurs at the earliest stages of its development, but is progressive, with new tendon markers either appearing, or becoming more concentrated in, the insertion site during the 2 weeks after birth. The fluorescence images were subjected to semi-quantitative analysis using the Zeiss Axiovision software for pixel intensity measurements. The data are shown for TNC and BGN in Figure 3E and J. High levels of staining in the adjacent tissues at early stages confounded quantitation of COMP expression.
FIG. 3.
Shows tenocyte markers expressed predominantly in the tibial insertion site. A–D show tenascin-C, F–J show biglycan, and K–N show COMP staining using specific antibodies. In each panel, red is specific stain, and blue shows cell nuclei stained using DAPI. Scale bar=100 μm. E and J show semi-quantitation of the antibody staining by measurement of pixel intensity. Scale bar=100 μm. Error bar represents standard error, and the asterisk represents statistical significance (p<0.05). COMP, cartilage oligomeric protein.
Signaling pathways active during patellar tendon differentiation
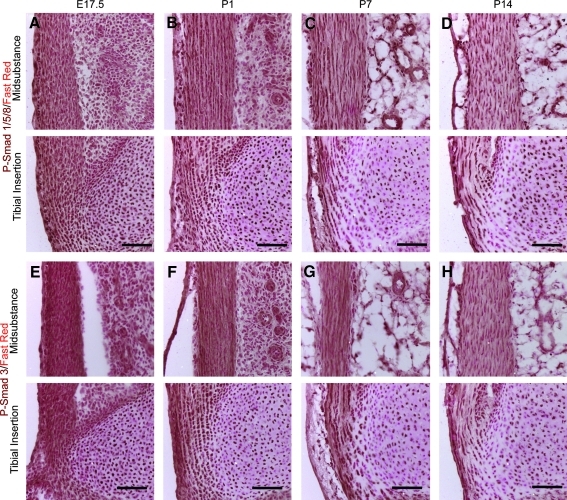
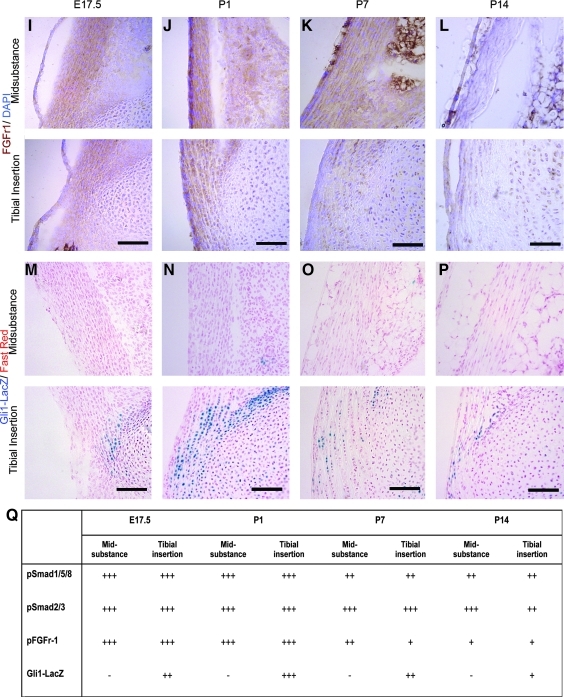
The activation of several major signaling pathways was examined, to identify any potential correlations between these and the temporo-spatial patterns seen in tendon differentiation. Cells responding to TGFβ and BMP signaling were identified using antibodies against phosphorylated Smad3 (PSmad3) and phosphorylated Smad1/5/8 (PSmad1/5/8) respectively. Cells responding to FGF signaling were identified using an antibody against the activated (phosphorylated) form of FGF receptor 1. Cells responding to Wnt signaling were identified using the TopGal strain of mice.22 Cells responding to hedgehog (Hh) signaling were identified using the Gli1-β-galactosidase strain of mice (Gli1-LacZ).23
Canonical Wnt signaling was not seen at any stage (data not shown). All cells in the tendon were found to be responding to TGFβ and BMP signaling at all stages examined (Fig. 4A–H, Q). FGF-responding cells were present in both regions of the tendon from E17.5 to P7. However, there was reduced antibody staining in the midsubstance at P14 and in the insertion site at P7 and P14. (Fig. 4I–L, Q), thus suggesting reduced or absent FGF signaling activity at later postnatal stages. Most interesting was the distribution of Hh-responding cells. At E17.5, a small number of cells at the insertion site were seen to be responding to Hh (Fig. 4M, Q). The staining increased in both intensity and in the number of positive cells at P1 (Fig. 4N, Q). At P7 and P14, the number of positive cells decreased to very low levels (Fig. 4O–Q). These data show that cells of the insertion site are responding to Hh signals at precisely the time at which we see the first molecular marker of the insertion site (TNC) appearing, and suggest a role for Hh signaling in this process.
FIG. 4.
Shows activated cell signaling pathways in the developing patellar tendon. A–D show cells responding to bone morphogenetic protein signaling (brown stain) using an antibody against pSmad1/5/8. Cell nuclei are stained using fast red. E–H show cells responding to transforming growth factor beta signaling using an antibody against pSmad2/3. Cell nuclei are stained with fast red. I–L show cells responding to FGF signaling (brown stain) using an antibody against pFGFr1. Nuclei are stained with DAPI (blue). M–P show cells responding to hedgehog signaling (blue stain), using the Gli1-LacZ reporter line. Cell nuclei are stained with fast red. Q show the summary of intensity levels of pSmad1/5/8, pSmad2/3, pFGFr-1, and Gli1-LacZ. Scale bar=100 μm. FGF, fibroblast growth factor.
The timing of cell proliferation during patellar tendon differentiation
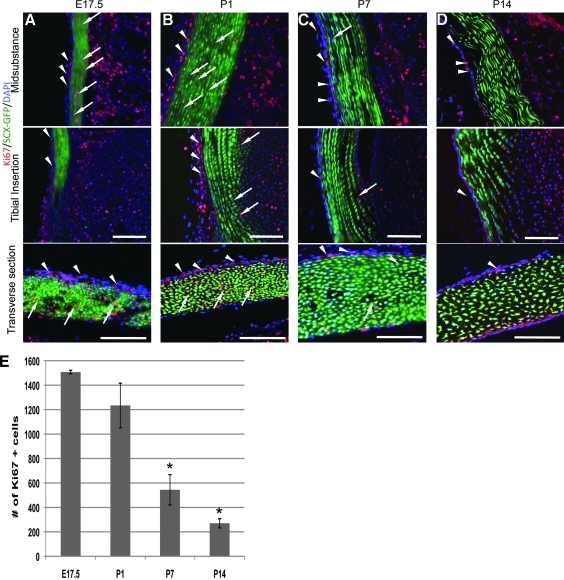
Tendon growth during late fetal and early postnatal life could be due to cell proliferation, the accumulation of extracellular matrix during differentiation, or both. To assay the spatiotemporal pattern of cell proliferation, we used the Ki67 antibody, which stains the nuclei of cells still in the cell cycle.22 To distinguish between tendon progenitor cells and connective tissue cells of the endo-, peri-, and epitenons, we used the ScxGFP reporter mouse, which expresses GFP in all cells that express the molecular marker of tendon precursor cells SCX.14 Ki67 positive cells were found in the mouse patellar tendon at all four time points. However, their number and distribution was different at each stage. There were more Ki67 positive cells in the tendon at E17.5 and P1 than at P7 and P14 (Fig. 5E). In addition, more of these cells were double positive (Ki67 and ScxGFP) (Fig.5A, B). By P7 and P14, most of the Ki67-positive, proliferating cells were not positive for ScxGFP, thus indicating that the cycling cells by P7 were not tenocytes (Fig. 5C, D). Most of the Ki67 cells at P7 and P14 were located in the epitenon. These data show that by P7 there are very few tenocytes (as defined by the expression of SCX) that are still in the cell cycle. This suggests either that they can re-enter the cycle after tendon injury, or that cells that repair tendons after injury arise from the connective tissue cells of the tendon, such as the epitenon, rather than the tenocytes themselves. The data also show that most, if not all, tenocytes, exit from the cell cycle during the time of major deposition of ECM, and, thus, cell differentiation.
FIG. 5.
Shows cells that are still in the cell cycle in the patellar tendon at E17.5 (A), P1 (B), P7 (C), and P14 (D), using an antibody against Ki67 (red stain). Nuclei are stained blue with DAPI, tenocytes are green, using the ScxGFP reporter mouse line. Cycling cells in the connective tissue of the epitenon are GFP-negative (arrowheads), whereas tenocytes in the cell cycle are GFP-positive (arrows). Scale bar=100 μm. (E) shows counts of cycling cells in the patellar tendon at each stage (mean±SEM) gained by counting Ki67-positive cells from 10 random sections from 3 knee joints at each stage. The asterisk represents statistical significance (p=0.3387 at E17.5, p=0.338 at P1, p=0.001 at P7, and p=0.0001 at P14). ScxGFP, scleraxis-green fluorescent protein.
Discussion
A fundamental understanding of normal tendon development is needed for any future biologically based treatments of tendon injuries. The current studies are aimed at producing information that will be useful in the identification of novel strategies for potential biological therapies. In particular, we have identified spatial and temporal landmarks of mouse patellar tendon proliferation and differentiation, which can be used to assess the degree of tendon healing, and some of the signaling pathways that may control these.
Tendons contain highly abundant ECM glycoproteins and proteoglycans, which are essential for both organization of the tendon and its healing. The timing and position of appearance of these in the tendon can be used as molecular markers for healing, or for attempts to develop tendons from stem cells in culture. In this work, we found that COL1, FMOD, and TNMD were present throughout the developing mouse patella tendon; whereas COMP, TNC, and BGN were concentrated at the insertion site. COL1 is the most abundant protein in the tendon and is important for its strength.24 Therefore, it is logical to see the dramatic increase in COL1 during the first 2 weeks after birth, when the patellar tendon becomes functional. FMOD and TNMD are thought to contribute to the distribution and patterning of the tendon collagen.25,26 Null mutations in Fmod mice have shown irregular fiber bundles and abnormal morphology in Achilles and tail tendons.27 Mice in which Tnmd had been targeted also developed abnormally organized collagen fibrils.25 These findings indicated that FMOD and TNMD regulate the composition of fibril bundles in tendons, and correlate well with the increase in both of these proteins at the same period as the increase in COL1. Although several studies showed that type II collagen (COL2) is present at the insertion site, we did not observe COL2 expression by immunocytochemistry during the period studied (data not shown). COL2 was expressed at high levels by adjacent articular cartilage cells, thus indicating that the anti-COL2 antibody used in the current study was able to detect COL2 protein. It may be that COL2 appears later in the differentiation of the patellar tendon.
The localization of BGN at the insertion site of the patella tendon was consistent with the previous findings using the Achilles tendon.28 COMP expression has been observed in the tendons/ligaments of both humans and different animal models29, whereas TNC is known to be expressed in many tissues during normal embryogenesis as well as in some cases after tendon rupture.30,31 However, to our knowledge, the current study is the first to show that the COMP and TNC are concentrated in the insertion site during normal tendon development at early postnatal stages. Interestingly, it has been shown that the expression of COMP and TNC are elevated during the healing process in the tendon-to-bone insertion site.30,32 This correlation between normal tendon development and healing suggests that COMP and TNC might play roles during the formation of tendon-to-bone insertion sites. In addition, ECM proteins such as FMOD, bigylcan, and tenasin-c have been suggested to be involved in the maintenance of stem cell niches.33,34 Therefore, COMP, BGN, and TNC might also play roles in regulating niches of tendon progenitor cells at insertion sites. However, how these might function in tendon cell generation is still unclear. It will be important to further investigate their functional roles in the formation of the insertion site. Further, it will be important to identify ECM components that have different expression patterns in the normal developing patellar tendon in culture models of tendon differentiation, because they may serve as markers for differentiation of mid-substance vs. insertion sites.
In the current study, we have identified the spatial and temporal expression patterns of activated cell signaling pathways during mouse patellar tendon development. Among these, Hh signaling is particular interesting. We observed that Gli1-LacZ -positive cells appeared in the tibial insertion site, thus indicating that the Hh signaling pathway was active in the insertion site. We also observed that the Gli1-LacZ positive cells were present at the cruciate ligament insertion sites (data not shown), thus suggesting the general importance of Hh signaling at tendon/ligament-to-cartilage insertion sites. The maturation of tendon-to-cartilage insertion sites requires contributions from both the tenocyte and chondrocyte.35 In addition, it is known that the differentiation and proliferation of chondrocytes is regulated by Hh signaling.36,37 Therefore, the localization of Hh-responding cells in the insertion site suggests that the Hh signaling pathway might control the chondrification of the tenocytes at the insertion site. There are also spatial and temporal correlations between the expression of BGN and TNC and Hh responding cells at the insertion site, thus suggesting that the Hh signaling pathway might regulate the expression of these ECM proteins in the developing insertion site of mouse patella tendon. Alternatively, the ECM glycoproteins and proteoglycans may actually control the function of the Hh signaling pathway in the tendon. It has been shown that proteoglycans such as BGN can bind to signaling ligands and control their range of action.38 It will be important to learn the relationship between these ECM components and the Hh signaling pathway in the insertion site. In addition, we also found that there were fewer Gli1-LacZ positive cells at P7 and P14 than there were at P1, thus suggesting that the activity of Hh signaling was decreased at the insertion site after P7. It is well known that the tendon-to-bone insertion is a common region for overuse injuries and is hard to repair. However, why exactly it is so difficult to reconstruct an injured insertion site is still unclear. We suggest two reasons: first, the loss of Hh signaling at the insertion site at later postnatal stages, and second, the ossification of the target cartilage to bone at later postnatal stages. It will be important to further explore the functional roles of Hh signaling pathway in the differentiation of the insertion site as well as its role in tissue repair. Two Hh ligands; sonic Hh and Indian Hh, have been shown to regulate the development of the musculoskeletal system.39 It will be important to learn which ligand is functional at the insertion site.
Many studies have shown that BMP, FGF, and TGFβ signaling are important during tendon development, including the initiation of tendon cell fate.40 However, how these signals regulate tendon differentiation is not fully understood. Here, we show that TGFβ and BMP signaling were active throughout the period studied and throughout the whole tendon. FGF signaling is responsible for the initiation of tendon development during embryogenesis.13,15,41 In the current study, we found that FGF signaling was decreased at P14 (Fig. 4E–H), thus suggesting that it may not be essential after the initial period of tendon differentiation is over. Several studies have shown that the healing process of tendons and ligaments can be enhanced by FGF,42–44 which might therefore re-awaken a developmental pathway in the tendon. Although the chief mechanism among these is not clear, it will be important to compare activation of FGF signaling during tendon healing with its timing of activation during the normal development of tendon.
Although we did not find any evidence to support the presence of canonical Wnt signaling in the developing mouse patellar tendon, our results could not exclude the involvement of noncanonical WNT signaling in regulation of tendon growth. Further analysis of noncanonical WNT signaling in tendon development is needed.
This study shows that Scx-expressing cells are still in the cell cycle at E17.5 and P1. However, by P7 and P14, most of the cycling cells were not ScxGFP positive, thus suggesting that most tenocytes have exited the cell cycle by this time. Occasional Scx-positive cells still in the cycle can be identified at the two later time points. It is not yet clear how this data might fit with the finding that cycling tenocytes are generated from tendon cell cultures, or with the suggestion that cells from the epitenon and endotenon might contribute to the healing process of tendon.45 It is not yet known whether cells in the connective tissues of the tendon can activate Scx expression and become tenocytes, It is also not clear whether the repair process of an injured tendon generates a fibrotic scar,46,47 or a tissue containing genuine tenocytes. It is most likely that healing cells in the tendon require signals that are present during normal development of the tendon for the generation of genuine tendon at the wound site. The markers identified in this study and the signaling pathways identified as active during normal development should be useful in gaining the answers to these important questions.
Acknowledgments
The authors are grateful to Dr. Ronen Shweitzer for the ScxGFP mice. They thank all the members of the Wylie-Heasman lab and the Butler lab. This work is supported by NIH grants AR46574-10, AR56943-02, and an IGERT training grant from NSF (#0333377).
Disclosure Statement
No competing financial interests exist.
References
- 1.Klepps S. Bishop J. Lin J. Cahlon O. Strauss A. Hayes P., et al. Prospective evaluation of the effect of rotator cuff integrity on the outcome of open rotator cuff repairs. Am J Sports Med. 2004;32:1716. doi: 10.1177/0363546504265262. [DOI] [PubMed] [Google Scholar]
- 2.Jost B. Pfirrmann C.W. Gerber C. Switzerland Z. Clinical outcome after structural failure of rotator cuff repairs. J Bone Joint Surg Am. 2000;82:304. doi: 10.2106/00004623-200003000-00002. [DOI] [PubMed] [Google Scholar]
- 3.Praemer A. Furner S. Rice D. Musculoskeletal Condition in the United States. Parke Ridge, IL: American Academy of Orthopaedic Surgeons; 1999. [Google Scholar]
- 4.Sharma P. Maffulli N. Tendon injury and tendinopathy: healing and repair. J Bone Joint Surg Am. 2005;87:187. doi: 10.2106/JBJS.D.01850. [DOI] [PubMed] [Google Scholar]
- 5.Riley G. Tendinopathy—from basic science to treatment. Nat Clin Pract Rheumatol. 2008;4:82. doi: 10.1038/ncprheum0700. [DOI] [PubMed] [Google Scholar]
- 6.Austin J.C. Phornphutkul C. Wojtys E.M. Loss of knee extension after anterior cruciate ligament reconstruction: effects of knee position and graft tensioning. J Bone Joint Surg Am. 2007;89:1565. doi: 10.2106/JBJS.F.00370. [DOI] [PubMed] [Google Scholar]
- 7.Petsche T.S. Hutchinson M.R. Loss of extension after reconstruction of the anterior cruciate ligament. J Am Acad Orthop Surg. 1999;7:119. doi: 10.5435/00124635-199903000-00005. [DOI] [PubMed] [Google Scholar]
- 8.Juncosa N. West J.R. Galloway M.T. Boivin G.P. Butler D.L. In vivo forces used to develop design parameters for tissue engineered implants for rabbit patellar tendon repair. J Biomech. 2003;36:483. doi: 10.1016/s0021-9290(02)00459-1. [DOI] [PubMed] [Google Scholar]
- 9.Butler D.L. Juncosa-Melvin N. Boivin G.P. Galloway M.T. Shearn J.T. Gooch C., et al. Functional tissue engineering for tendon repair: a multidisciplinary strategy using mesenchymal stem cells, bioscaffolds, and mechanical stimulation. J Orthop Res. 2008;26:1. doi: 10.1002/jor.20456. [DOI] [PubMed] [Google Scholar]
- 10.Juncosa-Melvin N. Matlin K.S. Holdcraft R.W. Nirmalanandhan V.S. Butler D.L. Mechanical stimulation increases collagen type I and collagen type III gene expression of stem cell-collagen sponge constructs for patellar tendon repair. Tissue Eng. 2007;13:1219. doi: 10.1089/ten.2006.0339. [DOI] [PubMed] [Google Scholar]
- 11.Kahane N. Cinnamon Y. Bachelet I. Kalcheim C. The third wave of myotome colonization by mitotically competent progenitors: regulating the balance between differentiation and proliferation during muscle development. Development. 2001;128:2187. doi: 10.1242/dev.128.12.2187. [DOI] [PubMed] [Google Scholar]
- 12.Brent A.E. Schweitzer R. Tabin C.J. A somitic compartment of tendon progenitors. Cell. 2003;113:235. doi: 10.1016/s0092-8674(03)00268-x. [DOI] [PubMed] [Google Scholar]
- 13.Brent A.E. Tabin C.J. FGF acts directly on the somitic tendon progenitors through the Ets transcription factors Pea3 and Erm to regulate scleraxis expression. Development. 2004;131:3885. doi: 10.1242/dev.01275. [DOI] [PubMed] [Google Scholar]
- 14.Schweitzer R. Chyung J.H. Murtaugh L.C. Brent A.E. Rosen V. Olson E.N., et al. Analysis of the tendon cell fate using Scleraxis, a specific marker for tendons and ligaments. Development. 2001;128:3855. doi: 10.1242/dev.128.19.3855. [DOI] [PubMed] [Google Scholar]
- 15.Edom-Vovard F. Bonnin M. Duprez D. Fgf8 transcripts are located in tendons during embryonic chick limb development. Mech Dev. 2001;108:203. doi: 10.1016/s0925-4773(01)00483-x. [DOI] [PubMed] [Google Scholar]
- 16.Pryce B.A. Watson S.S. Murchison N.D. Staverosky J.A. Dünker N. Schweitzer R. Recruitment and maintenance of tendon progenitors by TGFbeta signaling are essential for tendon formation. Development. 2009;136:1351. doi: 10.1242/dev.027342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brown D. Wagner D. Li X. Richardson J.A. Olson E.N. Dual role of the basic helix-loop-helix transcription factor scleraxis in mesoderm formation and chondrogenesis during mouse embryogenesis. Development. 1999;126:4317. doi: 10.1242/dev.126.19.4317. [DOI] [PubMed] [Google Scholar]
- 18.Ito Y. Toriuchi N. Yoshitaka T. Ueno-Kudoh H. Sato T. Yokoyama S., et al. The Mohawk homeobox gene is a critical regulator of tendon differentiation. Proc Natl Acad Sci USA. 2010;107:10538. doi: 10.1073/pnas.1000525107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Blitz E. Viukov S. Sharir A. Shwartz Y. Galloway J.L. Pryce B.A., et al. Bone ridge patterning during musculoskeletal assembly is mediated through SCX regulation of Bmp4 at the tendon-skeleton junction. Dev Cell. 2009;17:861. doi: 10.1016/j.devcel.2009.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pryce B.A. Brent A.E. Murchison N.D. Tabin C.J. Schweitzer R. Generation of transgenic tendon reporters, ScxGFP and ScxAP, using regulatory elements of the scleraxis gene. Dev Dyn. 2007;236:1677. doi: 10.1002/dvdy.21179. [DOI] [PubMed] [Google Scholar]
- 21.Lillie R.D. Histopathologic Technic and Practical Histochemistry. 3rd. New York: McGraw-Hill Book Co; 1965. [Google Scholar]
- 22.DasGupta R. Fuchs E. Multiple roles for activated LEF/TCF transcription complexes during hair follicle development and differentiation. Development. 1999;126:4557. doi: 10.1242/dev.126.20.4557. [DOI] [PubMed] [Google Scholar]
- 23.Bai C.B. Auerbach W. Lee J.S. Stephen D. Joyner A.L. Gli2, but not Gli1, is required for initial Shh signaling and ectopic activation of the Shh pathway. Development. 2002;129:4753. doi: 10.1242/dev.129.20.4753. [DOI] [PubMed] [Google Scholar]
- 24.Liu S.H. Yang R.S. al-Shaikh R. Lane J.M. Collagen in tendon, ligament, and bone healing. A current review. Clin Orthop Relat Res. 1995;318:265. [PubMed] [Google Scholar]
- 25.Docheva D. Hunziker E.B. Fassler R. Brandau O. Tenomodulin is necessary for tenocyte proliferation and tendon maturation. Mol Cell Biol. 2005;25:699. doi: 10.1128/MCB.25.2.699-705.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hedlund H. Mengarelli-Widholm S. Heinegard D. Reinholt F.P. Svensson O. Fibromodulin distribution and association with collagen. Matrix Biol. 1994;14:227. doi: 10.1016/0945-053x(94)90186-4. [DOI] [PubMed] [Google Scholar]
- 27.Svensson L. Aszodi A. Reinholt F.P. Fassler R. Heinegard D. Oldberg A. Fibromodulin-null mice have abnormal collagen fibrils, tissue organization, and altered lumican deposition in tendon. J Biol Chem. 1999;274:9636. doi: 10.1074/jbc.274.14.9636. [DOI] [PubMed] [Google Scholar]
- 28.Riley G. The pathogenesis of tendinopathy. A molecular perspective. Rheumatology (Oxford) 2004;43:131. doi: 10.1093/rheumatology/keg448. [DOI] [PubMed] [Google Scholar]
- 29.DiCesare P. Hauser N. Lehman D. Pasumarti S. Paulsson M. Cartilage oligomeric matrix protein (COMP) is an abundant component of tendon. FEBS Lett. 1994;354:237. doi: 10.1016/0014-5793(94)01134-6. [DOI] [PubMed] [Google Scholar]
- 30.Pajala A. Melkko J. Leppilahti J. Ohtonen P. Soini Y. Risteli J. Tenascin-C and type I and III collagen expression in total Achilles tendon rupture. An immunohistochemical study. Histol Histopathol. 2009;24:1207. doi: 10.14670/HH-24.1207. [DOI] [PubMed] [Google Scholar]
- 31.Riley G.P. Harrall R.L. Cawston T.E. Hazleman B.L. Mackie E.J. Tenascin-C and human tendon degeneration. Am J Pathol. 1996;149:933. [PMC free article] [PubMed] [Google Scholar]
- 32.Wurgler-Hauri C.C. Dourte L.M. Baradet T.C. Williams G.R. Soslowsky L.J. Temporal expression of 8 growth factors in tendon-to-bone healing in a rat supraspinatus model. J Shoulder Elbow Surg. 2007;16:S198. doi: 10.1016/j.jse.2007.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Garcion E. Halilagic A. Faissner A. ffrench-Constant C. Generation of an environmental niche for neural stem cell development by the extracellular matrix molecule tenascin C. Development. 2004;131:3423. doi: 10.1242/dev.01202. [DOI] [PubMed] [Google Scholar]
- 34.Bi Y. Ehirchiou D. Kilts T.M. Inkson C.A. Embree M.C. Sonoyama W.F. Identification of tendon stem/progenitor cells and the role of the extracellular matrix in their niche. Nat Med. 2007;13:1219. doi: 10.1038/nm1630. [DOI] [PubMed] [Google Scholar]
- 35.Thomopoulos S. Genin G.M. Galatz L.M. The development and morphogenesis of the tendon-to-bone insertion - what development can teach us about healing. J Musculoskelet Neuronal Interact. 2010;10:35. [PMC free article] [PubMed] [Google Scholar]
- 36.Mak K.K. Kronenberg H.M. Chuang P.T. Mackem S. Yang Y. Indian hedgehog signals independently of PTHrP to promote chondrocyte hypertrophy. Development. 2008;135:1947. doi: 10.1242/dev.018044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nie X. Luukko K. Kvinnsland I.H. Kettunen P. Developmentally regulated expression of Shh and Ihh in the developing mouse cranial base: comparison with Sox9 expression. Anat Rec A Discov Mol Cell Evol Biol. 2005;286:891. doi: 10.1002/ar.a.20231. [DOI] [PubMed] [Google Scholar]
- 38.Yoon J.H. Halper J. Tendon proteoglycans: biochemistry and function. J o Musculoskelet Neuronal Interact. 2005;5:22. [PubMed] [Google Scholar]
- 39.Mullor J.L. Sanchez P. Ruiz I. Altaba A. Pathways and consequences: Hedgehog signaling in human disease. Trends Cell Biol. 2002;12:562. doi: 10.1016/s0962-8924(02)02405-4. [DOI] [PubMed] [Google Scholar]
- 40.Liu C.F. Aschbacher-Smith L. Barthelery N.J. Dyment N. Butler D. Wylie C. What we should know before using tissue engineering techniques to repair injured tendons: a developmental biology perspective. Tissue Eng Part B Rev. 2011;17:165. doi: 10.1089/ten.teb.2010.0662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Edom-Vovard F. Schuler B. Bonnin M.A. Teillet M.A. Duprez D. Fgf4 positively regulates scleraxis and tenascin expression in chick limb tendons. Dev Biol. 2002;247:351. doi: 10.1006/dbio.2002.0707. [DOI] [PubMed] [Google Scholar]
- 42.Chan K.M. Fu S.C. Wong Y.P. Hui W.C. Cheuk Y.C. Wong M.W. Expression of transforming growth factor beta isoforms and their roles in tendon healing. Wound Repair Regen. 2008;16:399. doi: 10.1111/j.1524-475X.2008.00379.x. [DOI] [PubMed] [Google Scholar]
- 43.Fukui N. Katsuragawa Y. Sakai H. Oda H. Nakamura K. Effect of local application of basic fibroblast growth factor on ligament healing in rabbits. Rev Rhum Engl Ed. 1998;65:406. [PubMed] [Google Scholar]
- 44.Kobayashi D. Kurosaka M. Yoshiya S. Mizuno K. Effect of basic fibroblast growth factor on the healing of defects in the canine anterior cruciate ligament. Knee Surg Sports Traumatol Arthrosc. 1997;5:189. doi: 10.1007/s001670050049. [DOI] [PubMed] [Google Scholar]
- 45.Jones M.E. Mudera V. Brown R.A. Cambrey A.D. Grobbelaar A.O. McGrouther D.A. The early surface cell response to flexor tendon injury. J Hand Surg Am. 2003;28:221. doi: 10.1053/jhsu.2003.50044. [DOI] [PubMed] [Google Scholar]
- 46.Gelberman R.H. Manske P.R. Vande Berg J.S. Lesker P.A. Akeson W.H. Flexor tendon repair in vitro: a comparative histologic study of the rabbit, chicken, dog, and monkey. J Orthop Res. 1984;2:39. doi: 10.1002/jor.1100020107. [DOI] [PubMed] [Google Scholar]
- 47.Hatano I. Suga T. Diao E. Peimer C.A. Howard C. Adhesions from flexor tendon surgery: an animal study comparing surgical techniques. J Hand Surg Am. 2000;25:252. doi: 10.1053/jhsu.2000.jhsu25a0252. [DOI] [PubMed] [Google Scholar]