Abstract
The process of lipid peroxidation is widespread in biology and is mediated through both enzymatic and non-enzymatic pathways. A significant proportion of the oxidized lipid products are electrophilic in nature, the RLS (reactive lipid species), and react with cellular nucleophiles such as the amino acids cysteine, lysine and histidine. Cell signalling by electrophiles appears to be limited to the modification of cysteine residues in proteins, whereas non-specific toxic effects involve modification of other nucleophiles. RLS have been found to participate in several physiological pathways including resolution of inflammation, cell death and induction of cellular antioxidants through the modification of specific signalling proteins. The covalent modification of proteins endows some unique features to this signalling mechanism which we have termed the ‘covalent advantage’. For example, covalent modification of signalling proteins allows for the accumulation of a signal over time. The activation of cell signalling pathways by electrophiles is hierarchical and depends on a complex interaction of factors such as the intrinsic chemical reactivity of the electrophile, the intracellular domain to which it is exposed and steric factors. This introduces the concept of electrophilic signalling domains in which the production of the lipid electrophile is in close proximity to the thiol-containing signalling protein. In addition, we propose that the role of glutathione and associated enzymes is to insulate the signalling domain from uncontrolled electrophilic stress. The persistence of the signal is in turn regulated by the proteasomal pathway which may itself be subject to redox regulation by RLS. Cell death mediated by RLS is associated with bioenergetic dysfunction, and the damaged proteins are probably removed by the lysosome-autophagy pathway.
Keywords: electrophile-responsive proteome, Kelch-like ECH-associated protein 1 (Keap1), lipid peroxidation, nuclear factor-erythroid 2 related factor (Nrf2), protein modification, reactive lipid species (RLS)
Abbreviations: BLT, leukotriene B receptor; COX, cyclo-oxygenase; CysLT, cysteinyl leukotriene receptor; 15d-PGJ2, 15-deoxyprostaglandin J2; EpRE, electrophilic-response element; GST, glutathione transferase; HNE, 4-hydroxynonenal; HO-1, haem oxygenase-1; HSF, heat-shock factor; Hsp, heat-shock protein; Keap1, Kelch-like ECH-associated protein 1; LOX, lipoxygenase; LT, leukotriene; Nrf2, nuclear factor-erythroid 2-related factor; PG, prostaglandin; PPARγ, peroxisome-proliferator-activated receptor γ; PUFA, polyunsaturated fatty acid; RLS, reactive lipid species; RNS, reactive nitrogen species; ROS, reactive oxygen species
INTRODUCTION
The oxidation of PUFAs (polyunsaturated fatty acids), such as arachidonic acid, generates a broad range of oxidation products which historically have been used as markers of oxidative stress [1,2]. For example, the unique structural attributes of the non-specific oxidation products known as the isoprostanes have allowed for the development of accurate high-throughput assays for their measurement in complex biological systems [3]. Lipid peroxidation products have been detected in the blood, plasma, urine, and tissue samples of humans and animal models using an array of techniques, and, in many cases, their levels are elevated in pathological conditions [4–7]. The application of these analytical techniques has led to the concept that RLS (reactive lipid species) are mediators, not simply by-products, of multiple pathophysiological conditions [8–11]. The cell signalling mediated by RLS has some unique biochemical attributes. Importantly, many lipid peroxidation products are also electrophilic, which allows them to form stable covalent adducts with nucleophilic residues on proteins [12–14]. This is important since it is now well recognized that the thiol groups on cysteine residues act as redox switches controlling cell signalling and metabolism [15–17]. The cysteine thiol group is particularly versatile, and the concept has emerged that different thiol-reactive signalling molecules can selectively modulate protein function [16]. Specific mechanisms that have been shown to modify redox cell signalling include S-nitrosation, S-glutathionylation and Michael addition with biologically active electrophiles [15,18,19]. Other oxidative mechanisms mediated by either hydrogen peroxide or lipid peroxides to form sulfenic or sulfinic acids were initially thought to be markers of oxidative damage. However, a previous study suggested that they may also play a role in cell signalling [20]. Interestingly, although early studies implied that lipid peroxidation always results in damage, a more refined view of this process has evolved and suggests that oxidized lipids can elicit different cellular effects depending on the species present, their concentrations and their reactivity with protein targets [14,21–23].
Oxidized lipids can mediate biological responses through two diverse mechanisms: classic reversible binding and irreversible covalent modification of receptors [15,22,24–26]. Some oxidized lipids are ligands for specific receptors [e.g. PG (prostaglandin) receptors] and mediate biological effects through reversible receptor–ligand interactions [27,28]. This is best understood for the enzymatically produced PGs and LTs (leukotrienes) [29]. In contrast, some lipid peroxidation products modulate cellular activity through irreversible covalent modification of nucleophilic amino acid residues on proteins [15,30]. This concept was initially in conflict with the classical paradigms for cell signalling since to ‘turn-off’ the signal, the protein must be selectively degraded. However, signalling through the covalent modification of proteins is now accepted for a number of well-defined protein–lipid interactions, and selective degradation is mediated through the proteasome [31,32]. Interestingly, signalling though the covalent modification of proteins changes the relationship between the concentration of the ligand, in this case an oxidized lipid, and the resultant signal [33]. Since irreversible covalent modifications of proteins can accumulate over time and amplify a signal [33], even low levels of oxidized lipids initiate signalling. We have termed this concept ‘the covalent advantage’ [22].
In the present review, we will discuss: (i) the formation of RLS through both non-enzymatic and enzymatic processes; (ii) oxidized lipid signalling through classic receptor-mediated pathways and by covalent modification of protein targets; and (iii) susceptibility of thiols to modification by RLS. We will then relate these concepts to the ability of oxidized lipids to trigger adaptive and damaging biological effects with a focus on the role of subcellular localization.
FORMATION OF RLS
Much of the early research into mechanisms of lipid peroxidation was performed by scientists in the food industry. It was well appreciated that off odours and flavours could be attributed to lipid oxidation, and inhibiting this process results in products with a longer shelf life [34]. As the field evolved, it was recognized that lipid peroxidation is endogenous to living organisms and has multiple functions dependent on the site and mechanism of oxidation. For example, as shown in Figure 1(A), enzymatic sources of lipid peroxidation yield important biological mediators of inflammation such as the PGs from COXs (cyclo-oxygenases), and the LTs from LOXs (lipoxygenases) [35,36]. Importantly, both non-enzymatic and enzymatic oxidation of PUFAs results in the formation of RLS that are electrophilic (Figure 1B). A major substrate for lipid peroxidation is arachidonic acid, and its oxidation results in the formation of several products (Figure 1A). Of these lipid peroxidation products (Figure 1A), a subset are electrophilic in nature, and there are both structurally distinct species derived from either non-enzymatic or enzymatic lipid peroxidation (Figure 1B). Examples of electrophilic products of non-enzymatic lipid oxidation include aldehydes such as HNE (4-hydroxynonenal), malondialdehyde and acrolein as well as the J- and A-series isoprostanes. Other RLS include the isoketals, which result from the rearrangement of endoperoxide intermediates of the isoprostane pathway and have the potential to react with both proteins and lipids [37]. The approach to research with RLS has largely focused on defining the reactivity and biological effects of a candidate molecule. For this reason, we know a great deal about the behaviour of HNE, 15d-PGJ2 (15-deoxyprostaglandin J2) and nitroalkanes [21,38–44]. From these studies, two key facts have emerged: (i) the effects of all the RLS are dependent on the amount exposed to the cell with many exhibiting anti-inflammatory or cytoprotective effects over the lower concentration range; and (ii) the biological effects of the RLS vary according to the specific RLS and target cells [33,45]. The implications of these findings are that each RLS reacts with a specific family of proteins which we have called the electrophile-responsive proteome [12,22]. This concept will be explored in more depth throughout the present review.
Figure 1. Formation of lipid electrophiles via non-enzymatic and enzymatic lipid peroxidation.
(A) Arachidonic acid can be converted into several products through enzymatic and non-enzymatic lipid peroxidation. Both free-radical-catalysed as well as enzymatically controlled oxidation yields a subset of products that are electrophilic. 5-HPETE, 5-hydroperoxyeicosatetraenoic acid; LOOH, linoleic acid hydroperoxide. (B) Examples of RLS produced from arachidonic acid and their structures. For simplicity, stereochemistry is not indicated. TXA2, thromboxane A2; *reactive site.
NON-ENZYMATIC LIPID PEROXIDATION
PUFAs, such as arachidonic and linoleic acid, are targets for lipid peroxidation. Non-specific lipid peroxidation proceeds through a chain reaction composed of three main steps: initiation, propagation and termination. In enzymatic lipid peroxidation, initiation is controlled and stereospecific and propagation does not occur. The production of specific lipid oxidation signalling molecules is controlled by enzyme pathways and the release of non-enzyme-bound radical intermediates is minimized. Due to their unsaturated double bonds, the allylic hydrogen atoms in PUFAs are readily abstracted by initiating species such as ferryl radical, peroxynitrite (ONOO−), hydroperoxyl radicals (HO2•) and hydroxyl radical (OH•). This results in the formation of lipid radicals which react with oxygen if it is available. The products that are formed are diverse and depend on the substrate oxidized (e.g. arachidonic compared with linoleic acid) and the mechanism of oxidation (non-enzymatic or enzymatic). Once lipid peroxidation is initiated, lipid alkoxyl (LO•) and lipid peroxyl (LOO•) radicals are capable of abstracting a hydrogen atom from another fatty acid molecule, thus contributing to the propagation of lipid peroxidation [46]. In biological membranes, the presence of proteins can result in transfer of the lipid radicals to protein side chains and adduct formation [47,48]. In this setting, the proteins become active participants in the propagation of the lipid peroxidation reactions. Molecular oxygen (O2) is required for the propagation phase, and, for this reason, lipid peroxidation proceeds at a higher rate when oxygen concentrations are high [46]. Lipid peroxidation can be terminated by radical–radical reactions with other lipid radical species or with protein radicals. Termination can also occur by radical–radical reaction of a lipid radical species with the nitric oxide radical (NO•) [49].
Cardiovascular disease is a pathological condition in which predominantly non-specific lipid peroxidation occurs in vivo. For example, in atherosclerotic lesions, the lipid peroxidation products found are mostly those lacking stereospecificity which is a characteristic of the non-enzymatic pathways [50]. However, increases in inflammation do lead to production of low levels of stereospecific enzymatic lipid oxidation products in atherosclerosis [51,52]. Several factors may promote lipid peroxidation through non-enzymatic reactions in vivo [53]. For example, the production of ROS (reactive oxygen species) and RNS (reactive nitrogen species) in inflammation may result in damage to iron- or copper-containing proteins and the release of the metal from a protein environment in which radical reactions can be controlled. This can occur with the proteins myoglobin and haemoglobin [54,55]. Haem proteins then play an important role in lipid peroxidation by decomposing lipid hydroperoxides and facilitating the propagation phase [56]. However, unlike free haem, haem proteins can also initiate lipid peroxidation [57]. Interaction of hydrogen peroxide with metmyoglobin or methaemoglobin leads to the formation of an activated haem protein with a porphyrin cation radical (P+−Fe4+=O) [58]. This ferryl radical species is the initiator of lipid peroxidation rather than the hydroxyl radical [57]. Because of the ability of hydrogen peroxide to ‘activate’ these haem proteins to initiating species, myoglobin- and haemoglobin-mediated lipid peroxidation may be important for catalysing lipid peroxidation in biological systems where hydrogen peroxide is elevated [56,59].
Peroxynitrite (ONOO−), formed from the rapid reaction of NO• with superoxide has also been shown to promote lipid peroxidation [38,60,61], probably due to the reactivity of decomposition products hydroxyl radical and nitrogen dioxide (NO2•). These radical species are capable of abstracting a hydrogen atom from unsaturated fatty acids and this process is iron-independent [60]. iNOS (inducible nitric oxide synthase) and NADPH oxidases are cellular sources of NO• and superoxide respectively, and their expression is concomitantly increased in several pathologies and can form ONOO− [62,63]. The lipid peroxidation reactions initiated by ONOO− produce isoprostanes, aldehydes and oxysterols, but unique RLS such as nitrated lipids only occur with this mechanism of oxidation [64–66]. The interaction of lipid radicals with RNS such as nitrogen dioxide, or possibly nitrite, results in a family of electrophilic RLS known as the nitroalkenes [38,39,67].
ENZYMATIC LIPID PEROXIDATION
There are several enzymes that contribute to the controlled peroxidation of PUFAs and the activation of multiple biological pathways [36,68]. One of the best-studied enzymes is COX, which is responsible for the formation of PGs from arachidonic acid (Figure 1). Since COX acts predominantly on free fatty acids, in many cases the production of PGs is dependent upon phospholipase A2 [69]. COX contains two active sites including a COX domain and a peroxidase domain [70]. The COX site is responsible for oxygenating arachidonic acid to form hydroperoxide PGG2. The peroxidase site then reduces PGG2 to the alcohol PGH2, the final product of COX. There are two isoforms of COX in the cell [70]. COX-1 is constitutively expressed in all tissues; however, COX-2 is normally only detected in tissues with active inflammation except kidney and brain where COX-2 is constitutively expressed [71]. The protein expression of COX-2 is regulated by several transcription factors relevant to inflammation including NF-κB (nuclear factor κB), NF-IL-6 (nuclear factor for interleukin-6 expression) and CREB (cAMP-response-element-binding protein) [72,73]. Once expressed, COX's activity can also be regulated in a transcription-independent manner [74,75]. Several ROS are known to regulate COX-2 activity by regulating the levels of the lipid peroxide tone which is required for activation [75–77]. The major product of both COX-1 and COX-2 is PGH2, which can then be metabolized to other PGs through the action of PGD, PGE, PGF, and PGI synthases [78–83]. PGA2, PGJ2 and 15d-PGJ2 are examples of electrophilic PGs.
The COX enzymes generate several anti-inflammatory electrophilic RLS from arachidonic acid (e.g. cyclopentenones) as well as products of ω−3 fatty acids [e.g. DHA (docosahexaenoic acid) and EPA (eicosapentaenoic acid)] [40,84]. The latter products derived from COX-2 have been shown to be important anti-inflammatory mediators [84,85]. Interestingly, a subset of these are electrophilic, termed the EFOXs (electrophilic oxo-derivatives of ω−3 fatty acids) [84]. These enzymatically produced RLS may be important for the protection afforded by ω−3 supplementation.
Another important source of enzymatic lipid peroxidation products is through the action of LOXs. LTs and lipoxins are the products of this pathway and have been extensively studied in the field of immunology [29,86,87]. There are three LOX isoforms, with 5-, 12- and 15-LOX expressed in leucocytes, platelets and endothelial cells respectively [29,88]. The active site of LOX contains a non-haem iron which is critical to the enzyme's activity [89,90]. As with COX, LOX activity is also modulated by ROS through regulation of the enzyme's peroxide tone [91,92]. Among the LOXs, 5-LOX is the most well-studied in the context of cardiovascular disease [68]. It was originally found to contribute to asthma and was targeted with inhibitors developed to minimize airway inflammation [86]. It is now well-established that 5-LOX products also contribute to other inflammatory processes including the development of coronary artery disease [51,93]. As shown in Figure 1, following generation of LTA4 from LOX, the product LTB4 is formed by hydration, whereas the cysteinyl LTs, LTC4, LTD4 and LTE4, are produced by a specialized GST (glutathione transferase) enzyme, LTC4 synthase [94,95]. Aside from the known receptor-mediated effects of the LTs, one LT is known to be capable of receptor-independent effects through covalent modification. Because LTA4 is uniquely electrophilic owing to its epoxide group, it is capable of adducting to nucleophilic amino acids as well as DNA bases [96,97]. The nucleophilic attack of 5-LOX by LTA4 leads to the covalent modification and inactivation of the enzyme [98].
REVERSIBLE RECEPTOR-MEDIATED SIGNALLING BY OXIDIZED LIPIDS
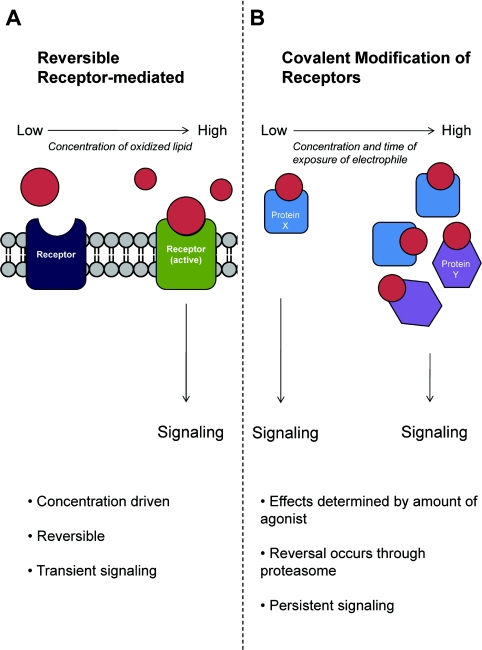
It is well appreciated that oxidized lipids including the PGs and LTs can act through reversible binding to cellular receptors [27,99] (Figure 2). The signalling responses vary depending on the spectrum of oxidized lipids formed, their concentration and which receptors are bound by the ligand. As shown in Figure 2(A), the signals arising from reversible receptor–ligand interactions are dependent on the concentration of specific lipid peroxidation products formed and are typically transient and saturable. Thus it will not be an effective agonist unless a product is present at a concentration close to the binding affinity of the receptor. In many cases, the biological lifetimes of the PGs are also extremely short, and this results in a transient signal.
Figure 2. Classical receptor-mediated signalling compared with signalling mediated through covalent modification.
(A) Classical receptor-mediated signalling only occurs when a ligand is present at concentrations which exceed the affinity constant (Km). This binding is reversible and quickly dissipates when ligand concentrations dip below the needed threshold. (B) Signalling via covalent modification can occur at low concentrations as well as high concentrations of the ligand. Low concentrations of electrophile accumulate over time, resulting in persistent signalling. Higher concentrations of electrophile may result in modification of more diverse protein targets and thus change the cellular response.
Many of the COX products act through G-protein-coupled receptors, including the EP receptors (EP1, EP2, EP3, and EP4 in humans) for PGE2, the DP receptor and the CRTH2 (chemoattractant receptor homologous molecule expressed on T helper type 2 cells) for PGD2 and its metabolites, the FP receptor for PGF2, and the IP receptor for PGI2 [100]. In contrast, electrophilic PGs can also participate in cell signalling through covalent modification of receptors [22,33], a signalling mechanism that will be discussed further below [23,101]. As with the PGs, LTS formed through LOX act through specific receptors [102]. BLT (LTB receptor) 1 and BLT2 mediate the pro-inflammatory effects of LTB4 [103]. The cysteinyl LTs (LTC4, LTD4 and LTE4) act through CysLT (cysteinyl LT receptor) 1 and CysLT2 [104]. CysLT1 is highly expressed in bronchial smooth muscle, and agonist binding induces smooth muscle contraction [105]. CysLT1 is also expressed in the spleen and platelets [105,106]. CysLT2 is expressed in the heart, adrenal gland, placenta, peripheral leucocytes, spleen, lymph nodes and central nervous system [107].
Other than through specific G-protein-coupled receptors, several lipid peroxidation products have been suggested to act through PPARγ (peroxisome-proliferator-activated receptor γ). This receptor is interesting since it appears that it can function through both reversible and irreversible binding to the receptor. For example, the reactive PGs (e.g. 15d-PGJ2), electrophilic fatty acids including the nitrated lipids such as nitro-arachidonic acid and nitro-oleic acid have also been shown to bind covalently to this receptor [39]. Binding to PPARγ is thought to be important for the anti-inflammatory effects of a number of RLS [39,41,108]. Upon binding to PPARγ, genes involved in metabolism, cellular differentiation and inflammation are up-regulated. PPARγ activation is anti-inflammatory and the protein is expressed on several cell types within the vasculature including endothelial cells, monocytes, macrophages and smooth muscle cells [109].
ACCUMULATION OF CELL SIGNALLING EFFECTS BY LOW CONCENTRATIONS OF RLS: THE COVALENT ADVANTAGE
Can the formation of a covalent bond between a receptor and a RLS (Figure 2B) elicit a biological response in vivo? This is a particularly important issue to address because the ‘free’ levels of RLS in biological systems are often reported to be in the nanomolar range and yet in vitro micromolar ranges are typically needed to elicit cell signalling [11,40,42,43,55,56,110]. For example, PPARγ activation may be the basis of signalling for some electrophilic lipids [39], but it has been suggested that they are present at concentrations insufficient to bind to and activate this receptor [111–113]. However, these RLS can bind covalently to a cysteine residue on the ligand-binding domain of the receptor [114,115]. In addition, quantitative estimation of RLS is confounded by the reactivity of the α,β-unsaturated ketone group, since substantial amounts of the lipid will be bound to proteins.
The fact that RLS can form covalent bonds with proteins may allow the accumulation of a signal over time. This is governed by several factors. The rate of activation will depend on the concentration of the activating electrophile and the receptor target. However, the net activation will depend on the amount of activating electrophile to which the receptor target is exposed to over time (Figure 2B). For example, the activation of a signalling pathway achieved by 10 pmol of electrophile will be half that achieved by 20 pmol in the cells that are exposed to the same volume. Using a cell culture model system and covalent adduct formation of 15d-PGJ2 with Keap1 (Kelch-like ECH-associated protein 1) as a model electrophile receptor, we have demonstrated experimentally that the covalent modification of Keap1 by 15d-PGJ2 accumulates over time [33]. Thus a low flux of an electrophilic RLS leads to full activation of a receptor even when the concentration is low (Figure 2B). In part specificity can then be attributed to the generation of a low flux of lipid electrophile, which favours reactions with cysteine residues and steric factors controlling the availability of the nucleophilic amino acid residue. Reversibility of the signal is then controlled by integration of the signalling pathway with the proteasome as will be discussed in a later section.
Thus the key feature of the covalent advantage signalling paradigm is that cysteine modification occurs in a specific manner. As such there are several factors which regulate susceptibility to thiol modification. Although cysteine is present in most proteins, only a small percentage of cysteine residues are susceptible to modification [116] as will be discussed in the next section.
POST-TRANSLATIONAL MODIFICATION OF PROTEINS BY RLS: PROTEIN DAMAGE COMPARED WITH CELL SIGNALLING
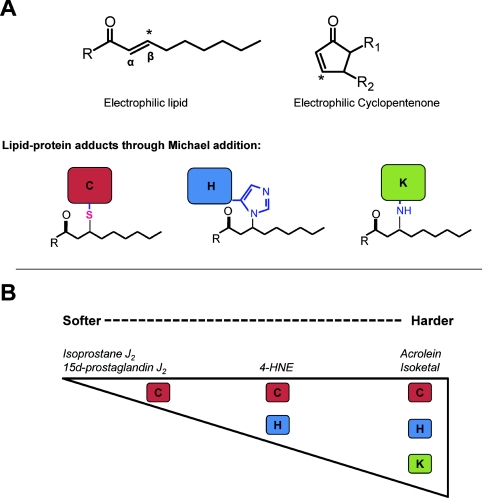
Protein adducts with RLS can occur by reaction with nucleophilic centres such as those shown in Figure 3(A) [117]. The RLS shown in this Figure have been extensively studied and include electrophilic lipid oxidation products such as acrolein and HNE as well as electrophilic cyclopentenone PGs and isoprostanes. Acrolein and HNE possess an aldehyde functional group and an α,β-unsaturated carbonyl functional group which allow for reaction by both Schiff's base formation and Michael addition respectively. The cyclopentenone structure reacts through Michael addition only. Michael addition occurs by reaction of the electrophile with the nucleophilic amino acids cysteine, lysine and histidine (Figure 3A).
Figure 3. RLS differ in their reactivity with specific amino acids.
(A) Basic structure of an electrophilic lipid and cylopentenone. The β-carbon of the α,β-unsaturated carbonyl is electrophilic, making the compound reactive with the nucleophilic amino acids cysteine, lysine and histidine. (B) The specificity and reactivity of lipid electrophiles differ depending on relative hardness. Although soft electrophiles such as the cyclopentenones 15d-PGJ2 and isoprostane J2 are largely reactive with cysteine residues, harder electrophiles including the isoketals show less specificity and react with many other nucleophilic targets.
Exposure of electrophiles to biological systems modifies a subset of proteins (termed the electrophile-responsive proteome) which, in concert, orchestrate the biological response. These proteomes are determined by a number of inter-related properties of both the electrophile and protein target. The chemical reactivity of pathologically relevant electrophiles and nucleophiles has been investigated in some depth [118]. As shown in Figure 3(B), an important property determining which nucleophilic amino acids are modified by an electrophile are governed by the hard/soft acid–base principle [118–120]. ‘Hard’ electrophiles include a number of mutagenic compounds and often react with the ‘hard’ nucleophilic centres in purine and pyrimidine bases. On the other hand, ‘soft’ electrophiles include many RLS [119] and react readily with ‘soft’ nucleophiles such as GSH and protein cysteinyl thiols [119,121]. In comparison with the cyclopentenone lipid electrophiles, α,β-unsaturated aldehydes including acrolein and isoketals are relatively harder [122], allowing them to adduct to harder nucleophiles including DNA and the amino groups on lysine and lipids. An example of a relatively soft electrophile is 15-PGJ2, which reacts with thiol groups on cysteine, but does not modify other nucleophilic amino acids [123–125]. Reactivity with soft lipid electrophiles occurs mostly through the modification of cysteine residues in the more reactive thiolate anion form [23,126,127]. These modifications are biologically significant since thiol residues are ‘redox sensors’, which are important in cell signalling. In addition to modification by RLS, signalling can also be initiated by several other thiol-dependent post-translational modifications including S-nitrosylation, S-glutathionylation, formation of sulfenic and sulfinic acids, and disulfide formation [128–133]. Importantly, the local protein environment can influence the pKa value of candidate thiols influencing their reactivity with electrophilic lipids as well as with other oxidants and will be discussed in more detail below [126–128].
The differences in reactivity of softer and harder electrophiles may explain, in part, the different cellular effects of RLS. For example, cyclopentenones react specifically with thiol groups on cysteine residues, whereas acrolein can react with other amino acids. This is important when considering the ability of RLS to modify proteins thus changing their function and the cellular response. It is now appreciated that site-specific modification of cysteine residues contributes to cell signalling through cysteine rich proteins, such as Keap1, whereas modification of lysine residues is associated with toxicity [21,122,134–137].
The development of lipid- and electrophile-tagging techniques has been crucial in the identification of several key metabolic and signalling proteins which are covalently modified by RLS at reactive cysteine residues [138–140]. Some selected proteins known to be modified by RLS and their cellular effects are given in Table 1. As can be seen, the signalling pathways regulated by electrophilic adduct formation are diverse in both their cellular location and their role in pathophysiology. These data imply that there is specificity in terms of RLS signalling with respect to both the electrophilic lipid and the protein targets. This concept is shown schematically in Figure 3. The amino acid residues on proteins can confer specificity in reaction with lipid electrophiles which, in turn, are important for determining whether the lipid peroxidation products formed elicit a cell signalling response or contribute to damage [37,124,136,141].
Table 1. Selected protein targets of RLS.
ANT, adenine nucleotide translocator; NF-κB, nuclear factor κB.
| Protein | Functional change | Reference |
|---|---|---|
| p50 subunit of NF-κB | Inhibition of NF-κB DNA binding | [183] |
| H-, N-, K-Ras | Activation of H-, N-, K-Ras | [124,184] |
| Keap1 | Release of Nrf2 | [136] |
| Thioredoxin | Serves as sensor for oxidative stress | [185] |
| Thioredoxin reductase | Disruption of the conformation of the tumour- suppressor protein p53 | [186] |
| c-Jun | Inhibition of AP1 DNA binding | [187] |
| β-Actin | Filament disruption | [188,189] |
| GSTP1-1 | Inactivation of GSTP1-1 | [30] |
| 26S proteasome | Inhibition of proteasome and impairment of proteasomal assembly | [190] |
| Hsp70 | Release of HSF1 to up-regulate the heat-shock response | [42,43] |
| Hsp90 | Release of HSF1 to up-regulate the heat-shock response | [42,43] |
| PPARγ | Activation of receptor; anti-inflammatory | [123,191] |
| ANT | Mitochondrial membrane permeabilization | [140,192] |
| ATP synthase | Inhibits ATP synthesis | [140,193] |
| Cytochrome c oxidase | Inhibited oxidase activity | [194–196] |
PROTEIN THIOL REACTIVITY AS A REGULATOR OF SIGNALLING BY RLS
The primary mechanism by which redox signalling occurs is through the post-translational modification of critical cysteine residues (thiols) in redox-sensitive proteins. This modification can then change the structure and/or function of the modified protein and alter downstream signalling [16]. There are multiple lines of evidence which demonstrate that redox signalling occurs in a regulated and specific manner and does not simply represent non-specific oxidative damage. Conserved cysteine residues occur in almost all classes of proteins and, in many cases, are important for protein function [142–144]. For example, there are 25 cysteine residues in the redox sensor Keap1 which are selectively modified by reactive species [145]. It is for this reason that thiols are poised to mediate diverse redox signalling responses to multiple stimuli.
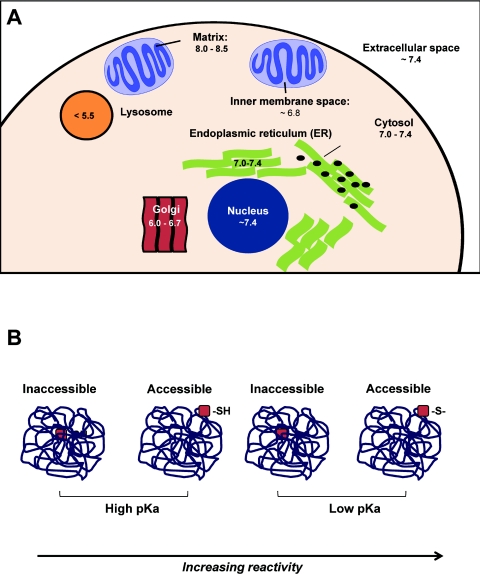
In Figure 4, we have integrated the factors which regulate susceptibility to thiol modification by RLS. The susceptibility of cysteine residues to modification by a defined RLS is dictated by a combination of factors including the pKa of the thiol and the local pH of the intracellular compartment (Figure 4A). Interestingly, the range of pH within the cell is likely to have a major impact on thiol reactivity. For example, the high intramitochondrial pH may be one reason mitochondrial protein thiols are particularly susceptible to modification and play a key role in cell signalling [146,147]. Other factors are the accessibility of the thiol within the protein structure, subcellular localization and the reactivity of the thiol-modifying agent. The pKa of a specific thiol is defined as the pH at which 50% of that thiol will be deprotonated. Thus a thiol having a pKa of 7.4 will be 50% deprotonated at physiological pH. Since deprotonated thiol (thiolate) is much more nucleophilic in nature, lower pKa thiols, which are more likely to be deprotonated at physiological pH, are favoured in their reaction with lipid electrophiles [148]. However, this factor alone is not sufficient to explain why electrophiles can activate cell signalling pathways independent of GSH.
Figure 4. Factors which determine susceptibility to thiol modification and cellular thiol targets.
Thiol residues have different susceptibilities to being modified by thiol-reactive agents. One important factor is accessibility (within the cell as within a protein) and the pKa value of the thiol target. (A) The pH of the environment is different, depending on the subcellular location. As shown, the average pH of the mitochondrial matrix is 8–8.5, whereas lysosomes are much more acidic, averaging a pH less than 5.5. The pH, along with the thiol pKa value determines whether a thiol is deprotonated to form thiolate anion. (B) The local protein environment is a very important determinant of thiol reactivity. For example, an inaccessible, high pKa protein thiol would be considered the least prone to modification. However, a low pKa accessible thiol would be a highly sensitive target.
Localization of thiol residues, either within a protein or within the cell, also seems to be important in dictating their relative susceptibilities to modification; however, these factors are less well characterized. This is shown in Figure 4(B), where the most accessible thiol residue is more likely to be modified than those less accessible. There is evidence demonstrating site-selective modification of cysteine residues within a single protein by two different RLS [124], although characterization of this type of regulation for a larger subset of proteins has not been examined to date. As shown in Figure 4(B), it is the combined properties of a low pKa thiol and accessibility which are the most consistent feature of proteins activated by electrophilic signalling.
The combination of steric and biochemical factors result in a functional hierarchy for the activation of cellular signalling pathways on exposure of cells to an electrophile. The first pathways to respond are those which are closest to the site of formation or exposure to the electrophile and with the most reactive protein thiol to the specific electrophile in question. The functional consequence of these factors is that the ‘first responders to electrophile exposure’ are not necessarily the most abundant thiol-containing proteins. For example, actin possesses several reactive thiol groups, but is modified by 15d-PGJ2 at concentrations higher than those required for modification of Keap1, which is a lower-abundance protein than actin [149]. The consequence for cell signalling is that the RLS-dependent signalling pathways are activated sequentially according to the site of formation, specific chemistry, available signalling proteins and characteristics of the specific electrophile.
GSH AND RELATED ENZYMES AS INSULATORS OF ELECTROPHILE SIGNALLING DOMAINS
In addition to protein thiols, GSH is an abundant low-molecular mass thiol, with a pKa value of 8.3, which is present at micromolar levels within the cell. Interestingly, electrophiles such as 15d-PGJ2 activate the Keap1 pathway at concentrations in which reaction with GSH is not detectable [150]. This lack of involvement of the GSH pathway in electrophile signalling probably occurs for two reasons. The first relates to the fact that the direct reaction of GSH with electrophiles, due to its high pKa value, is slow even though it is present in the cell in the micromolar range. It is often not appreciated that, although this may seem a high concentration, it may, in many cases, be lower than the protein thiol levels. For example, intramitochondrial protein thiol levels have been demonstrated to be 26-fold higher than GSH [151]. A further important factor is that the pH of intracellular compartments, which will modulate the amount of reactive thiols, shows great variability between cellular compartments as shown in Figure 4(A). In a compartment with elevated pH, such as the mitochondrion, electrophilic adduct formation with a reactive protein thiol on a signalling protein will be favoured over reactions with free GSH.
The major route of GSH reaction with electrophiles is catalysed by GSTs. The Km value for GSTs for electrophiles is typically in the micromolar range which suggests that if the RLS concentration is the nanomolar range, the metabolism of RLS through the GSH–GSTs pathway will be minimal [152]. In addition, the access to GSTs will be restricted by steric factors. This is important because the generation of a lipid electrophile is likely to occur in a hydrophobic environment inaccessible to water-soluble GSH or GSTs. Evidence is now emerging in which the potential site for an electrophile formation is in close proximity to the nucleophilic redox sensor. For example, a sub-population of Keap1 has been shown to be associated with the mitochondrion and may be a mechanism through which mitochondria regulate the Keap1/Nrf2 (nuclear factor-erythroid 2-related factor) system [153]. If this is the case, what is the role of the GSH–GSTs system in cells? We suggest that the conjugation of reactive electrophiles which are formed in an uncontrolled manner are deleterious to endogenous low-level signalling, and the GSH systems ‘insulate’ against high levels of electrophile production which may cause cellular damage.
The reactivity of the thiol-modifying agent itself imparts an important selective pressure for the subset of proteins (or subproteome) which will be modified. Similarly, the source of an electrophile often dictates its potential protein targets, as the site of generation may promote modification within a subcellular microdomain. Indeed, it is now becoming clear that the redox characteristics of intracellular compartments are not equivalent and differ widely in their key biochemical characteristics. As alluded to above, mitochondria have emerged as both a key site of RLS-mediated signalling and a target of RLS-dependent damage. Owing to the basic microenvironment in the mitochondrial matrix, mitochondrial protein thiols are more often in the thiolate form and are particularly sensitive to modification [151]. At low levels of electrophile, the interaction with mitochondria may result in adaptive cell signalling. For example, blocking mitochondrial thiols has been shown to attenuate HO-1 (haem oxygenase-1) induction by lipid electrophiles, suggesting that the mitochondrion plays a critical permissive role in this signalling pathway [125]. This raises the possibility that localized ROS production in mitochondria can induce the formation of reactive electrophiles that participate in cell signalling. Exogenous electrophilic lipids have also been shown to localize with the mitochondrion and modify proteins in this subcellular compartment [154]. At high concentrations this can result in the promotion of apoptosis, probably through inducing the permeability transition [140,155]. Some of the strongest evidence for a localized effect of electrophilic signalling in specific domains of the cell comes from a series of experiments with 15d-PGJ2. This electrophile activates the Keap1/Nrf2 system in the cytosol, as discussed below, but also targets mitochondria and induces mitochondrial ROS [154]. If a mitochondrial derivative of 15d-PGJ2 is used, the cytosolic signalling is essentially repressed and the dominant effect becomes mitochondrial dysfunction followed by apoptotic cell death [156]. Other studies have shown that cardiolipin (diphosphatidylglycerol) oxidation in the mitochondria can contribute to the release of cytochrome c from the organelle and the initiation of apoptosis [157]. This is one of the most direct examples in which mitochondrial lipid peroxidation has been linked to cell signalling.
LIPID ELECTROPHILES AND THE ADAPTIVE RESPONSE TO OXIDATIVE STRESS
One important protein target of electrophilic lipids mediating an adaptive response is Keap1, an adaptor protein normally bound to the transcription factor Nrf2. Modification of Keap1 by electrophilic lipids, including 15d-PGJ2 and HNE, results in the release of Nrf2 and translocation of the transcription factor to the nucleus (Figure 5A). In the nucleus, Nrf2 binds to the EpRE (electrophilic-response element) and genes responsible for the antioxidant proteins HO-1 and GCL (glutamate–cysteine ligase) are transcribed. The EpRE is also called the ARE (antioxidant-response element). It is thought that many of the protective effects of dietary electrophiles such as sulforaphane and resveratrol may be indirect and lie in their ability to up-regulate endogenous cellular antioxidants through the EpRE [158,159].
Figure 5. Modification of protein targets involved in the adaptive response.
(A) Modification of Keap1 leads to the release of the transcription factor Nrf2 and its translocation to the nucleus. Upon binding to the EpRE, several genes are up-regulated including HO-1, GCL (glutamate-cysteine ligase), GST and NQO1 (NADPH-quinone oxidoreductase). (B) The heat-shock response is also regulated by RLS. Normally present in monomeric form and bound to Hsp70 or Hsp90, HSF1 trimerizes upon exposure to several electrophilic lipids and up-regulates the expression of Hsps by binding to the heat-shock element (HSE). Ub, ubiquitin.
Electrophilic lipids have also been shown to increase protection through the up-regulation of Hsps (heat-shock proteins), particularly HSF (heat-shock factor) 1 [42,160]. As shown in Figure 5(B), the activation of HSF1 by oxidative stress is mediated by covalent modification of Hsp70 and Hsp90 [42]. These chaperone proteins normally maintain HSF1 in the cytosol [42,161–163]. Once HSF1 translocates to the nucleus, it is responsible for up-regulating Hsp110, Hsp90, Hsp70 and Hsp40, all of which are cytoprotective against toxic stressors [164]. Several oxidative stressors including hydrogen peroxide, ozone and metal toxicity have been shown to increase the heat-shock response [165–167]. It is likely that some of these pro-oxidants mediate their effects through the secondary production of RLS.
One important example of the activation of an adaptive protective response is cardiac ischaemic pre-conditioning. This describes the condition where several short periods (~5 min) of ischaemia and reperfusion protect the heart from longer ischaemic periods [168,169]. Importantly, the oxidants that have been hypothesized to play a role in the ischaemic pre-conditioning process may also cause lipid peroxidation [170,171]. It is clear that some of the protection afforded by ischaemic pre-conditioning occurs though increases in EpRE- and HSF-regulated genes (e.g. HO-1 or Hsp70) [172–179], and emerging literature suggests that electrophilic nitroalkanes can induce these protective mechanisms [160]. Understanding whether the modification of protein targets by RLS contribute to pre-conditioning may help direct the development of therapeutic agents in ischaemia/reperfusion injury.
FUTURE DIRECTIONS
A number of important questions remain to be addressed in our understanding of how RLS modulate cell function. The development of sophisticated mass spectrometry techniques for the identification of lipidomes will allow for a complete characterization of the oxylipidome, an important subset of the lipidome. The development of techniques to monitor specific lipid–protein adducts to define the electrophile-responsive proteome for specific RLS will be integral to investigate further this paradigm of covalent modification as a cell signalling mechanism. Relating the concentration-dependent biological responses to the intrinsic biochemical properties of the electrophile and the proteomes they modify is an important research problem. This has been most effectively demonstrated with the cyclopentenone 15d-PGJ2 [23,149,156,180].
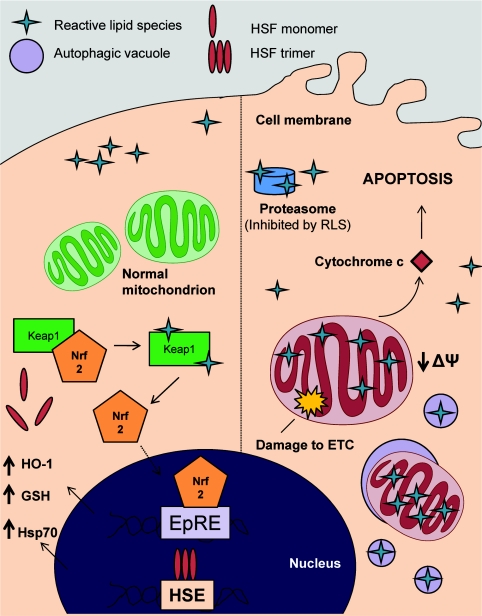
As markers of responses to physiology and pathophysiology, the isoprostanes have been particularly successful as indicators of oxidative stress in human subjects [7]. Perhaps surprisingly, it is now clear that the RLS react with a discrete electrophile-responsive proteome and this is domain-sensitive [23,124,156]. Although cytosolic targets, including the Hsps and Keap1, drive the adaptive response, mitochondrial targets govern apoptosis, ROS production and cellular respiration, and also participate in the adaptive response (Figure 6). For example, targeting an electrophile to the mitochondria suppresses activation of the Keap1/Nrf2 pathway and promotes mitochondria-dependent cell death [156]. This role of the mitochondrion in cell signalling is now becoming more prominent since it is a site for the controlled formation of ROS and plays an important role in the transcriptional regulation of HO-1 [125]. The reason the mitochondria plays such a central role is probably related to the extensive lipid environment within the inner mitochondrial membrane adjacent to redox active transition metals in the electron-transport chain. Is the mitochondrion the source of endogenous low levels of RLS for cell signalling? We have proposed that the mitochondrion can transduce hydrogen peroxide to form a reactive electrophilic lipid oxidation product [154,181]. Understanding the interface between the pathological effects of RLS and their cell signalling is challenging. An aspect we have not discussed in depth in the present review, but which is emerging as an important area, is the role autophagy and mitophagy play in the biological effects of RLS [182].
Figure 6. Subcellular localization of protein targets governs the biological response to RLS.
The diverse biological effects of electrophilic lipids is due, in part, to their subcellular targets. On the left-hand side, cytosolic targets predominate with the modification of proteins such as Keap1 and the Hsp70/Hsp90, leading to an increase in adaptive responses. On the right-hand side, mitochondrial targets control the response, leading to changes in cellular respiration and apoptotic cell death. In addition, RLS may mediate autophagy and/or mitophagy, leading to proteasome-independent degradation of adducted proteins. ETC, electron-transport chain; HSE, heat-shock element.
In summary, the current paradigm is that low levels of RLS can accumulate over time and specifically modify cysteinyl thiols to modulate protective cell signalling pathways. In contrast, high levels of RLS can modify other nucleophilic residues in a less specific manner resulting in protein damage and activation of GST-mediated GSH conjugation. Failure to decrease the RLS levels and subsequently repair or remove the damage is likely to lead to deleterious consequences for the cell and the development of pathology. It will be interesting to see how these concepts develop over the next few years as analytical techniques allow the identification of specific cellular targets for RLS in physiology and pathology.
FUNDING
The work described in the present review was supported by the National Institutes of Health [grant numbers ES10167, AA13395, DK075867 (to V.D.U.), HL096638 (to A.L.) and HL007918 (to A.R.D.)].
References
- 1.Pratico D., Lawson J. A., Rokach J., FitzGerald G. A. The isoprostanes in biology and medicine. Trends Endocrinol. Metab. 2001;12:243–247. doi: 10.1016/s1043-2760(01)00411-8. [DOI] [PubMed] [Google Scholar]
- 2.Gaut J. P., Heinecke J. W. Mechanisms for oxidizing low-density lipoprotein. Insights from patterns of oxidation products in the artery wall and from mouse models of atherosclerosis. Trends Cardiovasc. Med. 2001;11:103–112. doi: 10.1016/s1050-1738(01)00101-3. [DOI] [PubMed] [Google Scholar]
- 3.Milne G. L., Musiek E. S., Morrow J. D. F2-isoprostanes as markers of oxidative stress in vivo: an overview. Biomarkers. 2005;10:S10–S23. doi: 10.1080/13547500500216546. [DOI] [PubMed] [Google Scholar]
- 4.Marwah S. S., Blann A. D., Rea C., Phillips J. D., Wright J., Bareford D. Reduced vitamin E antioxidant capacity in sickle cell disease is related to transfusion status but not to sickle crisis. Am. J. Hematol. 2002;69:144–146. doi: 10.1002/ajh.10033. [DOI] [PubMed] [Google Scholar]
- 5.Yin H., Porter N. A. New insights regarding the autoxidation of polyunsaturated fatty acids. Antioxid. Redox Signaling. 2005;7:170–184. doi: 10.1089/ars.2005.7.170. [DOI] [PubMed] [Google Scholar]
- 6.Poon H. F., Calabrese V., Scapagnini G., Butterfield D. A. Free radicals: key to brain aging and heme oxygenase as a cellular response to oxidative stress. J. Gerontol. A Biol. Sci. Med. Sci. 2004;59:478–493. doi: 10.1093/gerona/59.5.m478. [DOI] [PubMed] [Google Scholar]
- 7.Morrow J. D. Quantification of isoprostanes as indices of oxidant stress and the risk of atherosclerosis in humans. Arterioscler. Thromb. Vasc. Biol. 2005;25:279–286. doi: 10.1161/01.ATV.0000152605.64964.c0. [DOI] [PubMed] [Google Scholar]
- 8.Volkel W., Sicilia T., Pahler A., Gsell W., Tatschner T., Jellinger K., Leblhuber F., Riederer P., Lutz W. K., Gotz M. E. Increased brain levels of 4-hydroxy-2-nonenal glutathione conjugates in severe Alzheimer's disease. Neurochem. Int. 2006;48:679–686. doi: 10.1016/j.neuint.2005.12.003. [DOI] [PubMed] [Google Scholar]
- 9.Musiek E. S., Breeding R. S., Milne G. L., Zanoni G., Morrow J. D., McLaughlin B. Cyclopentenone isoprostanes are novel bioactive products of lipid oxidation which enhance neurodegeneration. J. Neurochem. 2006;97:1301–1313. doi: 10.1111/j.1471-4159.2006.03797.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Patel M., Liang L. P., Hou H., Williams B. B., Kmiec M., Swartz H. M., Fessel J. P., Roberts L. J., II Seizure-induced formation of isofurans: novel products of lipid peroxidation whose formation is positively modulated by oxygen tension. J. Neurochem. 2008;104:264–270. doi: 10.1111/j.1471-4159.2007.04974.x. [DOI] [PubMed] [Google Scholar]
- 11.Chen J., Henderson G. I., Freeman G. L. Role of 4-hydroxynonenal in modification of cytochrome c oxidase in ischemia/reperfused rat heart. J. Mol. Cell. Cardiol. 2001;33:1919–1927. doi: 10.1006/jmcc.2001.1454. [DOI] [PubMed] [Google Scholar]
- 12.Landar A., Giles N. M., Zmijewski J. W., Watanabe N., Oh J. Y., Darley-Usmar V. M. Modification of lipids by reactive oxygen and nitrogen species: the oxy-nitroxy-lipidome and its role in redox cell signaling. Future Lipidol. 2006;1:203–211. [Google Scholar]
- 13.Codreanu S. G., Zhang B., Sobecki S. M., Billheimer D. D., Liebler D. C. Global analysis of protein damage by the lipid electrophile 4-hydroxy-2-nonenal. Mol. Cell. Proteomics. 2009;8:670–680. doi: 10.1074/mcp.M800070-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mochizuki M., Ishii Y., Itoh K., Iizuka T., Morishima Y., Kimura T., Kiwamoto T., Matsuno Y., Hegab A. E., Nomura A., et al. Role of 15-deoxyΔ(12,14) prostaglandin J2 and Nrf2 pathways in protection against acute lung injury. Am. J. Respir. Crit. Care Med. 2005;171:1260–1266. doi: 10.1164/rccm.200406-755OC. [DOI] [PubMed] [Google Scholar]
- 15.Rudolph T. K., Freeman B. A. Transduction of redox signaling by electrophile-protein reactions. Sci. Signaling. 2009;2:re7. doi: 10.1126/scisignal.290re7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cooper C. E., Patel R. P., Brookes P. S., Darley-Usmar V. M. Nanotransducers in cellular redox signaling: modification of thiols by reactive oxygen and nitrogen species. Trends Biochem. Sci. 2002;27:489–492. doi: 10.1016/s0968-0004(02)02191-6. [DOI] [PubMed] [Google Scholar]
- 17.Jones D. P. Radical-free biology of oxidative stress. Am. J. Physiol. Cell Physiol. 2008;295:C849–C868. doi: 10.1152/ajpcell.00283.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lopez-Sanchez L. M., Muntane J., de la Mata M., Rodriguez-Ariza A. Unraveling the S-nitrosoproteome: tools and strategies. Proteomics. 2009;9:808–818. doi: 10.1002/pmic.200800546. [DOI] [PubMed] [Google Scholar]
- 19.Mieyal J. J., Gallogly M. M., Qanungo S., Sabens E. A., Shelton M. D. Molecular mechanisms and clinical implications of reversible protein S-glutathionylation. Antioxid. Redox Signaling. 2008;10:1941–1988. doi: 10.1089/ars.2008.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rhee S. G., Chae H. Z., Kim K. Peroxiredoxins: a historical overview and speculative preview of novel mechanisms and emerging concepts in cell signaling. Free Radical Biol. Med. 2005;38:1543–1552. doi: 10.1016/j.freeradbiomed.2005.02.026. [DOI] [PubMed] [Google Scholar]
- 21.Ceaser E. K., Moellering D. R., Shiva S., Ramachandran A., Landar A., Venkartraman A., Crawford J., Patel R., Dickinson D. A., Ulasova E., et al. Mechanisms of signal transduction mediated by oxidized lipids: the role of the electrophile-responsive proteome. Biochem. Soc. Trans. 2004;32:151–155. doi: 10.1042/bst0320151. [DOI] [PubMed] [Google Scholar]
- 22.Dickinson D. A., Darley-Usmar V. M., Landar A. The covalent advantage: a new paradigm for cell signaling by thiol reactive lipid oxidation products. In: Dalle-Donne I., Scalone A., Butterfield D. A., editors. Redox Proteomics: from Protein Modifications to Cellular Dysfunction and Diseases. Indianapolis: John Wiley & Sons; 2006. pp. 345–367. [Google Scholar]
- 23.Stamatakis K., Perez-Sala D. Prostanoids with cyclopentenone structure as tools for the characterization of electrophilic lipid-protein interactomes. Ann. N.Y. Acad. Sci. 2006;1091:548–570. doi: 10.1196/annals.1378.096. [DOI] [PubMed] [Google Scholar]
- 24.Hong F., Sekhar K. R., Freeman M. L., Liebler D. C. Specific patterns of electrophile adduction trigger Keap1 ubiquitination and Nrf2 activation. J. Biol. Chem. 2005;280:31768–31775. doi: 10.1074/jbc.M503346200. [DOI] [PubMed] [Google Scholar]
- 25.Rachakonda G., Xiong Y., Sekhar K. R., Stamer S. L., Liebler D. C., Freeman M. L. Covalent modification at Cys151 dissociates the electrophile sensor Keap1 from the ubiquitin ligase CUL3. Chem. Res. Toxicol. 2008;21:705–710. doi: 10.1021/tx700302s. [DOI] [PubMed] [Google Scholar]
- 26.Tsujita T., Li L., Nakajima H., Iwamoto N., Nakajima-Takagi Y., Ohashi K., Kawakami K., Kumagai Y., Freeman B. A., Yamamoto M., Kobayashi M. Nitro-fatty acids and cyclopentenone prostaglandins share strategies to activate the Keap1–Nrf2 system: a study using green fluorescent protein transgenic zebrafish. Genes Cells. 2011;16:46–57. doi: 10.1111/j.1365-2443.2010.01466.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Breyer R. M., Bagdassarian C. K., Myers S. A., Breyer M. D. Prostanoid receptors: subtypes and signaling. Annu. Rev. Pharmacol. Toxicol. 2001;41:661–690. doi: 10.1146/annurev.pharmtox.41.1.661. [DOI] [PubMed] [Google Scholar]
- 28.Reilly M. P., Lawson J. A., FitzGerald G. A. Eicosanoids and isoeicosanoids: indices of cellular function and oxidant stress. J. Nutr. 1998;128:434S–438S. doi: 10.1093/jn/128.2.434S. [DOI] [PubMed] [Google Scholar]
- 29.Prigge S. T., Boyington J. C., Faig M., Doctor K. S., Gaffney B. J., Amzel L. M. Structure and mechanism of lipoxygenases. Biochimie. 1997;79:629–636. doi: 10.1016/s0300-9084(97)83495-5. [DOI] [PubMed] [Google Scholar]
- 30.Sanchez-Gomez F. J., Gayarre J., Avellano M. I., Perez-Sala D. Direct evidence for the covalent modification of glutathione-S-transferase P1-1 by electrophilic prostaglandins: implications for enzyme inactivation and cell survival. Arch. Biochem. Biophys. 2007;457:150–159. doi: 10.1016/j.abb.2006.10.032. [DOI] [PubMed] [Google Scholar]
- 31.Kobayashi M., Li L., Iwamoto N., Nakajima-Takagi Y., Kaneko H., Nakayama Y., Eguchi M., Wada Y., Kumagai Y., Yamamoto M. The antioxidant defense system Keap1–Nrf2 comprises a multiple sensing mechanism for responding to a wide range of chemical compounds. Mol. Cell. Biol. 2009;29:493–502. doi: 10.1128/MCB.01080-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Di Napoli M., Papa F. The proteasome system and proteasome inhibitors in stroke: controlling the inflammatory response. Curr. Opin. Invest. Drugs. 2003;4:1333–1342. [PubMed] [Google Scholar]
- 33.Oh J. Y., Giles N., Landar A., Darley-Usmar V. Accumulation of 15-deoxy-Δ(12,14)-prostaglandin J2 adduct formation with Keap1 over time: effects on potency for intracellular antioxidant defence induction. Biochem. J. 2008;411:297–306. doi: 10.1042/bj20071189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tappel A. L. The inhibition of hematin-catalyzed oxidations by α-tocopherol. Arch. Biochem. Biophys. 1953;47:223–225. doi: 10.1016/0003-9861(53)90454-8. [DOI] [PubMed] [Google Scholar]
- 35.Niki E., Yoshida Y., Saito Y., Noguchi N. Lipid peroxidation: mechanisms, inhibition, and biological effects. Biochem. Biophys. Res. Commun. 2005;338:668–676. doi: 10.1016/j.bbrc.2005.08.072. [DOI] [PubMed] [Google Scholar]
- 36.O'Donnell V. B., Maskrey B., Taylor G. W. Eicosanoids: generation and detection in mammalian cells. Methods Mol. Biol. 2009;462:5–23. [PubMed] [Google Scholar]
- 37.Davies S. S., Amarnath V., Roberts L. J., II Isoketals: highly reactive γ-ketoaldehydes formed from the H2–isoprostane pathway. Chem. Phys. Lipids. 2004;128:85–99. doi: 10.1016/j.chemphyslip.2003.10.007. [DOI] [PubMed] [Google Scholar]
- 38.O'Donnell V. B., Freeman B. A. Interactions between nitric oxide and lipid oxidation pathways: implications for vascular disease. Circ. Res. 2001;88:12–21. doi: 10.1161/01.res.88.1.12. [DOI] [PubMed] [Google Scholar]
- 39.Baker P. R., Lin Y., Schopfer F. J., Woodcock S. R., Groeger A. L., Batthyany C., Sweeney S., Long M. H., Iles K. E., Baker L. M., et al. Fatty acid transduction of nitric oxide signaling: multiple nitrated unsaturated fatty acid derivatives exist in human blood and urine and serve as endogenous peroxisome proliferator-activated receptor ligands. J. Biol. Chem. 2005;280:42464–42475. doi: 10.1074/jbc.M504212200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Scher J. U., Pillinger M. H. 15d-PGJ2: the anti-inflammatory prostaglandin? Clin. Immunol. 2005;114:100–109. doi: 10.1016/j.clim.2004.09.008. [DOI] [PubMed] [Google Scholar]
- 41.Trostchansky A., Souza J. M., Ferreira A., Ferrari M., Blanco F., Trujillo M., Castro D., Cerecetto H., Baker P. R., O'Donnell V. B., Rubbo H. Synthesis, isomer characterization, and anti-inflammatory properties of nitroarachidonate. Biochemistry. 2007;46:4645–4653. doi: 10.1021/bi602652j. [DOI] [PubMed] [Google Scholar]
- 42.Jacobs A. T., Marnett L. J. Systems analysis of protein modification and cellular responses induced by electrophile stress. Acc. Chem. Res. 2010;43:673–683. doi: 10.1021/ar900286y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vila A., Tallman K. A., Jacobs A. T., Liebler D. C., Porter N. A., Marnett L. J. Identification of protein targets of 4-hydroxynonenal using click chemistry for ex vivo biotinylation of azido and alkynyl derivatives. Chem. Res. Toxicol. 2008;21:432–444. doi: 10.1021/tx700347w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Petersen D. R., Doorn J. A. Reactions of 4-hydroxynonenal with proteins and cellular targets. Free Radical Biol. Med. 2004;37:937–945. doi: 10.1016/j.freeradbiomed.2004.06.012. [DOI] [PubMed] [Google Scholar]
- 45.Stamatakis K., Sanchez-Gomez F. J., Perez-Sala D. Identification of novel protein targets for modification by 15-deoxy-Δ12,14-prostaglandin J2 in mesangial cells reveals multiple interactions with the cytoskeleton. J. Am. Soc. Nephrol. 2006;17:89–98. doi: 10.1681/ASN.2005030329. [DOI] [PubMed] [Google Scholar]
- 46.Porter N. A., Caldwell S. E., Mills K. A. Mechanisms of free radical oxidation of unsaturated lipids. Lipids. 1995;30:277–290. doi: 10.1007/BF02536034. [DOI] [PubMed] [Google Scholar]
- 47.Gebicki J. M., Nauser T., Domazou A., Steinmann D., Bounds P. L., Koppenol W. H. Reduction of protein radicals by GSH and ascorbate: potential biological significance. Amino Acids. 2010;39:1131–1137. doi: 10.1007/s00726-010-0610-7. [DOI] [PubMed] [Google Scholar]
- 48.Fritz K. S., Petersen D. R. Exploring the biology of lipid peroxidation-derived protein carbonylation. Chem. Res. Toxicol. 2011;24:1411–1419. doi: 10.1021/tx200169n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.O'Donnell V. B., Chumley P. H., Hogg N., Bloodsworth A., Darley-Usmar V. M., Freeman B. A. Nitric oxide inhibition of lipid peroxidation: kinetics of reaction with lipid peroxyl radicals and comparison with α-tocopherol. Biochemistry. 1997;36:15216–15223. doi: 10.1021/bi971891z. [DOI] [PubMed] [Google Scholar]
- 50.Waddington E. I., Croft K. D., Sienuarine K., Latham B., Puddey I. B. Fatty acid oxidation products in human atherosclerotic plaque: an analysis of clinical and histopathological correlates. Atherosclerosis. 2003;167:111–120. doi: 10.1016/s0021-9150(02)00391-x. [DOI] [PubMed] [Google Scholar]
- 51.Qiu H., Gabrielsen A., Agardh H. E., Wan M., Wetterholm A., Wong C. H., Hedin U., Swedenborg J., Hansson G. K., Samuelsson B., et al. Expression of 5-lipoxygenase and leukotriene A4 hydrolase in human atherosclerotic lesions correlates with symptoms of plaque instability. Proc. Natl. Acad. Sci. U.S.A. 2006;103:8161–8166. doi: 10.1073/pnas.0602414103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cipollone F. COX-2 and prostaglandins in atherosclerosis. Lupus. 2005;14:756–759. doi: 10.1191/0961203305lu2215oa. [DOI] [PubMed] [Google Scholar]
- 53.Stocker R., Keaney J. F., Jr Role of oxidative modifications in atherosclerosis. Physiol. Rev. 2004;84:1381–1478. doi: 10.1152/physrev.00047.2003. [DOI] [PubMed] [Google Scholar]
- 54.Reeder B. J., Svistunenko D. A., Cooper C. E., Wilson M. T. The radical and redox chemistry of myoglobin and hemoglobin: from in vitro studies to human pathology. Antioxid. Redox Signaling. 2004;6:954–966. doi: 10.1089/ars.2004.6.954. [DOI] [PubMed] [Google Scholar]
- 55.Grunwald E. W., Richards M. P. Mechanisms of heme protein-mediated lipid oxidation using hemoglobin and myoglobin variants in raw and heated washed muscle. J. Agric. Food Chem. 2006;54:8271–8280. doi: 10.1021/jf061231d. [DOI] [PubMed] [Google Scholar]
- 56.Kanner J., German J. B., Kinsella J. E. Initiation of lipid peroxidation in biological systems. Crit. Rev. Food Sci. Nutr. 1987;25:317–364. doi: 10.1080/10408398709527457. [DOI] [PubMed] [Google Scholar]
- 57.Kanner J., Harel S. Lipid peroxidation and oxidation of several compounds by H2O2 activated metmyoglobin. Lipids. 1985;20:625–628. doi: 10.1007/BF02534290. [DOI] [PubMed] [Google Scholar]
- 58.Kanner J., Harel S. Initiation of membranal lipid peroxidation by activated metmyoglobin and methemoglobin. Arch. Biochem. Biophys. 1985;237:314–321. doi: 10.1016/0003-9861(85)90282-6. [DOI] [PubMed] [Google Scholar]
- 59.Galaris D., Sevanian A., Cadenas E., Hochstein P. Ferrylmyoglobin-catalyzed linoleic acid peroxidation. Arch. Biochem. Biophys. 1990;281:163–169. doi: 10.1016/0003-9861(90)90427-z. [DOI] [PubMed] [Google Scholar]
- 60.Radi R., Beckman J. S., Bush K. M., Freeman B. A. Peroxynitrite-induced membrane lipid peroxidation: the cytotoxic potential of superoxide and nitric oxide. Arch. Biochem. Biophys. 1991;288:481–487. doi: 10.1016/0003-9861(91)90224-7. [DOI] [PubMed] [Google Scholar]
- 61.Darley-Usmar V. M., Hogg N., O'Leary V. J., Wilson M. T., Moncada S. The simultaneous generation of superoxide and nitric oxide can initiate lipid peroxidation in human low density lipoprotein. Free Radical Res. Commun. 1992;17:9–20. doi: 10.3109/10715769209061085. [DOI] [PubMed] [Google Scholar]
- 62.Moncada S., Higgs E. A. The discovery of nitric oxide and its role in vascular biology. Br. J. Pharmacol. 2006;147:S193–S201. doi: 10.1038/sj.bjp.0706458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Al Ghouleh I., Khoo N. K., Knaus U. G., Griendling K. K., Touyz R. M., Thannickal V. J., Barchowsky A., Nauseef W. M., Kelley E. E., Bauer P. M., et al. Oxidases and peroxidases in cardiovascular and lung disease: new concepts in reactive oxygen species signaling. Free Radical Biol. Med. 2011;51:1271–1288. doi: 10.1016/j.freeradbiomed.2011.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Patel R. P., Diczfalusy U., Dzeletovic S., Wilson M. T., Darley-Usmar V. M. Formation of oxysterols during oxidation of low density lipoprotein by peroxynitrite, myoglobin, and copper. J. Lipid Res. 1996;37:2361–2371. [PubMed] [Google Scholar]
- 65.Moore K. P., Darley-Usmar V., Morrow J., Roberts L. J., II Formation of F2-isoprostanes during oxidation of human low-density lipoprotein and plasma by peroxynitrite. Circ. Res. 1995;77:335–341. doi: 10.1161/01.res.77.2.335. [DOI] [PubMed] [Google Scholar]
- 66.Cui T., Schopfer F. J., Zhang J., Chen K., Ichikawa T., Baker P. R., Batthyany C., Chacko B. K., Feng X., Patel R. P., et al. Nitrated fatty acids: endogenous anti-inflammatory signaling mediators. J. Biol. Chem. 2006;281:35686–35698. doi: 10.1074/jbc.M603357200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.O'Donnell V. B., Eiserich J. P., Chumley P. H., Jablonsky M. J., Krishna N. R., Kirk M., Barnes S., Darley-Usmar V. M., Freeman B. A. Nitration of unsaturated fatty acids by nitric oxide-derived reactive nitrogen species peroxynitrite, nitrous acid, nitrogen dioxide, and nitronium ion. Chem. Res. Toxicol. 1999;12:83–92. doi: 10.1021/tx980207u. [DOI] [PubMed] [Google Scholar]
- 68.Stables M. J., Gilroy D. W. Old and new generation lipid mediators in acute inflammation and resolution. Prog. Lipid Res. 2011;50:35–51. doi: 10.1016/j.plipres.2010.07.005. [DOI] [PubMed] [Google Scholar]
- 69.Ueno N., Murakami M., Tanioka T., Fujimori K., Tanabe T., Urade Y., Kudo I. Coupling between cyclooxygenase, terminal prostanoid synthase, and phospholipase A2. J. Biol. Chem. 2001;276:34918–34927. doi: 10.1074/jbc.M100429200. [DOI] [PubMed] [Google Scholar]
- 70.Garavito R. M., Mulichak A. M. The structure of mammalian cyclooxygenases. Annu. Rev. Biophys. Biomol. Struct. 2003;32:183–206. doi: 10.1146/annurev.biophys.32.110601.141906. [DOI] [PubMed] [Google Scholar]
- 71.Xie W. L., Chipman J. G., Robertson D. L., Erikson R. L., Simmons D. L. Expression of a mitogen-responsive gene encoding prostaglandin synthase is regulated by mRNA splicing. Proc. Natl. Acad. Sci. U.S.A. 1991;88:2692–2696. doi: 10.1073/pnas.88.7.2692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hinz B., Brune K. Cyclooxygenase-2: 10 years later. J. Pharmacol. Exp. Ther. 2002;300:367–375. doi: 10.1124/jpet.300.2.367. [DOI] [PubMed] [Google Scholar]
- 73.Pham H., Shafer L. M., Slice L. W. CREB-dependent cyclooxygenase-2 and microsomal prostaglandin E synthase-1 expression is mediated by protein kinase C and calcium. J. Cell. Biochem. 2006;98:1653–1666. doi: 10.1002/jcb.20899. [DOI] [PubMed] [Google Scholar]
- 74.Parfenova H., Balabanova L., Leffler C. W. Posttranslational regulation of cyclooxygenase by tyrosine phosphorylation in cerebral endothelial cells. Am. J. Physiol. 1998;274:C72–C81. doi: 10.1152/ajpcell.1998.274.1.C72. [DOI] [PubMed] [Google Scholar]
- 75.Upmacis R. K., Deeb R. S., Hajjar D. P. Regulation of prostaglandin H2 synthase activity by nitrogen oxides. Biochemistry. 1999;38:12505–12513. doi: 10.1021/bi983049e. [DOI] [PubMed] [Google Scholar]
- 76.Upmacis R. K., Deeb R. S., Hajjar D. P. Oxidative alterations of cyclooxygenase during atherogenesis. Prostaglandins Other Lipid Mediators. 2006;80:1–14. doi: 10.1016/j.prostaglandins.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 77.Schildknecht S., Bachschmid M., Ullrich V. Peroxynitrite provides the peroxide tone for PGHS-2-dependent prostacyclin synthesis in vascular smooth muscle cells. FASEB J. 2005;19:1169–1171. doi: 10.1096/fj.04-3465fje. [DOI] [PubMed] [Google Scholar]
- 78.Yamada T., Komoto J., Watanabe K., Ohmiya Y., Takusagawa F. Crystal structure and possible catalytic mechanism of microsomal prostaglandin E synthase type 2 (mPGES-2) J. Mol. Biol. 2005;348:1163–1176. doi: 10.1016/j.jmb.2005.03.035. [DOI] [PubMed] [Google Scholar]
- 79.Watanabe K., Urade Y., Mader M., Murphy C., Hayaishi O. Identification of beta-trace as prostaglandin D synthase. Biochem. Biophys. Res. Commun. 1994;203:1110–1116. doi: 10.1006/bbrc.1994.2297. [DOI] [PubMed] [Google Scholar]
- 80.Matsumoto H., Naraba H., Murakami M., Kudo I., Yamaki K., Ueno A., Oh-ishi S. Concordant induction of prostaglandin E2 synthase with cyclooxygenase-2 leads to preferred production of prostaglandin E2 over thromboxane and prostaglandin D2 in lipopolysaccharide-stimulated rat peritoneal macrophages. Biochem. Biophys. Res. Commun. 1997;230:110–114. doi: 10.1006/bbrc.1996.5894. [DOI] [PubMed] [Google Scholar]
- 81.Tokugawa Y., Kunishige I., Kubota Y., Shimoya K., Nobunaga T., Kimura T., Saji F., Murata Y., Eguchi N., Oda H., et al. Lipocalin-type prostaglandin D synthase in human male reproductive organs and seminal plasma. Biol. Reprod. 1998;58:600–607. doi: 10.1095/biolreprod58.2.600. [DOI] [PubMed] [Google Scholar]
- 82.Murakami M., Naraba H., Tanioka T., Semmyo N., Nakatani Y., Kojima F., Ikeda T., Fueki M., Ueno A., Oh S., Kudo I. Regulation of prostaglandin E2 biosynthesis by inducible membrane-associated prostaglandin E2 synthase that acts in concert with cyclooxygenase-2. J. Biol. Chem. 2000;275:32783–32792. doi: 10.1074/jbc.M003505200. [DOI] [PubMed] [Google Scholar]
- 83.Lin Y. Z., Deng H., Ruan K. H. Topology of catalytic portion of prostaglandin I2 synthase: identification by molecular modeling-guided site-specific antibodies. Arch. Biochem. Biophys. 2000;379:188–197. doi: 10.1006/abbi.2000.1892. [DOI] [PubMed] [Google Scholar]
- 84.Groeger A. L., Cipollina C., Cole M. P., Woodcock S. R., Bonacci G., Rudolph T. K., Rudolph V., Freeman B. A., Schopfer F. J. Cyclooxygenase-2 generates anti-inflammatory mediators from omega-3 fatty acids. Nat. Chem. Biol. 2010;6:433–441. doi: 10.1038/nchembio.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Musiek E. S., Brooks J. D., Joo M., Brunoldi E., Porta A., Zanoni G., Vidari G., Blackwell T. S., Montine T. J., Milne G. L., et al. Electrophilic cyclopentenone neuroprostanes are anti-inflammatory mediators formed from the peroxidation of the omega-3 polyunsaturated fatty acid docosahexaenoic acid. J. Biol. Chem. 2008;283:19927–19935. doi: 10.1074/jbc.M803625200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Leff A. R. Regulation of leukotrienes in the management of asthma: biology and clinical therapy. Annu. Rev. Med. 2001;52:1–14. doi: 10.1146/annurev.med.52.1.1. [DOI] [PubMed] [Google Scholar]
- 87.Rouzer C. A., Samuelsson B. On the nature of the 5-lipoxygenase reaction in human leukocytes: enzyme purification and requirement for multiple stimulatory factors. Proc. Natl. Acad. Sci. U.S.A. 1985;82:6040–6044. doi: 10.1073/pnas.82.18.6040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Yamamoto S. ‘Enzymatic’ lipid peroxidation: reactions of mammalian lipoxygenases. Free Radical Biol. Med. 1991;10:149–159. doi: 10.1016/0891-5849(91)90008-q. [DOI] [PubMed] [Google Scholar]
- 89.Suzuki H., Kishimoto K., Yoshimoto T., Yamamoto S., Kanai F., Ebina Y., Miyatake A., Tanabe T. Site-directed mutagenesis studies on the iron-binding domain and the determinant for the substrate oxygenation site of porcine leukocyte arachidonate 12-lipoxygenase. Biochim. Biophys. Acta. 1994;1210:308–316. doi: 10.1016/0005-2760(94)90234-8. [DOI] [PubMed] [Google Scholar]
- 90.Nelson M. J., Batt D. G., Thompson J. S., Wright S. W. Reduction of the active-site iron by potent inhibitors of lipoxygenases. J. Biol. Chem. 1991;266:8225–8229. [PubMed] [Google Scholar]
- 91.Weitzel F., Wendel A. Selenoenzymes regulate the activity of leukocyte 5-lipoxygenase via the peroxide tone. J. Biol. Chem. 1993;268:6288–6292. [PubMed] [Google Scholar]
- 92.Holzhutter H. G., Wiesner R., Rathmann J., Stosser R., Kuhn H. A kinetic model for the interaction of nitric oxide with a mammalian lipoxygenase. Eur. J. Biochem. 1997;245:608–616. doi: 10.1111/j.1432-1033.1997.00608.x. [DOI] [PubMed] [Google Scholar]
- 93.Spanbroek R., Grabner R., Lotzer K., Hildner M., Urbach A., Ruhling K., Moos M. P., Kaiser B., Cohnert T. U., Wahlers T., et al. Expanding expression of the 5-lipoxygenase pathway within the arterial wall during human atherogenesis. Proc. Natl. Acad. Sci. U.S.A. 2003;100:1238–1243. doi: 10.1073/pnas.242716099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Yokomizo T., Izumi T., Shimizu T. Leukotriene B4: metabolism and signal transduction. Arch. Biochem. Biophys. 2001;385:231–241. doi: 10.1006/abbi.2000.2168. [DOI] [PubMed] [Google Scholar]
- 95.Yokomizo T., Uozumi N., Takahashi T., Kume K., Izumi T., Shimizu T. Leukotriene A4 hydrolase and leukotriene B4 metabolism. J. Lipid Mediators Cell Signaling. 1995;12:321–332. doi: 10.1016/0929-7855(95)00041-n. [DOI] [PubMed] [Google Scholar]
- 96.Hankin J. A., Jones D. N., Murphy R. C. Covalent binding of leukotriene A4 to DNA and RNA. Chem. Res. Toxicol. 2003;16:551–561. doi: 10.1021/tx034018+. [DOI] [PubMed] [Google Scholar]
- 97.Orning L., Gierse J., Duffin K., Bild G., Krivi G., Fitzpatrick F. A. Mechanism-based inactivation of leukotriene A4 hydrolase/aminopeptidase by leukotriene A4. Mass spectrometric and kinetic characterization. J. Biol. Chem. 1992;267:22733–22739. [PubMed] [Google Scholar]
- 98.Lepley R. A., Fitzpatrick F. A. Irreversible inactivation of 5-lipoxygenase by leukotriene A4. Characterization of product inactivation with purified enzyme and intact leukocytes. J. Biol. Chem. 1994;269:2627–2631. [PubMed] [Google Scholar]
- 99.Goetzl E. J., An S., Smith W. L. Specificity of expression and effects of eicosanoid mediators in normal physiology and human diseases. FASEB J. 1995;9:1051–1058. doi: 10.1096/fasebj.9.11.7649404. [DOI] [PubMed] [Google Scholar]
- 100.Hata A. N., Breyer R. M. Pharmacology and signaling of prostaglandin receptors: multiple roles in inflammation and immune modulation. Pharm. Ther. 2004;103:147–166. doi: 10.1016/j.pharmthera.2004.06.003. [DOI] [PubMed] [Google Scholar]
- 101.Perez-Sala D., Cernuda-Morollon E., Pineda-Molina E., Canada F. J. Contribution of covalent protein modification to the antiinflammatory effects of cyclopentenone prostaglandins. Ann. N.Y. Acad. Sci. 2002;973:533–536. doi: 10.1111/j.1749-6632.2002.tb04695.x. [DOI] [PubMed] [Google Scholar]
- 102.Izumi T., Yokomizo T., Obinata H., Ogasawara H., Shimizu T. Leukotriene receptors: classification, gene expression, and signal transduction. J. Biochem. (Tokyo) 2002;132:1–6. doi: 10.1093/oxfordjournals.jbchem.a003185. [DOI] [PubMed] [Google Scholar]
- 103.Johansson A. S., Haeggstrom J. Z., Palmblad J. Commonly used leukotriene B4 receptor antagonists possess intrinsic activity as agonists in human endothelial cells: effects on calcium transients, adhesive events and mediator release. Prostaglandins Leukotrienes Essent. Fatty Acids. 2011;84:109–112. doi: 10.1016/j.plefa.2010.11.003. [DOI] [PubMed] [Google Scholar]
- 104.Ogasawara H., Ishii S., Yokomizo T., Kakinuma T., Komine M., Tamaki K., Shimizu T., Izumi T. Characterization of mouse cysteinyl leukotriene receptors mCysLT1 and mCysLT2: differential pharmacological properties and tissue distribution. J. Biol. Chem. 2002;277:18763–18768. doi: 10.1074/jbc.M109447200. [DOI] [PubMed] [Google Scholar]
- 105.Lynch K. R., O'Neill G. P., Liu Q., Im D. S., Sawyer N., Metters K. M., Coulombe N., Abramovitz M., Figueroa D. J., Zeng Z., et al. Characterization of the human cysteinyl leukotriene CysLT1 receptor. Nature. 1999;399:789–793. doi: 10.1038/21658. [DOI] [PubMed] [Google Scholar]
- 106.Hasegawa S., Ichiyama T., Hashimoto K., Suzuki Y., Hirano R., Fukano R., Furukawa S. Functional expression of cysteinyl leukotriene receptors on human platelets. Platelets. 2010;21:253–259. doi: 10.3109/09537101003615394. [DOI] [PubMed] [Google Scholar]
- 107.Takasaki J., Kamohara M., Matsumoto M., Saito T., Sugimoto T., Ohishi T., Ishii H., Ota T., Nishikawa T., Kawai Y., et al. The molecular characterization and tissue distribution of the human cysteinyl leukotriene CysLT2 receptor. Biochem. Biophys. Res. Commun. 2000;274:316–322. doi: 10.1006/bbrc.2000.3140. [DOI] [PubMed] [Google Scholar]
- 108.Diab A., Deng C., Smith J. D., Hussain R. Z., Phanavanh B., Lovett-Racke A. E., Drew P. D., Racke M. K. Peroxisome proliferator-activated receptor-γ agonist 15-deoxy-Δ(12,14)-prostaglandin J2 ameliorates experimental autoimmune encephalomyelitis. J. Immunol. 2002;168:2508–2515. doi: 10.4049/jimmunol.168.5.2508. [DOI] [PubMed] [Google Scholar]
- 109.Madrazo J. A., Kelly D. P. The PPAR trio: regulators of myocardial energy metabolism in health and disease. J. Mol. Cell. Cardiol. 2008;44:968–975. doi: 10.1016/j.yjmcc.2008.03.021. [DOI] [PubMed] [Google Scholar]
- 110.Schopfer F. J., Cipollina C., Freeman B. A. Formation and signaling actions of electrophilic lipids. Chem. Rev. 2011;111:5997–6021. doi: 10.1021/cr200131e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Bell-Parikh L. C., Ide T., Lawson J. A., McNamara P., Reilly M., FitzGerald G. A. Biosynthesis of 15-deoxy-Δ12,14-PGJ2 and the ligation of PPARγ. J. Clin. Invest. 2003;112:945–955. doi: 10.1172/JCI18012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Ide T., Egan K., Bell-Parikh L. C., FitzGerald G. A. Activation of nuclear receptors by prostaglandins. Thromb. Res. 2003;110:311–315. doi: 10.1016/s0049-3848(03)00418-3. [DOI] [PubMed] [Google Scholar]
- 113.Powell W. S. 15-Deoxy-Δ12,14-PGJ2: endogenous PPARγ ligand or minor eicosanoid degradation product? J. Clin. Invest. 2003;112:828–830. doi: 10.1172/JCI19796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Schopfer F. J., Cole M. P., Groeger A. L., Chen C. S., Khoo N. K., Woodcock S. R., Golin-Bisello F., Motanya U. N., Li Y., Zhang J., et al. Covalent peroxisome proliferator-activated receptor γ adduction by nitro-fatty acids: selective ligand activity and anti-diabetic signaling actions. J. Biol. Chem. 2010;285:12321–12333. doi: 10.1074/jbc.M109.091512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Li Y., Zhang J., Schopfer F. J., Martynowski D., Garcia-Barrio M. T., Kovach A., Suino-Powell K., Baker P. R., Freeman B. A., Chen Y. E., Xu H. E. Molecular recognition of nitrated fatty acids by PPARγ. Nat. Struct. Mol. Biol. 2008;15:865–867. doi: 10.1038/nsmb.1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Landar A., Oh J. Y., Giles N. M., Isom A., Kirk M., Barnes S., Darley-Usmar V. A sensitive method for the quantitative measurement of protein thiol modification in response to oxidative stress. Free Radical Biol. Med. 2006;40:459–468. doi: 10.1016/j.freeradbiomed.2005.08.046. [DOI] [PubMed] [Google Scholar]
- 117.Schaur R. J. Basic aspects of the biochemical reactivity of 4-hydroxynonenal. Mol. Aspects Med. 2003;24:149–159. doi: 10.1016/s0098-2997(03)00009-8. [DOI] [PubMed] [Google Scholar]
- 118.Lopachin R. M., Gavin T., Decaprio A., Barber D. S. Application of the hard and soft, acids and bases (HSAB) theory to toxicant–target interactions. Chem. Res. Toxicol. 2011 doi: 10.1021/tx2003257. doi:10.1021/tx2003257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Carlson R. M. Assessment of the propensity for covalent binding of electrophiles to biological substrates. Environ. Health Perspect. 1990;87:227–232. doi: 10.1289/ehp.9087227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Schultz T. W., Carlson R. E., Cronin M. T., Hermens J. L., Johnson R., O'Brien P. J., Roberts D. W., Siraki A., Wallace K. B., Veith G. D. A conceptual framework for predicting the toxicity of reactive chemicals: modeling soft electrophilicity. SAR QSAR Environ. Res. 2006;17:413–428. doi: 10.1080/10629360600884371. [DOI] [PubMed] [Google Scholar]
- 121.Qing W. G., Powell K. L., MacLeod M. C. Kinetics of the reaction of a potential chemopreventive agent, 2,6-dithiopurine, and its major metabolite, 2,6-dithiouric acid, with multiple classes of electrophilic toxicants. Chem. Res. Toxicol. 1996;9:1298–1304. doi: 10.1021/tx960088n. [DOI] [PubMed] [Google Scholar]
- 122.Schultz T. W., Yarbrough J. W. Trends in structure-toxicity relationships for carbonyl-containing α,β-unsaturated compounds. SAR QSAR Environ. Res. 2004;15:139–146. doi: 10.1080/10629360410001665839. [DOI] [PubMed] [Google Scholar]
- 123.Sanchez-Gomez F. J., Cernuda-Morollon E., Stamatakis K., Perez-Sala D. Protein thiol modification by 15-deoxy-Δ12,14-prostaglandin J2 addition in mesangial cells: role in the inhibition of pro-inflammatory genes. Mol. Pharmacol. 2004;66:1349–1358. doi: 10.1124/mol.104.002824. [DOI] [PubMed] [Google Scholar]
- 124.Renedo M., Gayarre J., Garcia-Dominguez C. A., Perez-Rodriguez A., Prieto A., Canada F. J., Rojas J. M., Perez-Sala D. Modification and activation of Ras proteins by electrophilic prostanoids with different structure are site-selective. Biochemistry. 2007;46:6607–6616. doi: 10.1021/bi602389p. [DOI] [PubMed] [Google Scholar]
- 125.Ricart K. C., Bolisetty S., Johnson M. S., Perez J., Agarwal A., Murphy M. P., Landar A. The permissive role of mitochondria in the induction of haem oxygenase-1 in endothelial cells. Biochem. J. 2009;419:427–436. doi: 10.1042/BJ20081350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Liu S., Hanzlik R. P. Structure–activity relationships for inhibition of papain by peptide Michael acceptors. J. Med. Chem. 1992;35:1067–1075. doi: 10.1021/jm00084a012. [DOI] [PubMed] [Google Scholar]
- 127.Liu X. W., Sok D. E. Inactivation of protein disulfide isomerase by alkylators including α,β-unsaturated aldehydes at low physiological pHs. Biol. Chem. 2004;385:633–637. doi: 10.1515/BC.2004.078. [DOI] [PubMed] [Google Scholar]
- 128.Ying J., Clavreul N., Sethuraman M., Adachi T., Cohen R. A. Thiol oxidation in signaling and response to stress: detection and quantification of physiological and pathophysiological thiol modifications. Free Radical Biol. Med. 2007;43:1099–1108. doi: 10.1016/j.freeradbiomed.2007.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Mallis R. J., Buss J. E., Thomas J. A. Oxidative modification of H-ras: S-thiolation and S-nitrosylation of reactive cysteines. Biochem. J. 2001;355:145–153. doi: 10.1042/0264-6021:3550145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Martinez-Ruiz A., Lamas S. Detection and proteomic identification of S-nitrosylated proteins in endothelial cells. Arch. Biochem. Biophys. 2004;423:192–199. doi: 10.1016/j.abb.2003.12.006. [DOI] [PubMed] [Google Scholar]
- 131.Saurin A. T., Neubert H., Brennan J. P., Eaton P. Widespread sulfenic acid formation in tissues in response to hydrogen peroxide. Proc. Natl. Acad. Sci. U.S.A. 2004;101:17982–17987. doi: 10.1073/pnas.0404762101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Biswas S., Chida A. S., Rahman I. Redox modifications of protein-thiols: emerging roles in cell signaling. Biochem. Pharmacol. 2006;71:551–564. doi: 10.1016/j.bcp.2005.10.044. [DOI] [PubMed] [Google Scholar]
- 133.Moran L. K., Gutteridge J. M., Quinlan G. J. Thiols in cellular redox signalling and control. Curr. Med. Chem. 2001;8:763–772. doi: 10.2174/0929867013372904. [DOI] [PubMed] [Google Scholar]
- 134.Lopachin R. M., Barber D. S., Geohagen B. C., Gavin T., He D., Das S. Structure–toxicity analysis of type-2 alkenes: in vitro neurotoxicity. Toxicol. Sci. 2007;95:136–146. doi: 10.1093/toxsci/kfl127. [DOI] [PubMed] [Google Scholar]
- 135.Chan K., Poon R., O'Brien P. J. Application of structure–activity relationships to investigate the molecular mechanisms of hepatocyte toxicity and electrophilic reactivity of α,β-unsaturated aldehydes. J. Appl. Toxicol. 2008;28:1027–1039. doi: 10.1002/jat.1369. [DOI] [PubMed] [Google Scholar]
- 136.Levonen A. L., Landar A., Ramachandran A., Ceaser E. K., Dickinson D. A., Zanoni G., Morrow J. D., Darley-Usmar V. M. Cellular mechanisms of redox cell signalling: role of cysteine modification in controlling antioxidant defences in response to electrophilic lipid oxidation products. Biochem. J. 2004;378:373–382. doi: 10.1042/BJ20031049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Hong F., Freeman M. L., Liebler D. C. Identification of sensor cysteines in human Keap1 modified by the cancer chemopreventive agent sulforaphane. Chem. Res. Toxicol. 2005;18:1917–1926. doi: 10.1021/tx0502138. [DOI] [PubMed] [Google Scholar]
- 138.Zhang B., Shi Z., Duncan D. T., Prodduturi N., Marnett L. J., Liebler D. C. Relating protein adduction to gene expression changes: a systems approach. Mol. Biosyst. 2011;7:2118–2127. doi: 10.1039/c1mb05014a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Kim H. Y., Tallman K. A., Liebler D. C., Porter N. A. An azido-biotin reagent for use in the isolation of protein adducts of lipid-derived electrophiles by streptavidin catch and photorelease. Mol. Cell. Proteomics. 2009;8:2080–2089. doi: 10.1074/mcp.M900121-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Landar A., Shiva S., Levonen A. L., Oh J. Y., Zaragoza C., Johnson M. S., Darley-Usmar V. M. Induction of the permeability transition and cytochrome c release by 15-deoxy-Δ12,14-prostaglandin J2 in mitochondria. Biochem. J. 2006;394:185–195. doi: 10.1042/BJ20051259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Roberts L. J., II, Fessel J. P. The biochemistry of the isoprostane, neuroprostane, and isofuran pathways of lipid peroxidation. Chem. Phys. Lipids. 2004;128:173–186. doi: 10.1016/j.chemphyslip.2003.09.016. [DOI] [PubMed] [Google Scholar]
- 142.Trachootham D., Lu W., Ogasawara M. A., Nilsa R. D., Huang P. Redox regulation of cell survival. Antioxid. Redox Signaling. 2008;10:1343–1374. doi: 10.1089/ars.2007.1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Satoh T., Lipton S. A. Redox regulation of neuronal survival mediated by electrophilic compounds. Trends Neurosci. 2007;30:37–45. doi: 10.1016/j.tins.2006.11.004. [DOI] [PubMed] [Google Scholar]
- 144.Claiborne A., Yeh J. I., Mallett T. C., Luba J., Crane E. J., III, Charrier V., Parsonage D. Protein-sulfenic acids: diverse roles for an unlikely player in enzyme catalysis and redox regulation. Biochemistry. 1999;38:15407–15416. doi: 10.1021/bi992025k. [DOI] [PubMed] [Google Scholar]
- 145.McMahon M., Lamont D. J., Beattie K. A., Hayes J. D. Keap1 perceives stress via three sensors for the endogenous signaling molecules nitric oxide, zinc, and alkenals. Proc. Natl. Acad. Sci. U.S.A. 2010;107:18838–18843. doi: 10.1073/pnas.1007387107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Murphy M. P. Mitochondrial thiols in antioxidant protection and redox signalling: distinct roles for glutathionylation and other thiol modifications. Antioxid. Redox Signaling. 2011 doi: 10.1089/ars.2011.4289. doi:10.1089/ars.2011.4289. [DOI] [PubMed] [Google Scholar]
- 147.Wong H. L., Liebler D. C. Mitochondrial protein targets of thiol-reactive electrophiles. Chem. Res. Toxicol. 2008;21:796–804. doi: 10.1021/tx700433m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Meng T. C., Fukada T., Tonks N. K. Reversible oxidation and inactivation of protein tyrosine phosphatases in vivo. Mol. Cell. 2002;9:387–399. doi: 10.1016/s1097-2765(02)00445-8. [DOI] [PubMed] [Google Scholar]
- 149.Diers A. R., Dranka B. P., Ricart K. C., Oh J. Y., Johnson M. S., Zhou F., Pallero M. A., Bodenstine T. M., Murphy-Ullrich J. E., Welch D. R., Landar A. Modulation of mammary cancer cell migration by 15-deoxy-Δ(12,14)-prostaglandin J2: implications for anti-metastatic therapy. Biochem. J. 2010;430:69–78. doi: 10.1042/BJ20091193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Levonen A. L., Dickinson D. A., Moellering D. R., Mulcahy R. T., Forman H. J., Darley-Usmar V. M. Biphasic effects of 15-deoxy-Δ(12,14)-prostaglandin J2 on glutathione induction and apoptosis in human endothelial cells. Arterioscler. Thromb. Vasc. Biol. 2001;21:1846–1851. doi: 10.1161/hq1101.098488. [DOI] [PubMed] [Google Scholar]
- 151.Requejo R., Hurd T. R., Costa N. J., Murphy M. P. Cysteine residues exposed on protein surfaces are the dominant intramitochondrial thiol and may protect against oxidative damage. FEBS J. 2010;277:1465–1480. doi: 10.1111/j.1742-4658.2010.07576.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Sheehan D., Meade G., Foley V. M., Dowd C. A. Structure, function and evolution of glutathione transferases: implications for classification of non-mammalian members of an ancient enzyme superfamily. Biochem. J. 2001;360:1–16. doi: 10.1042/0264-6021:3600001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Niture S. K., Jaiswal A. K. Inhibitor of Nrf2 (INrf2 or Keap1) degrades Bcl-xL via phosphoglycerate mutase 5 and controls cellular apoptosis. J. Biol. Chem. 2011;286:44542–44556. doi: 10.1074/jbc.M111.275073. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 154.Landar A., Zmijewski J. W., Dickinson D. A., Le Goffe C., Johnson M. S., Milne G. L., Zanoni G., Vidari G., Morrow J. D., Darley-Usmar V. M. Interaction of electrophilic lipid oxidation products with mitochondria in endothelial cells and formation of reactive oxygen species. Am. J. Physiol. Heart Circ. Physiol. 2006;290:H1777–H1787. doi: 10.1152/ajpheart.01087.2005. [DOI] [PubMed] [Google Scholar]
- 155.Kondo M., Shibata T., Kumagai T., Osawa T., Shibata N., Kobayashi M., Sasaki S., Iwata M., Noguchi N., Uchida K. 15-Deoxy-Δ(12,14)-prostaglandin J2: the endogenous electrophile that induces neuronal apoptosis. Proc. Natl. Acad. Sci. U.S.A. 2002;99:7367–7372. doi: 10.1073/pnas.112212599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Diers A. R., Higdon A. N., Ricart K. C., Johnson M. S., Agarwal A., Kalyanaraman B., Landar A., Darley-Usmar V. M. Mitochondrial targeting of the electrophilic lipid 15-deoxy-Δ12,14-prostaglandin J2 increases apoptotic efficacy via redox cell signalling mechanisms. Biochem. J. 2010;426:31–41. doi: 10.1042/BJ20091293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Kagan V. E., Tyurin V. A., Jiang J., Tyurina Y. Y., Ritov V. B., Amoscato A. A., Osipov A. N., Belikova N. A., Kapralov A. A., Kini V., et al. Cytochrome c acts as a cardiolipin oxygenase required for release of proapoptotic factors. Nat. Chem. Biol. 2005;1:223–232. doi: 10.1038/nchembio727. [DOI] [PubMed] [Google Scholar]
- 158.Morimitsu Y., Nakagawa Y., Hayashi K., Fujii H., Kumagai T., Nakamura Y., Osawa T., Horio F., Itoh K., Iida K., et al. A sulforaphane analogue that potently activates the Nrf2-dependent detoxification pathway. J. Biol. Chem. 2002;277:3456–3463. doi: 10.1074/jbc.M110244200. [DOI] [PubMed] [Google Scholar]
- 159.Li Y., Cao Z., Zhu H. Upregulation of endogenous antioxidants and phase 2 enzymes by the red wine polyphenol, resveratrol in cultured aortic smooth muscle cells leads to cytoprotection against oxidative and electrophilic stress. Pharmacol. Res. 2006;53:6–15. doi: 10.1016/j.phrs.2005.08.002. [DOI] [PubMed] [Google Scholar]
- 160.Kansanen E., Jyrkkanen H. K., Volger O. L., Leinonen H., Kivela A. M., Hakkinen S. K., Woodcock S. R., Schopfer F. J., Horrevoets A. J., Yla-Herttuala S., et al. Nrf2-dependent and -independent responses to nitro-fatty acids in human endothelial cells: identification of heat shock response as the major pathway activated by nitro-oleic acid. J. Biol. Chem. 2009;284:33233–33241. doi: 10.1074/jbc.M109.064873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Powers M. V., Workman P. Inhibitors of the heat shock response: biology and pharmacology. FEBS Lett. 2007;581:3758–3769. doi: 10.1016/j.febslet.2007.05.040. [DOI] [PubMed] [Google Scholar]
- 162.Mosser D. D., Theodorakis N. G., Morimoto R. I. Coordinate changes in heat shock element-binding activity and HSP70 gene transcription rates in human cells. Mol. Cell. Biol. 1988;8:4736–4744. doi: 10.1128/mcb.8.11.4736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Shi Y., Mosser D. D., Morimoto R. I. Molecular chaperones as HSF1-specific transcriptional repressors. Genes Dev. 1998;12:654–666. doi: 10.1101/gad.12.5.654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Fujimoto M., Nakai A. The heat shock factor family and adaptation to proteotoxic stress. FEBS J. 2010;277:4112–4125. doi: 10.1111/j.1742-4658.2010.07827.x. [DOI] [PubMed] [Google Scholar]
- 165.Bocci V., Aldinucci C., Mosci F., Carraro F., Valacchi G. Ozonation of human blood induces a remarkable upregulation of heme oxygenase-1 and heat stress protein-70. Mediators Inflammation. 2007;2007:26785. doi: 10.1155/2007/26785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Yu Z., Yang X., Wang K. Metal ions induced heat shock protein response by elevating superoxide anion level in HeLa cells transformed by HSE-SEAP reporter gene. Toxicology. 2006;223:1–8. doi: 10.1016/j.tox.2006.02.021. [DOI] [PubMed] [Google Scholar]
- 167.Barrett W. C., DeGnore J. P., Konig S., Fales H. M., Keng Y. F., Zhang Z. Y., Yim M. B., Chock P. B. Regulation of PTP1B via glutathionylation of the active site cysteine 215. Biochemistry. 1999;38:6699–6705. doi: 10.1021/bi990240v. [DOI] [PubMed] [Google Scholar]
- 168.Murry C. E., Jennings R. B., Reimer K. A. Preconditioning with ischemia: a delay of lethal cell injury in ischemic myocardium. Circulation. 1986;74:1124–1136. doi: 10.1161/01.cir.74.5.1124. [DOI] [PubMed] [Google Scholar]
- 169.Reimer K. A., Murry C. E., Jennings R. B. Cardiac adaptation to ischemia. Ischemic preconditioning increases myocardial tolerance to subsequent ischemic episodes. Circulation. 1990;82:2266–2268. doi: 10.1161/01.cir.82.6.2266. [DOI] [PubMed] [Google Scholar]
- 170.Starkopf J., Andreasen T. V., Bugge E., Ytrehus K. Lipid peroxidation, arachidonic acid and products of the lipoxygenase pathway in ischaemic preconditioning of rat heart. Cardiovasc. Res. 1998;37:66–75. doi: 10.1016/s0008-6363(97)00240-x. [DOI] [PubMed] [Google Scholar]
- 171.Tritto I., Ambrosio G. Role of oxidants in the signaling pathway of preconditioning. Antiox. Redox Signaling. 2001;3:3–10. doi: 10.1089/152308601750100425. [DOI] [PubMed] [Google Scholar]
- 172.Xu B., Gao X., Xu J., Lei S., Xia Z. Y., Xu Y., Xia Z. Ischemic postconditioning attenuates lung reperfusion injury and reduces systemic proinflammatory cytokine release via heme oxygenase 1. J. Surg. Res. 2011;166:e157–e164. doi: 10.1016/j.jss.2010.11.902. [DOI] [PubMed] [Google Scholar]
- 173.Patel A., van de Poll M. C., Greve J. W., Buurman W. A., Fearon K. C., McNally S. J., Harrison E. M., Ross J. A., Garden O. J., Dejong C. H., Wigmore S. J. Early stress protein gene expression in a human model of ischemic preconditioning. Transplantation. 2004;78:1479–1487. doi: 10.1097/01.tp.0000144182.27897.1e. [DOI] [PubMed] [Google Scholar]
- 174.Jancso G., Cserepes B., Gasz B., Benko L., Borsiczky B., Ferenc A., Kurthy M., Racz B., Lantos J., Gal J., et al. Expression and protective role of heme oxygenase-1 in delayed myocardial preconditioning. Ann. N.Y. Acad. Sci. 2007;1095:251–261. doi: 10.1196/annals.1397.029. [DOI] [PubMed] [Google Scholar]
- 175.Zou Y., Zhu W., Sakamoto M., Qin Y., Akazawa H., Toko H., Mizukami M., Takeda N., Minamino T., Takano H., et al. Heat shock transcription factor 1 protects cardiomyocytes from ischemia/reperfusion injury. Circulation. 2003;108:3024–3030. doi: 10.1161/01.CIR.0000101923.54751.77. [DOI] [PubMed] [Google Scholar]
- 176.Liu Y., Kato H., Nakata N., Kogure K. Temporal profile of heat shock protein 70 synthesis in ischemic tolerance induced by preconditioning ischemia in rat hippocampus. Neuroscience. 1993;56:921–927. doi: 10.1016/0306-4522(93)90138-6. [DOI] [PubMed] [Google Scholar]
- 177.Tsuruma T., Yagihashi A., Matsuno T., Zou X. M., Asanuma K., Sasaki K., Hirata K. The heat-shock protein 70 family reduces ischemia/reperfusion injury in small intestine. Transplant. Proc. 1996;28:2629–2630. [PubMed] [Google Scholar]
- 178.Kume M., Yamamoto Y., Saad S., Gomi T., Kimoto S., Shimabukuro T., Yagi T., Nakagami M., Takada Y., Morimoto T., Yamaoka Y. Ischemic preconditioning of the liver in rats: implications of heat shock protein induction to increase tolerance of ischemia–reperfusion injury. J. Lab. Clin. Med. 1996;128:251–258. doi: 10.1016/s0022-2143(96)90026-8. [DOI] [PubMed] [Google Scholar]
- 179.Pagel P. S. Induction of heat shock protein 70 and preconditioning by sevoflurane: a potent protective interaction against myocardial ischemia–reperfusion injury. Anesth. Analg. 2008;107:742–745. doi: 10.1213/ane.0b013e31817f6d40. [DOI] [PubMed] [Google Scholar]
- 180.Straus D. S., Glass C. K. Cyclopentenone prostaglandins: new insights on biological activities and cellular targets. Med. Res. Rev. 2001;21:185–210. doi: 10.1002/med.1006. [DOI] [PubMed] [Google Scholar]
- 181.Watanabe N., Zmijewski J. W., Takabe W., Umezu-Goto M., Le Goffe C., Sekine A., Landar A., Watanabe A., Aoki J., Arai H., et al. Activation of mitogen-activated protein kinases by lysophosphatidylcholine-induced mitochondrial reactive oxygen species generation in endothelial cells. Am. J. Pathol. 2006;168:1737–1748. doi: 10.2353/ajpath.2006.050648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Hill B. G., Haberzettl P., Ahmed Y., Srivastava S., Bhatnagar A. Unsaturated lipid peroxidation-derived aldehydes activate autophagy in vascular smooth-muscle cells. Biochem. J. 2008;410:525–534. doi: 10.1042/BJ20071063. [DOI] [PubMed] [Google Scholar]
- 183.Cernuda-Morollon E., Pineda-Molina E., Canada F. J., Perez-Sala D. 15-Deoxy-Δ12,14-prostaglandin J2 inhibition of NF-κB-DNA binding through covalent modification of the p50 subunit. J. Biol. Chem. 2001;276:35530–35536. doi: 10.1074/jbc.M104518200. [DOI] [PubMed] [Google Scholar]
- 184.Oliva J. L., Perez-Sala D., Castrillo A., Martinez N., Canada F. J., Bosca L., Rojas J. M. The cyclopentenone 15-deoxy-Δ12,14-prostaglandin J2 binds to and activates H-Ras. Proc. Natl. Acad. Sci. U.S.A. 2003;100:4772–4777. doi: 10.1073/pnas.0735842100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Shibata T., Yamada T., Ishii T., Kumazawa S., Nakamura H., Masutani H., Yodoi J., Uchida K. Thioredoxin as a molecular target of cyclopentenone prostaglandins. J. Biol. Chem. 2003;278:26046–26054. doi: 10.1074/jbc.M303690200. [DOI] [PubMed] [Google Scholar]
- 186.Cassidy P. B., Edes K., Nelson C. C., Parsawar K., Fitzpatrick F. A., Moos P. J. Thioredoxin reductase is required for the inactivation of tumor suppressor p53 and for apoptosis induced by endogenous electrophiles. Carcinogenesis. 2006;27:2538–2549. doi: 10.1093/carcin/bgl111. [DOI] [PubMed] [Google Scholar]
- 187.Perez-Sala D., Cernuda-Morollon E., Canada F. J. Molecular basis for the direct inhibition of AP-1 DNA binding by 15-deoxy-Δ12,14-prostaglandin J2. J. Biol. Chem. 2003;278:51251–51260. doi: 10.1074/jbc.M309409200. [DOI] [PubMed] [Google Scholar]
- 188.Gayarre J., Sanchez D., Sanchez-Gomez F. J., Terron M. C., Llorca O., Perez-Sala D. Addition of electrophilic lipids to actin alters filament structure. Biochem. Biophys. Res. Commun. 2006;349:1387–1393. doi: 10.1016/j.bbrc.2006.09.005. [DOI] [PubMed] [Google Scholar]
- 189.Aldini G., Dalle-Donne I., Vistoli G., Maffei Facino R., Carini M. Covalent modification of actin by 4-hydroxy-trans-2-nonenal (HNE): LC-ESI–MS/MS evidence for Cys374 Michael adduction. J. Mass Spectrom. 2005;40:946–954. doi: 10.1002/jms.872. [DOI] [PubMed] [Google Scholar]
- 190.Ishii T., Sakurai T., Usami H., Uchida K. Oxidative modification of proteasome: identification of an oxidation-sensitive subunit in 26 S proteasome. Biochemistry. 2005;44:13893–13901. doi: 10.1021/bi051336u. [DOI] [PubMed] [Google Scholar]
- 191.Waku T., Shiraki T., Oyama T., Fujimoto Y., Maebara K., Kamiya N., Jingami H., Morikawa K. Structural insight into PPARγ activation through covalent modification with endogenous fatty acids. J. Mol. Biol. 2009;385:188–199. doi: 10.1016/j.jmb.2008.10.039. [DOI] [PubMed] [Google Scholar]
- 192.Vieira H. L., Belzacq A. S., Haouzi D., Bernassola F., Cohen I., Jacotot E., Ferri K. F., El Hamel C., Bartle L. M., Melino G., et al. The adenine nucleotide translocator: a target of nitric oxide, peroxynitrite, and 4-hydroxynonenal. Oncogene. 2001;20:4305–4316. doi: 10.1038/sj.onc.1204575. [DOI] [PubMed] [Google Scholar]
- 193.Yarian C. S., Rebrin I., Sohal R. S. Aconitase and ATP synthase are targets of malondialdehyde modification and undergo an age-related decrease in activity in mouse heart mitochondria. Biochem. Biophys. Res. Commun. 2005;330:151–156. doi: 10.1016/j.bbrc.2005.02.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Kaplan P., Tatarkova Z., Racay P., Lehotsky J., Pavlikova M., Dobrota D. Oxidative modifications of cardiac mitochondria and inhibition of cytochrome c oxidase activity by 4-hydroxynonenal. Redox Rep. 2007;12:211–218. doi: 10.1179/135100007X200308. [DOI] [PubMed] [Google Scholar]
- 195.Chen J., Robinson N. C., Schenker S., Frosto T. A., Henderson G. I. Formation of 4-hydroxynonenal adducts with cytochrome c oxidase in rats following short-term ethanol intake. Hepatology. 1999;29:1792–1798. doi: 10.1002/hep.510290611. [DOI] [PubMed] [Google Scholar]
- 196.Chen J., Schenker S., Frosto T. A., Henderson G. I. Inhibition of cytochrome c oxidase activity by 4-hydroxynonenal (HNE). Role of HNE adduct formation with the enzyme subunits. Biochim. Biophys. Acta. 1998;1380:336–344. doi: 10.1016/s0304-4165(98)00002-6. [DOI] [PubMed] [Google Scholar]