Abstract
Selective small-molecule inhibitors represent powerful tools for the dissection of complex biological processes. ESI (eeyarestatin I) is a novel modulator of ER (endoplasmic reticulum) function. In the present study, we show that in addition to acutely inhibiting ERAD (ER-associated degradation), ESI causes production of mislocalized polypeptides that are ubiquitinated and degraded. Unexpectedly, our results suggest that these non-translocated polypeptides promote activation of the UPR (unfolded protein response), and indeed we can recapitulate UPR activation with an alternative and quite distinct inhibitor of ER translocation. These results suggest that the accumulation of non-translocated proteins in the cytosol may represent a novel mechanism that contributes to UPR activation.
Keywords: eeyarestatin, endoplasmic reticulum, non-translocated protein, Sec61, unfolded protein response
Abbreviations: CHX, cycloheximide; cpd A, translocation inhibitor compound A; DMEM, Dulbecco's modified Eagle's medium; DTT, dithiothreitol; eIF2α, eukaryotic initiation factor 2α; EndoH, endoglycosidase H; ER, endoplasmic reticulum; EDEM-1, ER degradation-enhancing α-mannosidase-like 1; ERAD, ER-associated degradation; ES, eeyarestatin; HEK, human embryonic kidney; IP, immunoprecipitation; IRE1, inositol-requiring enzyme 1; PDI, protein disulfide-isomerase; PERK, PKR (double-stranded-RNA-dependent protein kinase)-like ER kinase; PS2, proteasome inhibitor 2; RT, reverse transcription; TCRα, T-cell receptor α subunit; UPR, unfolded protein response; UPS, ubiquitin–proteasome system; XBP1, X-box-binding protein 1
INTRODUCTION
In eukaryotic cells, the ER (endoplasmic reticulum) is the major site for the synthesis of membrane and secretory proteins. These are co-translationally translocated across or integrated into the membrane through a proteinaceous channel, the Sec61 complex [1]. Once exposed to the ER lumen, polypeptides are folded, assembled and may undergo post-translational modification, such as N-linked glycosylation, prior to being transported to their site of function. However, folding is an intrinsically error-prone process, and polypeptides may also fail to attain their native state due to errors in translation, genetic mutation, absence of partner subunits or unfavourable environmental conditions. Such misfolded or misassembled polypeptides are potentially harmful, and in order to prevent their deployment within the cell, they are retained in the ER by a quality control system [2]. To avoid congestion and potential interference with productive protein folding, misfolded proteins must be cleared from the ER, and this is achieved via a process known as ERAD (ER-associated degradation) [3]. ERAD is mediated by the cytosolic proteasome, which means that proteins selected for ERAD must be retrotranslocated back across the ER membrane into the cytosol prior to degradation [3]. Ubiquitin ligases at the cytosolic face of the ER membrane ubiquitinate the ERAD substrate, directing it to the UPS (ubiquitin–proteasome system). Ubiquitination also attracts p97, an ATPase that facilitates extraction of many ERAD substrates from the ER membrane. The ERAD substrate is then deglycosylated and deubiquitinated, before entering the catalytic core of the proteasome where it is broken down into small peptides [3].
Situations that perturb the balance between protein synthesis, folding and degradation pathways (termed proteostasis [4]), can lead to a build-up of misfolded proteins in the ER, causing ER stress. Eukaryotic cells possess a signalling network known as the UPR (unfolded protein response), to detect and manage such imbalances in ER proteostasis [5]. UPR signalling pathways function to reduce the rate of protein synthesis and up-regulate the expression of chaperones and ERAD factors, thereby reducing levels of misfolded proteins in the ER and restoring homoeostasis. If homoeostasis is not re-established, prolonged activation of the UPR can initiate programmed cell death [5]. Disturbances in ER proteostasis and UPR signalling are implicated in the pathogenesis of a diverse range of diseases, including diabetes mellitus, cancer, neurodegeneration, and bone and joint diseases, as well as classical ER protein folding diseases such as cystic fibrosis [6]. As a result, there is considerable interest in the therapeutic potential of selective small molecules to manipulate these pathways in order to ameliorate disease [4].
The small molecules ESI (eeyarestatin I) and ESII (eeyarestatin II) are ERAD inhibitors, first identified in a high-throughput screen based on stabilization of GFP (green fluorescent protein)-tagged MHC I heavy chain, which is degraded via an ERAD-like pathway in cells expressing the viral protein US11 [7]. The degradation of a conventional ERAD substrate, TCRα (T-cell receptor α subunit), was also inhibited by ESI [7], and it was suggested that the compound acts at an early stage in the ERAD pathway, prior to retrotranslocation. Subsequent work provided evidence that a later stage in ERAD, namely the p97 ATPase and an associated deubquitinating activity, may also be inhibited by ESI [8,9]. ESI has also been shown to activate the UPR and induce cell death [10]. Intriguingly, the cytotoxic activity of ESI appears to be particularly effective against cancer cells, and synergizes with that of bortezomib (Velcade), a proteasome inhibitor with proven anticancer properties [10]. Together with accumulating evidence that UPR activation plays a role in a variety of cancers [11], these observations highlight a potential therapeutic application for ESI-related compounds. Both activation of the UPR and perturbation of cellular ubiquitin homoeostasis have been proposed to cause ESI cytotoxicity [10]. However, the underlying molecular mechanism(s) through which ESI activates the UPR and exerts its effects is not yet clear.
We have previously shown that ESI inhibits Sec61-mediated translocation of a range of proteins across the ER membrane, albeit to various degrees depending on the precursor studied [12]. The consequences of ESI-dependent perturbations of ER translocation in cells are not known, raising the possibility that inhibition of translocation contributes to the potent cellular effects of ESI. In the present study we show that ESI treatment results in the production of mislocalized membrane and secretory proteins, and demonstrate that accumulation of these species correlates with activation of the UPR. Using an alternative inhibitor of translocation, we provide evidence that accumulation of non-translocated polypeptides in the cytosol promotes activation of the UPR.
EXPERIMENTAL
Materials
The M2 anti-FLAG antibody was from Sigma–Aldrich. The P4D1 anti-ubiquitin antibody was from Santa Cruz Biotechnology. Derlin-1 and PDI (protein disulfide-isomerase) polyclonal rabbit antisera were made by Eurogentec. Anti-p97 antiserum was from Professor Bernhard Dobberstein (Center for Molecular Biology, University of Heidelberg, Heidelberg, Germany). Plasmids encoding TCRα were from Professor Ron Kopito (Department of Biology, Stanford University, Stanford, CA, U.S.A.) and Professor Emmanuel Wiertz (Leiden University, Leiden, The Netherlands). Bovine opsin cDNA was subcloned from the pZEO vector into pcDNA5/FRT/TO (Invitrogen). The preprolactin–mCherry construct was from Professor Viki Allan (Faculty of Life Sciences, University of Manchester, Manchester, U.K.).
Synthesis of ESI, ESII and ESR35
Synthesis of the ESs (see the Supplementary methods section at http://www.BiochemJ.org/bj/442/bj4420639add.htm) commenced with nitroso-compound 1 (see Supplementary Figure S1 at http://www.BiochemJ.org/bj/442/bj4420639add.htm), which was prepared in a straightforward fashion by the reaction of iso-butene with nitrosyl chloride (generated in situ from iso-amyl nitrite and HCl) [13]. The reaction of compound 1 with methyl glycinate proceeded smoothly to give a very good yield of oximino ester 2 [14]. Exposure of compound 2 to two equivalents of 4-chlorophenyl isocyanate or 1-naphthyl isocyanate gave the bis-adducts 3a and 3b [3], which were subsequently converted into their corresponding acyl hydrazides 4a and 4b by treatment with an aqueous methanolic solution of hydrazine (acyl hydrazide 4a is the compound also referred to as inactive analogue ESR35). Finally, condensation of hydrazides 4a and 4b with E-3-(5-nitro-2-furyl)acrylaldehyde in methanol gave the corresponding ESs (ESI and ESII) in excellent yields. The double-bond geometry of the final products was indicated as E by the magnitude of the vicinal coupling constant between the olefinic protons (15.8 Hz for ESI and 15.4 Hz for ESII).
Cell culture and transfection
HeLa cells were cultured in DMEM (Dulbecco's modified Eagle's medium) containing 10% (v/v) FBS (fetal bovine serum) and 2 mM L-glutamine. Cells were transfected using Lipofectamine™ 2000 (Invitrogen) and used in experiments after 24 h.
Treatment with compounds
Stock solutions (10 mM) of ESR35, ESII, ESI, cpd A (translocation inhibitor compound A) and PS2 (proteasome inhibitor 2; benzyloxycarbonyl-Leu-Leu-Phe-aldehyde; Calbiochem) were made up in DMSO. ESR35, ESII and ESI were added to a final concentration of 8 μM, PS2 and cpd A to 10 μM, and CHX (cycloheximide) to 100 μg/ml, then cells were incubated at 37°C.
Metabolic labelling and IP (immunoprecipitation)
Cells were incubated in DMEM lacking methionine and cysteine (Invitrogen) for 20 min, and were labelled with 22 μCi/ml EasyTag [35S]Met/Cys (PerkinElmer) for 40 min at 37°C. Cells were washed twice in PBS and either harvested immediately or chased in DMEM supplemented with 2.5 mM unlabelled methionine/cysteine plus the indicated compounds. Cells were harvested in 50 μl of ice-cold IP buffer (140 mM NaCl, 10 mM Tris/HCl, pH 7.5, 1 mM EDTA and 1% Triton X-100) plus protease inhibitor cocktail (Sigma). For IP, lysates were denatured in 1% SDS, then 5 vol. of IP buffer containing 10 mM non-radioactive methionine/cysteine, 1 mM PMSF and 4% pansorbin (Calbiochem) was added. Samples were rotated for 1 h at 37°C, centrifuged at 15000 g for 10 min, and the supernatant was incubated overnight at 4°C with an anti-FLAG antibody. Immune complexes were collected on Protein A–Sepharose beads, washed with IP buffer, eluted in reducing SDS/PAGE sample buffer and resolved by SDS/PAGE. Radioactive gels were analysed by FLA-3000 phosphorimaging (Fuji) and quantified using AIDA v3.52 software (Raytest Isotopenmessgerate).
Western blotting
To detect polyubiquitinated material, cells cultured in 12-well dishes were lysed in 50 μl of IP buffer and then denatured in SDS/PAGE sample buffer at 70°C for 10 min. Half of the sample was run on SDS/10% PAGE gels, analysed by blotting with an anti-ubiquitin antibody and then visualized by enhanced chemiluminescence.
XBP1 (X-box-binding protein 1) splicing
Total RNA was extracted using TRIzol® reagent (Invitrogen), and first-strand cDNA was synthesized with an oligo-p(dT)15 primer. cDNA was used as a template for PCR using primers flanking the XBP-1 intron (FWD, 5′-ACAGCGCTTGGGGATGGATG-3′; REV, 5′-TGACTGGGTCCAAGTTGTCC-3′), and PCR products were analysed on 2% agarose gels.
Fluorescence microscopy
HeLa cells incubated with 8 μM ESI in the medium for 0–8 h were fixed in methanol for 4 min at −20°C, then probed with primary antibodies against PDI and fluorophore-conjugated secondary antibodies (Molecular Probes). For visualization of preprolactin–mCherry, HeLa cells transfected using JetPEI reagent (Peqlab), were incubated with 100 μg/ml CHX for 3 h at 37°C. Cells were washed three times for 1 min with 1 ml of PBS, then treated with 8 μM ESI, 10 μM PS2 or 8 μM ESI and 10 μM PS2 for 6 h. Following treatment, cells were washed in PBS and fixed in 3% paraformaldehyde in PBS for 25 min at room temperature (21–23°C). Images were obtained using an Olympus BX60 upright microscope with a MicroMax cooled charge-coupled device camera (Roper Scientific) driven by Metamorph software (Universal Imaging Corporation).
Subcellular fractionation and EndoH (endoglycosidase H) treatment
Cells in 10 cm dishes were rinsed twice in PBS, harvested by scraping and resuspended in 100 μl of hypotonic buffer (20 mM Hepes, pH 7.6, 5 mM KCl, 2.5 mM EDTA and 1 mM PMSF) on ice. Cells were sonicated for three 10 s pulses in a sonicating waterbath. Lysates were centrifuged at 50000 g for 30 min at 4°C. The membrane pellet was resuspended in 100 μl of 100 mM Na2CO3, pH 11.5, incubated on ice for 20 min, then centrifuged at 100000 g for 1 h at 4°C. The supernatant was removed, and the pellet was solubilized in 100 μl of IP buffer for 10 min at 4°C, followed by a final centrifugation step at 100000 g for 1 h at 4°C. The supernatant was removed and the Triton X-100-insoluble pellet was resuspended in IP buffer containing 0.1% SDS. Opsin was immunoprecipitated from the carbonate supernatant, Triton X-100 supernatant and Triton X-100-insoluble fractions, and was resolved by SDS/PAGE. Where indicated, immunoprecipitated material was incubated with 500 units of EndoH (New England Biolabs) for 2 h at 37°C.
RESULTS
ESI induces accumulation of polyubiquitinated proteins and induces ER stress
ESI, ESII and a related compound ESR35, designed to provide a negative control treatment (cf. [9]), were synthesized according to the scheme outlined in Supplementary Figure S1. The inhibitory effect of ESI on the ERAD pathway was confirmed in two different human cell lines (see Supplementary Figure S2 at http://www.BiochemJ.org/bj/442/bj4420639add.htm). Notably, we found that acute (<1 h) treatment of cells with ESI inhibited degradation of the model substrate TCRα (Supplementary Figure S2A), showing that the action of ESI on ERAD is rapid and does not require prolonged treatment. In contrast, ESII treatment did not reproducibly delay TCRα degradation, and ESR35 had no detectable inhibitory effect (Supplementary Figure S2). ESI treatment also resulted in the appearance of substantial amounts of higher-molecular-mass forms of the model ERAD substrate (Supplementary Figure S2B). Less pronounced high-molecular-mass species were also visible in cells treated with a proteasome inhibitor (PS2) and at the start of the chase in untreated cells (Supplementary Figure S2B), suggesting that they might represent polyubiquitinated forms of TCRα en route to degradation. Consistent with this interpretation, the total levels of polyubiquitinated proteins detected in cells were also markedly increased upon ESI treatment (Figure 1A, lane 4, and Supplementary Figure S2C). A similar, although less pronounced, effect was observed following treatment with ESII and PS2 (Figure 1A, lanes 3 and 5), whereas ESR35 had no discernable effect (Figure 1A, lane 2). These high-molecular-mass polyubiquitin conjugates accumulated with time, and became apparent after 1 h of exposure to ESI (Figure 1B). Increased levels of polyubiquitinated material following ESI treatment have been observed previously and have been proposed to reflect an inhibition of deubiquitination [8].
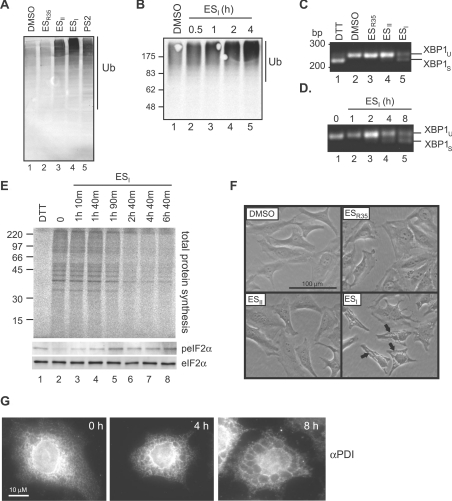
Figure 1. Cellular effects of ES compounds.
(A) HEK cells were treated for 8 h with DMSO, 8 μM ESR35, ESII or ESI, or 10 μM PS2, and lysates were analysed by blotting with an anti-ubiquitin (Ub) antibody. (B) HeLa cells were treated with DMSO for 4 h or 8 μM ESI for the time indicated, and lysates were analysed by blotting with an anti-ubiquitin antibody. (C) HeLa cells were treated with 2 mM DTT for 2 h, DMSO, or 8 μM ESR35, ESII or ESI for 8 h. Xbp1 mRNA splicing was determined by RT–PCR. PCR products corresponding to unspliced (u) and spliced (s) Xbp1 mRNA are indicated. (D) HeLa cells were treated with 8 μM ESI for the time indicated and Xbp1 splicing was determined as above. (E) HeLa cells treated with 10 mM DTT for 30 min or 8 μM ESI for the time indicated were labelled with [35S]Met/Cys for 40 min. Lysates were analysed by phosphorimaging (top panel), or blotting with anti-peIF2α (phospho-eIF2α) or anti-eIF2α antibodies (bottom two panels). In (B and E) the molecular mass in kDa is indicated on the left-hand side. (F) HeLa cells were treated with DMSO or 8 μM ESR35, ESII or ESI for 8 h then visualized by phase-contrast microscopy. Arrows indicate vacuolar structures. (G) HeLa cells were treated with 8 μM ESI for the time indicated, fixed, stained with anti-PDI and fluorescently labelled secondary antibodies, and examined by fluorescence microscopy.
Consistent with previous work [10], we also found that ESI induced ER stress in cultured cells. Splicing of Xbp1 mRNA, an early event in the IRE1 (inositol-requiring enzyme 1) branch of the UPR can be measured using RT (reverse transcription)–PCR. A single PCR product, representing the unspliced mRNA, was observed in DMSO-treated cells (Figure 1C, lane 2). A faster-migrating product, representing spliced Xbp1 mRNA, was apparent in cells treated with the reducing agent DTT (dithothreitol) to perturb oxidative protein folding (Figure 1C, lane 1). ESI also induced Xbp1 mRNA splicing (Figure 1C, lane 5), showing that treatment with this compound resulted in activation of IRE1. In contrast, no spliced Xbp1 was detected in cells treated with ESII or ESR35 (Figure 1C, lanes 3 and 4), consistent with the minor effect of these compounds on ERAD and polyubiquitination (Supplementary Figure S2 and Figure 1A). The response to ESI did not require prolonged treatment with ESI, and spliced Xbp1 was evident after 4 h (Figure 1D). Activation of the PERK [PKR (double-stranded-RNA-dependent protein kinase)-like ER kinase] branch of the UPR results in phosphorylation of eIF2α (eukaryotic initiation factor 2α), resulting in a decrease in the rate of protein synthesis under conditions of ER stress, as observed following treatment with the reducing agent DTT to perturb oxidative protein folding (Figure 1E, cf. lanes 1 and 2). Phosphorylation of eIF2α occurred rapidly upon ESI treatment (Figure 1E, bottom two panels), and this effect correlated with a decrease in total protein synthesis (Figure 1E, top panel, lanes 3–8), showing that the PERK branch of the UPR was also activated. Similar effects of ESI were also observed in HEK (human embryonic kidney)-293 cells (see Supplementary Figure S3 at http://www.BiochemJ.org/bj/442/bj4420639add.htm). Interestingly, ESI treatment resulted in a more pronounced phosphorylation of eIF2α and greater translational attenuation in HEK cells than in HeLa cells (Supplementary Figure S3), suggesting that sensitivity to ESI can vary between cell types (cf. [10]).
Our recent work suggests that ESI delays certain intracellular trafficking pathways [13], and we therefore examined the effect of the ES compounds on cell morphology. Interestingly, large vacuole-like structures developed within 4–8 h of treatment with ESI (Figure 1F, arrows), and these were also apparent by immunofluorescence microscopy using an antibody specific for ER-localized proteins (Figure 1G), suggesting that these structures were derived from the ER. ESR35 had no obvious effect on cell morphology (Figure 1F), and although no effect of ESII was visible within 8 h (Figure 1F), prolonged treatment (12–16 h) did cause the appearance of such vacuoles (results not shown). A similar swelling of the ER occurs during paraptosis [14], a non-apoptotic form of cell death, and has also been observed under conditions of chronic ER stress [15,16]. Hence these changes in ER structure may reflect ESI-induced ER stress and/or commitment to cell death.
Inhibition of translocation promotes ER stress
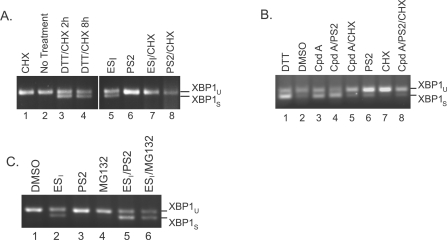
The mechanisms through which ESI induces ER stress are not known, although inhibition of ERAD and/or perturbation of ubiquitin homoeostasis could potentially play a role [10]. Indeed, inhibition of the proteasome is able to induce activation of the UPR in a variety of cell types (e.g. [17]). Interestingly, however, treatment of HeLa cells with PS2 for 8 h failed to induce Xbp1 mRNA splicing (Figure 2A, lane 3), despite causing a clear build-up of polyubiquitinated species and profound inhibition of ERAD (Figure 1A and Supplementary Figure S2). Similarly, Xbp1 splicing was not observed in cells treated with an alternative proteasome inhibitor MG132 (Figure 2A, lane 4), or in response to a small molecule that inhibits protein deubiquitination (NSC687852) (Figure 2A, lane 5), and causes a pronounced accumulation of polyubiquitin adducts [18,19] (see Supplementary Figure S4A at http://www.BiochemJ.org/bj/442/bj4420639add.htm). Thus neither blockade of ERAD nor perturbation of global ubiquitin homoeostasis (using a variety of different inhibitors) activated the UPR in HeLa cells over this time course. In contrast, parallel treatment with ESI induced a robust ER stress response (Figure 2A, lane 2). These results are significant as they suggest that ESI does not induce ER stress solely by blocking degradation of ERAD substrates or disrupting the UPS [10]. We therefore considered the alternative possibility that the ability of ESI to activate UPR was related to its ability to perturb protein translocation [12]. In order to test this hypothesis, we used a second small-molecule inhibitor of co-translational translocation (referred to in the present study as cpd A), derived from a naturally occurring cyclic depsipeptide [20]. Like ESI, the cyclodepsipeptide inhibitors cause a wide-ranging inhibition of translocation when used at low micromolar concentrations [21,22], but are structurally distinct and target a different stage of Sec61-mediated protein translocation across the ER membrane [20,23]. Strikingly, treatment of HeLa cells with this translocation inhibitor also induced Xbp1 mRNA splicing on a timescale comparable with ESI treatment (cf. Figures 1C and 2B). In addition, cpd A induced phosphorylation of eIF2α and reduced the rate of translation (Figures 2C and 2D), demonstrating that both the IRE1 and PERK branches of the UPR were activated. This effect of the cyclodepsipeptide inhibitors has not previously been reported and supports the view that inhibiting Sec61-mediated translocation promotes ER stress. This effect could potentially be caused by the reduced biogenesis of specific ER factors such as chaperones, some of which are known to be relatively short-lived [24,25]. However, the global inhibition of protein synthesis using CHX did not induce Xbp1 splicing (Figure 2E, lane 5), suggesting that a lack of ER components resulting from a defect in co-translational translocation is unlikely to underlie the ability of ESI and cpd A to activate the UPR. These observations raised the possibility that the induction of ER stress by these translocation inhibitors might instead be related to the production of non-translocated forms of membrane and secretory proteins. In order to address this issue further, we examined the fate of various ER-targeted proteins in the presence of ESI.
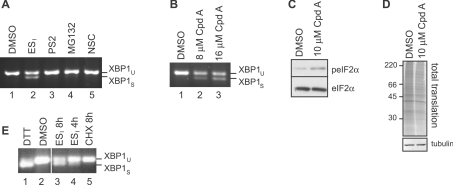
Figure 2. Inhibition of translocation promotes ER stress.
(A) HeLa cells were incubated with DMSO, 8 μM ESI, 10 μM PS2, 10 μM MG132 or 0.4 μM of the deubiquitinase inhibitor NSC687852 (NSC) for 8 h. Xbp1 mRNA splicing was determined by RT–PCR. Products corresponding to unspliced (u) and spliced (s) Xbp1 mRNA are indicated. (B) HeLa cells were incubated with DMSO or the indicated concentration of the translocation inhibitor cpd A for 8 h, and Xbp1 splicing was determined as above. (C) HeLa cells were incubated with DMSO or 10 μM cpd A for 8 h, and lysates were analysed by blotting with anti-peIF2α (phospho-eIF2α) or anti-eIF2α antibodies. (D) HeLa cells were incubated with DMSO or 10 μM cpd A for 8 h, pulse-labelled with [35S]Met/Cys for 40 min, and lysates were analysed by phosphorimaging (top panel) or blotting with an anti-tubulin antibody (bottom panel). Molecular mass in kDa is indicated on the left-hand side. (E) HeLa cells were incubated with 2 mM DTT for 2 h, DMSO for 8 h, 8 μM ESI for 4 or 8 h, or 100 μg/ml CHX for 8 h. Xbp1 splicing was determined as above.
ESI induces mislocalization and degradation of membrane and secretory proteins in cultured cells
Addition of N-linked glycans to polypeptides occurs within the ER lumen and was thus used to provide a readout of TCRα insertion into the ER (cf. [12,22]). In DMSO- and ESR35-treated cells, FLAG–TCRα was efficiently N-glycosylated and migrated as a species of approximately 48 kDa (Figure 3A, lanes 1 and 2). As observed previously [12], levels of the N-glycosylated form of TCRα were reduced in ESI-treated cells (Figure 3A, lane 4). In addition, a more rapidly migrating species of the size expected for the unglycosylated protein was found to accumulate after 8 h of treatment with ESI (Figure 3A, lane 4). A similar, although less pronounced, effect was seen upon ESII treatment (Figure 3A, lane 3). These observations indicate that ESI treatment results in the production of a form of TCRα lacking N-glycosylation.
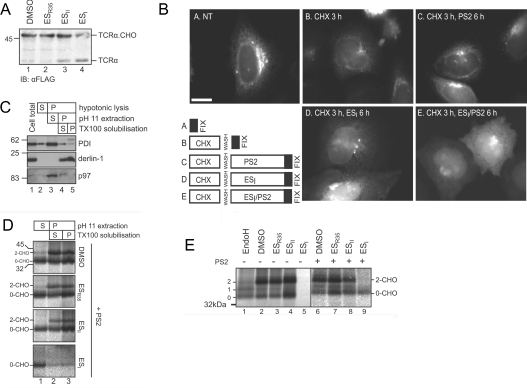
Figure 3. ESI causes mislocalization of membrane and secretory proteins.
(A) HeLa cells expressing FLAG–TCRα were treated with DMSO or 8 μM ESR35, ESII or ESI for 8 h. Cell lysates were analysed by blotting with an anti-FLAG antibody. The position of glycosylated (TCRα.CHO) and unglycosylated (TCRα) FLAG–TCRα are indicated. (B) HeLa cells expressing ppl–mCherry were left untreated (NT) or incubated with CHX for 3 h, then washed and treated with 8 μM ESI, 10 μM PS2, or 8 μM ESI and 10 μM PS2 for 6 h. Cells were fixed in methanol and visualized by fluorescence microscopy. Scale bar, 10 μm. (C) HeLa cells were lysed in hypotonic buffer, total membranes were isolated and extracted with NaCO3, pH 11, and the resulting membrane fraction was solubilized with Triton X-100 (TX100). Samples of the total lysate (cell total), and the supernatant (S) and pellet (P) fractions at each step were analysed by blotting with anti-PDI, anti-derlin-1 and anti-p97 antibodies. In (A and C), the molecular mass in kDa is indicated on the left-hand side. (D) HeLa cells transiently expressing opsin were treated for 1 h with DMSO or 8 μM ESR35, ESII or ESI, plus 10 μM PS2, then labelled with [35S]Met/Cys for 40 min. Cells were fractionated as in (B), and opsin was immunoprecipitated from each fraction and analysed by phosphorimaging. (E) HeLa cells transiently expressing opsin were treated with DMSO or 8 μM ESR35, ESII or ESI for 1 h in the absence (−) or presence (+) of 10 μM PS2, then labelled with [35S]Met/Cys. Opsin was immunoprecipitated and analysed by phosphorimaging.
We next examined the effect of ESI on the subcellular localization of a chimaeric secretory protein composed of fluorescent mCherry fused to the ER-targeting sequence of preprolactin (ppl–mCherry). In untreated cells, ppl–mCherry was predominantly localized to the secretory pathway, as demonstrated by the reticular and ribbon-like pattern of fluorescence, typical of the ER and Golgi apparatus respectively (Figure 3B, panel A). Cells were then treated with CHX to prevent further protein synthesis and allow existing protein to exit the ER. After washing to remove CHX, cells were incubated with ESI, PS2 or a combination of ESI and PS2 for a further 6 h, prior to imaging. This treatment regime (outlined in Figure 3B) allowed us to determine the localization of ppl–mCherry synthesized in the presence of ESI. ppl–mCherry fluorescence was considerably lower after 3 h of CHX treatment, confirming that much of the protein had been secreted from the cells (Figure 3B, panel B). In cells treated with PS2 alone following CHX treatment, the ppl–mCherry distribution was similar to that in untreated cells (Figure 3B, cf. panels A and C), suggesting that the majority of the protein synthesized under conditions of proteasome inhibition was correctly targeted to the ER. A different pattern of fluorescence was seen in cells treated with ESI (Figure 3B, panel D). Under these conditions, the reticular distribution of ppl–mCherry was replaced by more diffuse fluorescence distributed throughout the cell, suggesting that a proportion of the newly synthesized protein had not entered the ER, but was instead located in the cytosol. A variable amount of mCherry fluorescent protein was also observed in punctate structures adjacent to the nucleus (Figure 3B, panels D and E). Co-treatment with PS2 resulted in a much clearer accumulation of fluorescent protein in the cytosol (Figure 3B, panel E), suggesting that ppl–mCherry which failed to enter the ER in the presence of ESI was degraded via the proteasome.
To validate these data, we took a complementary biochemical approach to explore the fate of a polytopic membrane protein synthesized in the presence of ESI. Membranes isolated from HeLa cells transiently expressing opsin were extracted with sodium carbonate, pH>11, to remove peripherally associated proteins [26]. Membrane-integrated proteins are resistant to such treatment, and were re-isolated by centrifugation, then solubilized in Triton X-100. This method allowed separation of the ER lumenal chaperone PDI (Figure 3C, lane 3), from the integral ER membrane protein derlin-1, which remained in the membrane-associated pellet after carbonate extraction (Figure 3C, lanes 4 and 5). p97, which is peripherally associated with the ER membrane, was removed in the carbonate supernatant (Figure 3C, lane 3), confirming that this treatment effectively strips non-integral proteins from the ER membrane. Next, cells treated with DMSO, ESI, ESII or ESR35 for 1 h were pulse-labelled in the continued presence of the compounds plus PS2, prior to carbonate extraction and opsin IP. In untreated cells, the large majority of the radiolabelled opsin was present in the carbonate-resistant pellet (Figure 3D, lanes 2 and 3, DMSO), demonstrating that it was stably integrated into the ER membrane. A smaller proportion of the radiolabelled non-glycosylated opsin was extracted in the carbonate supernatant (Figure 3D, lane 1, DMSO), probably reflecting protein that failed to be correctly integrated into the ER membrane. A similar distribution of opsin was seen in ESII- and ESR35-treated cells (Figure 3D, ESII and ESR35). In the presence of ESI, however, radiolabelled opsin was almost exclusively found in the carbonate supernatant and was non-glycosylated (Figure 3D, lane 1, ESI). Virtually no radiolabelled opsin could be detected in the sodium carbonate-extracted pellet and there was no indication of any glycosylated protein chains (Figure 3D, lanes 2 and 3, ESI), suggesting that ESI inhibited the integration of opsin into the ER membrane, leading to production of non-glycosylated protein. Notably, the population of non-glycosylated opsin was only readily apparent in HeLa cells when PS2 was included during the pulse-labelling period (Figure 3E, cf. lanes 5 and 9), showing that proteins prevented from entering the ER upon ESI treatment are normally subjected to proteasomal degradation.
Taken together, these results support a model whereby an inhibition of translocation by ESI leads to the production of mislocalized membrane and secretory proteins in cultured cells. The non-translocated forms of ppl–mCherry and opsin that failed to enter the ER in the presence of ESI were degraded via the proteasome, suggesting that such polypeptides are targeted to the UPS. We therefore postulated that the polyubiquitinated species observed in ESI-treated cells (Figure 1A; see also [8]), may include a variety of polypeptides that had failed to translocate correctly across the ER membrane and were en route to proteasomal degradation. Indeed, treatment of cells with ESII, which has only a minor effect on protein translocation [12], caused a less pronounced accumulation of polyubiquitinated material (Figure 1A and Supplementary Figure 2C). Furthermore, we found that treatment with the cyclodepsipeptide translocation inhibitor cpd A also resulted in some accumulation of high-molecular-mass polyubiquitinated material (Figure 4A, lane 3). This previously unreported effect of cpd A provides additional evidence that inhibition of ER translocation in live cells promotes a build-up of polyubiquitin conjugates. Significantly, co-treatment of cells with CHX completely prevented the accumulation of polyubiquitin adducts caused by both cpd A and ESI (Figure 4A, lane 4, and Figure 4B, lanes 4 and 5). This requirement for ongoing protein synthesis suggests that these species represent proteins newly synthesized in the presence of the translocation inhibitors. In contrast, PS2 still caused accumulation of polyubiquitinated proteins in the presence of CHX (Figure 4B, lanes 8 and 9), suggesting that a significant proportion of these adducts were derived from pre-existing proteins. We also found that treatment with ESI led to the appearance of higher-molecular-mass forms of EDEM-1 (ER degradation-enhancing α-mannosidase-like 1), a model ER-resident protein (Figure 4C, lane 2). These species migrated as diffuse bands near the top of the polyacrylamide gel, comparable with the polyubiquitin conjugates detected with anti-ubiquitin antibodies (Figure 4B). However, they were not observed in cells treated with PS2 (Figure 4C, lane 3), showing that they were not generated solely as a consequence of inhibiting proteasomal degradation. Accumulation of these higher-molecular-mass forms of EDEM-1 was prevented by co-treatment with CHX (Figure 4C, lane 6), showing that they represent polypeptides synthesized in the presence of ESI and not an ESI-dependent modification of a pre-existing pool of EDEM-1 (cf. [27]). On the basis of these results, we conclude that the polyubiquitinated species induced by ESI and cpd A most probably include non-translocated membrane and secretory proteins targeted for proteasomal degradation. Supporting this interpretation, endogenous ESI-induced polyubiquitin conjugates were eliminated by cells following addition of CHX and a subsequent chase period (Figure 4D). However, since PS2 rapidly induces accumulation of polyubiquitinated proteins even in the presence of CHX (Supplementary Figure S4B), it was not possible to demonstrate that the loss of ESI-induced polyubiquitin conjugates was proteasome-dependent. Therefore we cannot exclude the possibility that this effect is the result of deubiquitination of the substrates rather than degradation.
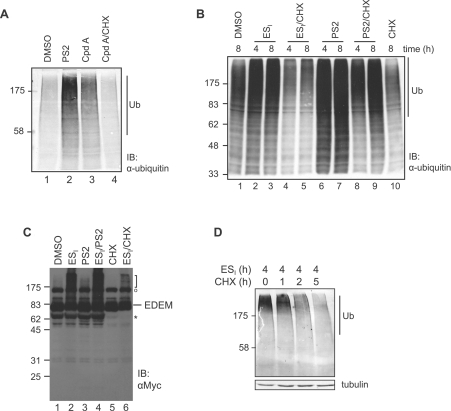
Figure 4. Translocation inhibitors induce polyubiquitination of newly synthesized proteins.
(A) HeLa cells were treated with DMSO, 10 μM PS2, 10 μM translocation inhibitor cpd A, or 10 μM cpd A and 100 μg/ml CHX for 8 h. Lysates were analysed by blotting with an anti-ubiquitin (Ub) antibody. (B) HeLa cells were treated with DMSO, 8 μM ESI or 10 μM PS2 in the presence or absence of 100 μg/ml CHX for 4–8 h. Lysates were analysed by blotting with the anti-ubiquitin antibody. (C) HeLa cells expressing EDEM1–Myc were treated with DMSO, 8 μM ESI, 10 μM PS2, 8 μM ESI and 10 μM PS2, 100 μg/ml CHX, or 8 μM ESI and 100 μg/ml CHX for 8 h. Lysates were analysed by blotting with an anti-Myc antibody. The position of full-length EDEM is indicated, and a Myc-cross-reacting species is indicated by O. Additional smaller product(s) marked * disappeared when cells were treated with CHX and may represent unstable fragments of EDEM–Myc. (D) HeLa cells were incubated with 8 μM ESI for 4 h, washed and then incubated with 10 μg/ml CHX for 1–5 h to prevent further protein synthesis. Equal quantities of cell lysate were analysed by blotting with the anti-ubiquitin antibody. IB, immunoblot. The molecular mass in kDa is indicated on the left-hand side.
Inhibition of protein translocation contributes to the cellular actions of ESI
Non-translocated membrane and secretory proteins have potentially harmful properties, such as hydrophobic transmembrane domains and/or uncleaved signal peptides. Therefore we wondered whether the accumulation of such aberrant polypeptides could contribute to the cellular effects of ESI and cpd A. To address this issue, we exploited the observation that non-translocated polypeptides accumulated to a greater extent when proteasome activity is inhibited (Figure 3), whereas their production is effectively blocked by CHX (Figure 4). This allowed us to examine whether the ability of ESI and cpd A to induce ER stress was altered by conditions that promoted or inhibited the accumulation of non-translocated polypeptides. When protein synthesis was prevented by co-treatment with CHX, both Xbp1 mRNA splicing and the ER vacuolization induced by ESI were completely abolished (Figure 5A, lanes 5 and 7, and Supplementary Figure 4C). Similarly, CHX also substantially diminished Xbp1 splicing in response to cpd A (Figure 5B, lanes 3 and 5). In contrast, the ability of DTT to induce Xbp1 splicing was not prevented by CHX (Figure 5A, lanes 3 and 4), demonstrating that activation of this pathway is not dependent on the continued import of cargo proteins into the ER, nor mechanistically on protein synthesis itself. Thus activation of the UPR by both ESI and cpd A requires ongoing translation. This is an important finding as it suggests that polypeptides synthesized in the presence of these compounds are responsible for their ability to induce ER stress. In contrast, combinatorial treatment with ESI and proteasome inhibitors more effectively induced Xbp1 splicing (Figure 5C, lanes 5 and 6, and Supplementary Figure S4D) and enhanced ER vacuolization (Supplementary Figure S4E) when compared with ESI alone. The ability of cpd A to induce Xbp1 splicing was also increased by proteasome inhibition (Figure 5B, lanes 3 and 4). Thus conditions that inhibit the efficient degradation of non-translocated proteins (Figure 3) enhance the ability of ESI and cpd A to activate the UPR, supporting the hypothesis that accumulation of polyubiquitinated non-translocated proteins promotes activation of the UPR. Consistent with this interpretation, the accumulation of polyubiquitinated proteins in ESI-treated cells correlated well with the induction of ER stress (cf. Figures 1B and 1E).
Figure 5. Correlation between accumulation of mistranslocated polypeptides and induction of ER stress.
(A) HeLa cells were treated with 100 μg/ml CHX for 8 h, 2 mM DTT and CHX for 2 h or 8 h, or with 8 μM ESI or 10 μM PS2 in the presence or absence of 100 μg/ml CHX for 8 h. Xbp1 splicing was determined by RT–PCR. PCR products corresponding to unspliced (u) and spliced (s) Xbp1 mRNA are indicated. (B) HeLa cells were treated with DMSO, 2 mM DTT, or the indicated combination of 10 μM translocation inhibitor cpd A, 10 μM PS2 and 100 μg/ml CHX for 8 h. Xbp1 splicing was determined by RT–PCR. (C) HeLa cells were incubated with DMSO, 8 μM ESI, 10 μM PS2, 10 μM MG132, 8 μM ESI and 10 μM PS2, or 8 μM ESI and 10 μM PS2 for 8 h. Xbp1 splicing was determined by RT–PCR.
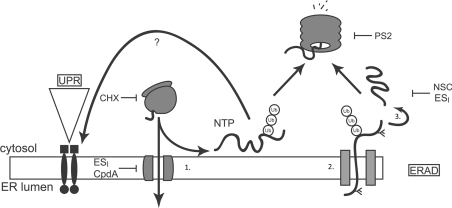
On the basis of these findings, we propose a working model for the cellular actions of ESI. In this model (Figure 6), a proportion of nascent membrane and secretory proteins fail to enter the ER in the presence of ESI or cpd A, and are instead polyubiquitinated and may be subjected to proteasomal degradation. Since ESI and cpd A only activate Xbp1 splicing if protein synthesis is ongoing, we suggest that accumulation of these non-translocated proteins in the cytosol contributes to activation of the ER stress response.
Figure 6. Proposed model for cellular actions of ESI.
Proteins possessing an ER signal sequence are targeted to the ER and translocated through the Sec61 translocon (1). Properly folded proteins exit the ER and move along the secretory pathway. Misfolded proteins are retained by the ER quality control machinery (ER-QC) (2), handed over to the retrotranslocation machinery, polyubiquitinated and retrotranslocated (2). The retrotranslocated protein is deubiquitinated and deglycosylated (3) before being degraded by the proteasome. Accumulation of misfolded proteins in the ER lumen activates UPR signalling. ESI inhibits translocation of proteins across the ER membrane, leading to the production of non-translocated proteins (NTP) at the cytosolic face of the ER membrane. These aberrant species are degraded by the UPS and also activate the UPR through an unknown mechanism. ESI may also inhibit one or more steps of the ERAD pathway, including retrotranslocation or deubiquitination. Also shown are the points of action of CHX, PS2 and NSC687852 (NSC), a deubiquitinase inhibitor that induces accumulation of polyubiquitin in cells [18].
DISCUSSION
ER protein folding and degradation pathways have emerged as exciting targets for the treatment of a diverse array of diseases, from cancer and stroke to classical protein folding diseases. As a result, there is much interest in the therapeutic potential of selective small molecules to modulate ER protein folding homoeostasis and the ER stress response. In the present study we describe the preparative synthesis and cellular activity of the ERAD inhibitor ESI and two related compounds ESII and ESR35. We confirm that ESI inhibits ERAD of TCRα, causes accumulation of polyubiquitinated proteins, and rapidly induces ER stress. We also show that ESI reduces the efficiency of protein translocation at the ER in cultured cells, and provide evidence that membrane and secretory proteins that fail to enter the ER in the presence of ESI are subjected to proteasomal degradation. Using a second small-molecule inhibitor of co-translational translocation, we examine the consequences of reduced translocation efficiency in cultured cells. Intriguingly, our results suggest that the accumulation of non-translocated membrane and secretory proteins promotes activation of the ER stress response.
Originally identified as an ERAD inhibitor, ESI was subsequently found to impair Sec61-mediated protein translocation at the ER for a range of substrates [12]. In the present study, we have employed different techniques to show that ESI also perturbs the entry of a variety of proteins into the ER of living cells. Thus the N-glycosylation of TCRα, the membrane integration and N-glycosylation of opsin, and the ER localization of ppl–mCherry were all impaired by treatment of cells with ESI. In the presence of ESI, non-glycosylated TCRα, non-integrated opsin and non-ER-localized ppl–mCherry continued to be synthesized, and could be observed in ESI-treated cells. However, the amount of these non-translocated species that accumulated in cells following ESI treatment was somewhat variable. These variations may reflect differences in the response of different cell types to ESI treatment, for example greater translational attenuation in HEK-293 cells, but could also be due to differences in the degree of translocation inhibition caused by ESI. Indeed, non-translocated forms of membrane proteins, including TCRα and MHC class I heavy chain, were not observed in previous studies [7,9], suggesting that ESI-mediated inhibition of translocation is incomplete and/or can be influenced by experimental variables such as the duration of drug treatment.
The non-translocated species we observed in ESI-treated cells were labile, and inhibitor studies indicate that they are degraded by the proteasome. Similarly, the cytosolic form of the VCAM1 (vascular cell adhesion molecule 1) precursor generated in the presence of translocation inhibitor cpd A is also normally degraded via a proteasome-dependent pathway [20]. Other non-translocated proteins generated under various conditions of reduced translocation efficiency are also rapidly removed by the proteasome [22,28,29]. Taken together, these observations strongly support the existence of an efficient cellular quality control system that functions to eliminate proteins that fail to be properly translocated across the ER membrane. Indeed, recent work has begun to identify the cytosolic machinery involved in such a pathway [30]. Non-translocated species will typically expose hydrophobic transmembrane domains and/or uncleaved signal peptides, and it is easy to envisage that their accumulation in the cytosol might be particularly harmful. Indeed, aberrant interactions of non-translocated prion protein have been proposed to contribute to neurodegeneration in prion diseases [31,32]. Our results suggest that non-translocated polypeptides may also participate in cellular dysfunction by promoting ER stress. Thus we find that both ESI and another small-molecule inhibitor of co-translational translocation induce accumulation of polyubiquitinated proteins and activate the UPR. Strikingly, co-treatment with CHX, which blocks accumulation of polyubiquitin adducts, also abolishes the ER stress response induced by these compounds. This requirement for ongoing translation suggests that it is actually the polyubiquitinated non-translocated polypeptides synthesized in the presence of the inhibitors that promote activation of the UPR. In support of this model, the ability of both ESI and cpd A to induce ER stress is enhanced under conditions of proteasome inhibition that prevent efficient degradation of non-translocated proteins. Our working model (Figure 6) is consistent with the known actions of ESI and cpd A, and would also explain the previously observed synergy between ESI and the proteasome inhibitor bortezomib [10]. Although we cannot formally exclude the possibility that ESI and cpd A activate the UPR by ‘off-target’ (ie. non-translocon) effects, the facts that they are structurally distinct and inhibit different stages of co-translational translocation suggest that this is unlikely. Furthermore, the observation that CHX prevents UPR activation in response to ESI and cpd A, but does not affect DTT-induced UPR, suggests that the translocation inhibitors do not induce ER stress through a direct effect on ER chaperones, ERAD machinery or UPR components [7,27,33]. In addition to perturbing ER translocation, ESI also inhibits p97 and an associated deubiquitinase activity [9,10]. Interfering with p97 function is known to inhibit ERAD and can also induce swelling of the ER (cf. [34]). It thus seems likely that reduced p97 function may also contribute to the pronounced ER stress phenotype induced by ESI. However, the ability of a second small-molecule inhibitor of the Sec61 translocon to recapitulate many features of ESI-induced UPR activation supports a model whereby products of failed translocation promote ER stress.
Since the UPR is typically triggered by the presence of misfolded proteins in the ER lumen, it is not immediately obvious how the generation of non-translocated polypeptides in the cytosol might promote this pathway (Figure 6). One potential explanation is that polyubiquitination of these species reduces the availability of free ubiquitin, thereby indirectly inhibiting removal of misfolded proteins from the ER and inducing the UPR. This would be consistent with the suggestion that inhibition of deubiquitination by ESI causes a global ubiquitin deficiency [10]. However, several distinct inhibitors of the UPS, namely PS2, MG132 and NSC687852, failed to induce Xbp1 mRNA splicing despite causing comparable accumulation of polyubiqitinated material. On this basis, we believe that perturbation of ubiquitin homoeostasis is not the underlying cause of ESI-induced UPR. Non-translocated proteins could also disrupt protein homoeostasis in the cytosol, for example by sequestering molecular chaperones. Cytosolic chaperones such as Hsp70 (heat-shock protein 70) play an important role in the biogenesis of some membrane proteins [35], and therefore interfering with their function may perturb folding at the ER and promote ER stress. Alternatively, the accumulation of non-translocated polypeptides, which have a high propensity to aggregate and initiate non-native interactions, at the cytosolic face of the ER could potentially interfere with the removal of misfolded proteins from the ER. This would result in a build-up of misfolded proteins in the lumen and activation of the UPR, and is consistent with the suggestion that ESI inhibits retrotranslocation [7]. Indeed, certain misfolded cytosolic proteins have been shown to engage in inappropriate interactions with cytosolically oriented components of the ERAD machinery, thereby inhibiting retrotranslocation and activating the UPR [36,37]. We speculate that perturbation of such an early stage in the ERAD process may have a particularly severe effect on levels of misfolded proteins in the ER lumen and hence ER stress. In contrast, inhibition of the proteasome does not always block retrotranslocation, and several ERAD substrates can still exit the ER when the proteasome is inhibited [38–41]. Alternatively, non-translocated polypeptides may also activate UPR signalling through some other, as yet undefined, mechanism [33,42].
The possibility that modulating protein translocation at the ER might have therapeutic potential is further supported by the finding that apratoxin A, a cyclodepsipeptide with anticancer properties [43], inhibits co-translational translocation [44]. In this case, depletion of cancer-associated transmembrane receptors was proposed to underlie the cytotoxic activity of apratoxin A towards cancer cells [44]. Thus the results of the present study further extend the prospect that inhibitors of ER translocation may offer novel therapeutic opportunities in the future, and underline the potential for using small-molecule inhibitors to understand the consequence of defects in membrane translocation events at the cellular level [45].
Online data
AUTHOR CONTRIBUTION
Eileithyia Swanton and Stephen High conceived and supervised the project. Eileithyia Swanton wrote the paper. Craig McKibbin, Alina Mares, Peristera Roboti, Marjo Puumalainen and Karolina Lesiak-Mieczkowska performed the experiments, analysed the data, and contributed to preparation of the Figures and paper. Anna Callan performed pilot experiments. Michela Piacenti and Helen Williams performed chemical syntheses and wrote synthetic protocols. Roger Whitehead and Sabine Flitsch conceived and supervised the synthetic chemistry. Hanna Harant and Stig Linder provided reagents and contributed to analysis and interpretation of the results.
ACKNOWLEDGEMENTS
We thank Novartis for generously providing the translocation inhibitor cpd A, Ray Boot-Handford for the use of equipment, and Martin Pool and Ray Boot-Handford for valuable comments on the paper prior to submission.
FUNDING
This work was supported by a Selective Chemical Intervention in Biological Systems (SCIBS) initiative project grant from the Biotechnology and Biological Sciences Research Council (BBSRC); a targeted priority Ph.D. studentship from the BBSRC (to H.W.); a Wellcome Trust Ph.D. studentship (to P.R.); and a collaborative grant from the Medical Research Council (MRC) [grant number G0501725].
References
- 1.Cross B. C., Sinning I., Luirink J., High S. Delivering proteins for export from the cytosol. Nat. Rev. Mol. Cell Biol. 2009;10:255–264. doi: 10.1038/nrm2657. [DOI] [PubMed] [Google Scholar]
- 2.Ellgaard L., Helenius A. Quality control in the endoplasmic reticulum. Nat. Rev. Mol. Cell Biol. 2003;4:181–191. doi: 10.1038/nrm1052. [DOI] [PubMed] [Google Scholar]
- 3.Vembar S. S., Brodsky J. L. One step at a time: endoplasmic reticulum-associated degradation. Nat. Rev. Mol. Cell Biol. 2008;9:944–957. doi: 10.1038/nrm2546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Balch W. E., Morimoto R. I., Dillin A., Kelly J. W. Adapting proteostasis for disease intervention. Science. 2008;319:916–919. doi: 10.1126/science.1141448. [DOI] [PubMed] [Google Scholar]
- 5.Ron D., Walter P. Signal integration in the endoplasmic reticulum unfolded protein response. Nat. Rev. Mol. Cell Biol. 2007;8:519–529. doi: 10.1038/nrm2199. [DOI] [PubMed] [Google Scholar]
- 6.Lin J. H., Walter P., Yen T.S. B. Endoplasmic reticulum stress in disease pathogenesis. Annu. Rev. Path. Mech. Dis. 2008;3:399–425. doi: 10.1146/annurev.pathmechdis.3.121806.151434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fiebiger E., Hirsch C., Vyas J. M., Gordon E., Ploegh H. L., Tortorella D. Dissection of the dislocation pathway for type I membrane proteins with a new small molecule inhibitor, eeyarestatin. Mol. Biol. Cell. 2004;15:1635–1646. doi: 10.1091/mbc.E03-07-0506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang Q., Li L., Ye Y. Inhibition of p97-dependent protein degradation by eeyarestatin I. J. Biol. Chem. 2008;283:7445–7454. doi: 10.1074/jbc.M708347200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang Q., Shinkre B. A., Lee J. G., Weniger M. A., Liu Y., Chen W., Wiestner A., Trenkle W. C., Ye Y. The ERAD inhibitor eeyarestatin I is a bifunctional compound with a membrane-binding domain and a p97/VCP inhibitory group. PLoS ONE. 2011;5:e15479. doi: 10.1371/journal.pone.0015479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang Q., Mora-Jensen H., Weniger M. A., Perez-Galan P., Wolford C., Hai T., Ron D., Chen W., Trenkle W., Wiestner A., Ye Y. ERAD inhibitors integrate ER stress with an epigenetic mechanism to activate BH3-only protein NOXA in cancer cells. Proc. Natl. Acad. Sci. U.S.A. 2009;106:2200–2205. doi: 10.1073/pnas.0807611106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Moenner M., Pluquet O., Bouchecareilh M., Chevet E. Integrated endoplasmic reticulum stress responses in cancer. Cancer Res. 2007;67:10631–10634. doi: 10.1158/0008-5472.CAN-07-1705. [DOI] [PubMed] [Google Scholar]
- 12.Cross B. C., McKibbin C., Callan A. C., Roboti P., Piacenti M., Rabu C., Wilson C. M., Whitehead R., Flitsch S. L., Pool M. R., et al. Eeyarestatin I inhibits Sec61-mediated protein translocation at the endoplasmic reticulum. J. Cell Sci. 2009;122:4393–4400. doi: 10.1242/jcs.054494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Aletrari M. O., McKibbin C., Williams H., Pawar V., Pietroni P., Lord J. M., Flitsch S. L., Whitehead R., Swanton E., High S., Spooner R. A. Eeyarestatin 1 interferes with both retrograde and anterograde intracellular trafficking pathways. PLoS ONE. 2011;6:e22713. doi: 10.1371/journal.pone.0022713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kar R., Singha P. K., Venkatachalam M. A., Saikumar P. A novel role for MAP1 LC3 in nonautophagic cytoplasmic vacuolation death of cancer cells. Oncogene. 2009;28:2556–2568. doi: 10.1038/onc.2009.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lin T. Y., Wang S. M., Fu W. M., Chen Y. H., Yin H. S. Toxicity of tunicamycin to cultured brain neurons: ultrastructure of the degenerating neurons. J. Cell. Biochem. 1999;74:638–647. doi: 10.1002/(sici)1097-4644(19990915)74:4<638::aid-jcb13>3.3.co;2-3. [DOI] [PubMed] [Google Scholar]
- 16.Roboti P., Swanton E., High S. Differences in endoplasmic-reticulum quality control determine the cellular response to disease-associated mutants of proteolipid protein. J. Cell Sci. 2009;122:3942–3953. doi: 10.1242/jcs.055160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Choy M. S., Chen M. J., Manikandan J., Peng Z. F., Jenner A. M., Melendez A. J., Cheung N. S. Up-regulation of endoplasmic reticulum stress-related genes during the early phase of treatment of cultured cortical neurons by the proteasomal inhibitor lactacystin. J. Cell. Physiol. 2011;226:494–510. doi: 10.1002/jcp.22359. [DOI] [PubMed] [Google Scholar]
- 18.Berndtsson M., Beaujouin M., Rickardson L., Havelka A. M., Larsson R., Westman J., Liaudet-Coopman E., Linder S. Induction of the lysosomal apoptosis pathway by inhibitors of the ubiquitin-proteasome system. Int. J. Cancer. 2009;124:1463–1469. doi: 10.1002/ijc.24004. [DOI] [PubMed] [Google Scholar]
- 19.D'Arcy P., Brnjic S., Olofsson M. H., Fryknas M., Lindsten K., De Cesare M., Perego P., Sadeghi B., Hassan M., Larsson R., Linder S. Inhibition of proteasome deubiquitinating activity as a new cancer therapy. Nat. Med. 2011;17:1636–1640. doi: 10.1038/nm.2536. [DOI] [PubMed] [Google Scholar]
- 20.Besemer J., Harant H., Wang S., Oberhauser B., Marquardt K., Foster C. A., Schreiner E. P., de Vries J. E., Dascher-Nadel C., Lindley I. J. Selective inhibition of cotranslational translocation of vascular cell adhesion molecule 1. Nature. 2005;436:290–293. doi: 10.1038/nature03670. [DOI] [PubMed] [Google Scholar]
- 21.Harant H., Wolff B., Schreiner E. P., Oberhauser B., Hofer L., Lettner N., Maier S., de Vries J. E., Lindley I. J. Inhibition of vascular endothelial growth factor cotranslational translocation by the cyclopeptolide CAM741. Mol. Pharmacol. 2007;71:1657–1665. doi: 10.1124/mol.107.034249. [DOI] [PubMed] [Google Scholar]
- 22.Kang S. W., Rane N. S., Kim S. J., Garrison J. L., Taunton J., Hegde R. S. Substrate-specific translocational attenuation during ER stress defines a pre-emptive quality control pathway. Cell. 2006;127:999–1013. doi: 10.1016/j.cell.2006.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.MacKinnon A. L., Garrison J. L., Hegde R. S., Taunton J. Photo-leucine incorporation reveals the target of a cyclodepsipeptide inhibitor of cotranslational translocation. J. Am. Chem. Soc. 2007;129:14560–14561. doi: 10.1021/ja076250y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Calì T., Galli C., Olivari S., Molinari M. Segregation and rapid turnover of EDEM1 by an autophagy-like mechanism modulates standard ERAD and folding activities. Biochem. Biophys. Res. Commun. 2008;371:405–410. doi: 10.1016/j.bbrc.2008.04.098. [DOI] [PubMed] [Google Scholar]
- 25.Wu Y., Termine D. J., Swulius M. T., Moremen K. W., Sifers R. N. Human endoplasmic reticulum mannosidase I is subject to regulated proteolysis. J. Biol. Chem. 2007;282:4841–4849. doi: 10.1074/jbc.M607156200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fujiki Y., Hubbard A. L., Fowler S., Lazarow P. B. Isolation of intracellular membranes by means of sodium carbonate treatment: application to endoplasmic reticulum. J. Cell Biol. 1982;93:97–102. doi: 10.1083/jcb.93.1.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ernst R., Claessen J. H., Mueller B., Sanyal S., Spooner E., van der Veen A. G., Kirak O., Schlieker C. D., Weihofen W. A., Ploegh H. L. Enzymatic blockade of the ubiquitin-proteasome pathway. PLoS Biol. 2011;8:e1000605. doi: 10.1371/journal.pbio.1000605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lakkaraju A. K., Mary C., Scherrer A., Johnson A. E., Strub K. SRP keeps polypeptides translocation-competent by slowing translation to match limiting ER-targeting sites. Cell. 2008;133:440–451. doi: 10.1016/j.cell.2008.02.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Orsi A., Fioriti L., Chiesa R., Sitia R. Conditions of endoplasmic reticulum stress favor the accumulation of cytosolic prion protein. J. Biol. Chem. 2006;281:30431–30438. doi: 10.1074/jbc.M605320200. [DOI] [PubMed] [Google Scholar]
- 30.Hessa T., Sharma A., Mariappan M., Eshleman H. D., Gutierrez E., Hegde R. S. Protein targeting and degradation are coupled for elimination of mislocalized proteins. Nature. 2011;475:394–397. doi: 10.1038/nature10181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chakrabarti O., Hegde R. S. Functional depletion of mahogunin by cytosolically exposed prion protein contributes to neurodegeneration. Cell. 2009;137:1136–1147. doi: 10.1016/j.cell.2009.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rane N. S., Chakrabarti O., Feigenbaum L., Hegde R. S. Signal sequence insufficiency contributes to neurodegeneration caused by transmembrane prion protein. J. Cell Biol. 2010;188:515–526. doi: 10.1083/jcb.200911115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wiseman R. L., Zhang Y., Lee K. P., Harding H. P., Haynes C. M., Price J., Sicheri F., Ron D. Flavonol activation defines an unanticipated ligand-binding site in the kinase-RNase domain of IRE1. Mol. Cell. 2010;38:291–304. doi: 10.1016/j.molcel.2010.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dalal S., Rosser M.F. N., Cyr D. M., Hanson P. I. Distinct roles for the AAA ATPases NSF and p97 in the secretory pathway. Mol. Biol. Cell. 2004;15:637–648. doi: 10.1091/mbc.E03-02-0097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Meacham G. C., Lu Z., King S., Sorscher E., Tousson A., Cyr D. M. The Hdj-2/Hsc70 chaperone pair facilitates early steps in CFTR biogenesis. EMBO J. 1999;18:1492–1505. doi: 10.1093/emboj/18.6.1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Duennwald M. L., Lindquist S. Impaired ERAD and ER stress are early and specific events in polyglutamine toxicity. Genes Dev. 2008;22:3308–3319. doi: 10.1101/gad.1673408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nishitoh H., Kadowaki H., Nagai A., Maruyama T., Yokota T., Fukutomi H., Noguchi T., Matsuzawa A., Takeda K., Ichijo H. ALS-linked mutant SOD1 induces ER stress- and ASK1-dependent motor neuron death by targeting Derlin-1. Genes Dev. 2008;22:1451–1464. doi: 10.1101/gad.1640108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ashok A., Hegde R. S. Retrotranslocation of prion proteins from the endoplasmic reticulum by preventing GPI signal transamidation. Mol. Biol. Cell. 2008;19:3463–3476. doi: 10.1091/mbc.E08-01-0087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Huppa J. B., Ploegh H. L. The α chain of the T cell antigen receptor is degraded in the cytosol. Immunity. 1997;7:113–122. doi: 10.1016/s1074-7613(00)80514-2. [DOI] [PubMed] [Google Scholar]
- 40.Johnston J. A., Ward C. L., Kopito R. R. Aggresomes: a cellular response to misfolded proteins. J. Cell Biol. 1998;143:1883–1898. doi: 10.1083/jcb.143.7.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.VanSlyke J. K., Musil L. S. Dislocation and degradation from the ER are regulated by cytosolic stress. J. Cell Biol. 2002;157:381–394. doi: 10.1083/jcb.200111045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chakrabarti O., Rane N. S., Hegde R. S. Cytosolic aggregates perturb the degradation of nontranslocated secretory and membrane proteins. Mol. Biol. Cell. 2011;22:1625–1637. doi: 10.1091/mbc.E10-07-0638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Luesch H., Chanda S. K., Raya R. M., DeJesus P. D., Orth A. P., Walker J. R., Izpisua Belmonte J. C., Schultz P. G. A functional genomics approach to the mode of action of apratoxin A. Nat. Chem. Biol. 2006;2:158–167. doi: 10.1038/nchembio769. [DOI] [PubMed] [Google Scholar]
- 44.Liu Y., Law B. K., Luesch H. Apratoxin a reversibly inhibits the secretory pathway by preventing cotranslational translocation. Mol. Pharmacol. 2009;76:91–104. doi: 10.1124/mol.109.056085. [DOI] [PubMed] [Google Scholar]
- 45.Hasson S. A., Damoiseaux R., Glavin J. D., Dabir D. V., Walker S. S., Koehler C. M. Substrate specificity of the TIM22 mitochondrial import pathway revealed with small molecule inhibitor of protein translocation. Proc. Natl. Acad. Sci. U.S.A. 2010;107:9578–9583. doi: 10.1073/pnas.0914387107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.