Abstract
The canonical pathway of regulation of the GCK (germinal centre kinase) III subgroup member, MST3 (mammalian Sterile20-related kinase 3), involves a caspase-mediated cleavage between N-terminal catalytic and C-terminal regulatory domains with possible concurrent autophosphorylation of the activation loop MST3(Thr178), induction of serine/threonine protein kinase activity and nuclear localization. We identified an alternative ‘non-canonical’ pathway of MST3 activation (regulated primarily through dephosphorylation) which may also be applicable to other GCKIII (and GCKVI) subgroup members. In the basal state, inactive MST3 co-immunoprecipitated with the Golgi protein GOLGA2/gm130 (golgin A2/Golgi matrix protein 130). Activation of MST3 by calyculin A (a protein serine/threonine phosphatase 1/2A inhibitor) stimulated (auto)phosphorylation of MST3(Thr178) in the catalytic domain with essentially simultaneous cis-autophosphorylation of MST3(Thr328) in the regulatory domain, an event also requiring the MST3(341–376) sequence which acts as a putative docking domain. MST3(Thr178) phosphorylation increased MST3 kinase activity, but this activity was independent of MST3(Thr328) phosphorylation. Interestingly, MST3(Thr328) lies immediately C-terminal to a STRAD (Sterile20-related adaptor) pseudokinase-like site identified recently as being involved in binding of GCKIII/GCKVI members to MO25 scaffolding proteins. MST3(Thr178/Thr328) phosphorylation was concurrent with dissociation of MST3 from GOLGA2/gm130 and association of MST3 with MO25, and MST3(Thr328) phosphorylation was necessary for formation of the activated MST3–MO25 holocomplex.
Keywords: cardiac myocyte, germinal centre kinase, mammalian Sterile20-related kinase 3 (MST3), MO25, phosphorylation, regulation
Abbreviations: AdV, adenovirus; CCM3, cerebral cavernous malformation 3; ECL, enhanced chemiluminescence; GCK, germinal centre kinase; GOLGA2, golgin A2; gm130, Golgi matrix protein 130; GST, glutathione transferase; HA, haemagglutinin; LKB1, liver kinase B1; mAb, monoclonal antibody; MASK, MST3 and SOK1-related kinase; MBP, myelin basic protein; MST, mammalian Sterile20-related kinase; NLS, nuclear localization signal; OXSR1, oxidative-stress responsive 1; PAK, p21-activated kinase; PASK, proline-alanine-rich Sterile20-related kinase; PDCD10, programmed cell death 10; PKA, cAMP-dependent protein kinase; PP, protein serine/threonine phosphatase; SOK1, Sterile20/oxidant stress response kinase1; SPAK, Sterile20/Sps1-related proline/alanine-rich kinase; Ste20, Sterile20; STRAD, Ste20-related adapter; TBST, Tris-buffered saline plus Tween 20; YSK1, yeast Sps1/Ste20-related kinase 1
INTRODUCTION
The human genome encodes 28 mammalian Ste20 (Sterile20)- related protein kinases (so-called because of their homology with Saccharomyces cerevisiae Ste20p or Sps1p) comprising 22 GCKs (germinal centre kinases) and six PAKs (p21-activated kinases) [1,2]. GCKs possess an N-terminal ‘catalytic’ domain of approximately 275 residues and a C-terminal ‘regulatory’ domain, whereas the domain order is reversed in PAKs. GCKs are subdivided into eight subgroups (GCKI–VIII) on the basis of sequence motifs in the regulatory domains and the identity of a residue (cysteine, phenylalanine or tyrosine) in the P+1 loop of subdomain VIII. In addition, there are two STRAD (Ste20-related adapter) ‘pseudokinases’ which are most closely related to the GCKVI subgroup [3,4]. The GCKIII subgroup [5] is the simplest in terms of the complexity of the regulatory domain and comprises MST (mammalian Ste20-related protein kinase) 3 (also known as Stk24) [6], SOK1 (Ste20/oxidant stress response kinase 1)/YSK1 (yeast Sps1/Ste20-related kinase 1; also known as Stk25) [7,8], and MST4 [also known as MASK (MST3 and SOK1-related kinase)] [9,10]. MST3 transcripts are most abundant in the heart, placenta, skeletal muscle and pancreas [6], SOK1/YSK1 transcripts are most abundant in the testis (and possibly the brain) [7,8], and MST4 transcripts are most abundant in placenta [9,10], although all of the tested tissues probably express each GCKIII to a degree. Interest in GCKIII members is likely to increase because of their putative roles in pathologies such as cerebral cavernous malformations (see [11] for a review of this disease) and those associated with cardiac development [12].
The 431 residue MST3 ‘short’ isoform (also called MST3a by Irwin et al. [13] to avoid confusion with the longer MST3b isoform [14]) was first cloned from a HeLa cell library [6]. For simplicity, we refer to this isoform as ‘MST3’. Subsequently, brain-restricted MST3b (443 amino acid residues) was identified [14]. As shown in Supplementary Figure S1 (at http://www.BiochemJ.org/bj/442/bj4420595add.htm), there is considerable confusion about MST3 nomenclature in the literature with the nomenclature of the 431- and 443-residue isoforms frequently being reversed (see [15] for a recent example). MST3b may be identical with a protein kinase (confusingly known as protein kinase N) first identified in 1987 [16–18]. The two MST3 isoforms arise from translation of alternative 5′ coding exons: exon 1 (42 nt) encodes the 14 N-terminal residues in MST3, whereas exon 2 (78 nt) encodes the 26 N-terminal residues in MST3b. Otherwise, the sequences of MST3 and MST3b are identical. The 431 residue MST3 is activated by autophosphorylation of the activation loop Thr178 [19]. MST3b is additionally phosphorylated at Thr18 (absent from MST3) by PKA (cAMP-dependent protein kinase) [14], but the role of this phosphorylation in the regulation of MST3b activity is ambiguous. The only established MST3 ‘activator’ [6] is the PP (protein serine/threonine phosphatase) 1/PP2A inhibitor calyculin A [20], which presumably simply inhibits the dephosphorylation step of a constitutively active phosphorylation/dephosphorylation cycle. In contrast, ‘physiological regulators’ of MST3b have been identified. In PC12 cells, MST3b is activated by nerve growth factor or inosine and is inhibited by 6-thioguanine [13]. In addition to its possible involvement in systemic pathologies [12], MST3 may be involved in the regulation of cell motility [19], in the regulation of other protein kinases [14,21–23] and caspase-dependent [24] or -independent apoptosis [24,25]. The biological functions of MST3b may include stimulation of axon outgrowth [13,26], and its activation by inosine is potentially of therapeutic benefit in restoring limb usage following cerebral focal ischaemia (‘stroke’) [27].
Only one pathway of MST3 activation (the ‘canonical’ pathway) has thus far been characterized in detail and this is involved in Jurkat cell apoptosis [24]. Anti-Fas antibodies or staurosporine induce caspase-dependent cleavage of MST3 at Asp313 at the boundary between the catalytic and regulatory domains [24]. This reveals a bipartite NLS (nuclear localization signal) in the C-terminus of the catalytic domain, as well as possibly removing a nuclear export signal in the regulatory domain [28]. Cleaved MST3 translocates into the nucleus and is at least partly responsible for subsequent apoptosis [24,28], although whether the cleaved MST3 is autophosphorylated and activated seems unclear. In contrast, on the basis of incomplete inhibition of staurosporine-induced apoptosis by the caspase inhibitor Z-DEVD-FMK (benzyloxycarbonyl-DL-Asp-Glu-Val-DL-Asp-fluoromethylketone), a caspase-independent involvement of MST3 in HeLa cell apoptosis has been identified [25]. This involves loss of mitochondrial membrane potential, and the subsequent nuclear import of apoptosis-inducing factor and endonuclease G released from the mitochondria [25]. Although the canonical pathway of MST3 regulation is widely accepted, in the present paper we describe a second non-canonical pathway of MST3 activation in cardiac myocytes which does not involve caspase-dependent cleavage.
EXPERIMENTAL
Antibodies and proteins
Antibodies were used at a 1:1000 dilution except when stated otherwise. Mouse mAbs (monoclonal antibodies) against human MST3 (residues 275–393 as immunogen) were from BD Transduction Laboratories (catalogue number 611056). A variety of FLAG/OctA antibodies were used: rabbit polyclonal antibodies were from Sigma–Aldrich (catalogue number F7425) or Santa Cruz Biotechnology (catalogue number sc-807, lot K1607), and mouse mAbs were from Santa Cruz Biotechnology (catalogue number sc-166355). Anti-HA (haemagglutinin) antibodies (rabbit polyclonal catalogue number sc-805, mouse mAb catalogue number sc-7392) were from Santa Cruz Biotechnology. Mouse monoclonal EZview™ red anti-FLAG M2 affinity gel was from Sigma–Aldrich (catalogue number F2426). Rabbit polyclonal antibodies against GST (glutathione transferase) were from Abcam (catalogue number ab9085-200). Rabbit polyclonal antibodies against GOLGA2/gm130 (golgin A2/Golgi matrix protein 130; catalogue number 11308-1-AP) and CCM3/PDCD10 (cerebral cavernous malformation 3/programmed cell death 10; catalogue number 10294-2-AP) were from Proteintech Group. Rabbit mAbs against MO25α (catalogue number 2716) and antibodies against cleaved (activated) caspase 3(Asp175) (catalogue number 9661) were from Cell Signaling Technology. Sheep polyclonal antibodies against MO25β were gifts from Professor Dario Alessi and Dr Paola de los Heros (MRC Protein Phosphorylation Unit, University of Dundee, Dundee, U.K.) and were used at a 1:400 dilution.
Horseradish peroxidase-conjugated anti-mouse (catalogue number P0260) and anti-rabbit (catalogue number P0448) secondary antibodies were from Dako, and Pierce horseradish peroxidase-conjugated anti-sheep antibodies were from ThermoFisher (catalogue number 31627). The Alexa Fluor® 488-conjugated anti-mouse secondary antibody (catalogue number A11001) and Texas Red–phalloidin (catalogue number T7471) were from Invitrogen.
The catalytic domain of human recombinant PP2A was from Cayman Chemicals (catalogue number CAY10011237) and was obtained via Cambridge Bioscience. MBP (myelin basic protein) was from Millipore (catalogue number 13-104).
Cardiac myocyte isolation and culture
Rats were housed and work was under taken in accordance with the local institutional animal care committee and the U.K. Animals (Scientific Procedures) Act 1986. Primary cultures of ventricular cardiac myocytes were prepared from 1–2-day-old Sprague–Dawley rats by collagenase and pancreatin digestion of ventricles using an adaptation of the method of Iwaki et al. [29] as described recently in detail [30]. Except for immunofluorescent staining experiments, cardiac myocytes were plated on gelatin-precoated Primaria™ surface-modified polystyrene culture dishes [BD Biosciences, catalogue numbers 353801 (35 mm) and 353802 (60 mm)] at densities of 2×106 myocytes per 35 mm dish or 4×106 myocytes per 60 mm dish. After 18 h, myocytes were confluent and beating spontaneously. Unless they were to be infected with AdV (adenoviral) vectors, serum was then withdrawn and myocytes were incubated in maintenance medium {Dulbecco's modified Eagle's medium/medium 199 [4:1 (v/v)] and penicillin and streptomycin (100 units/ml for each)} for a further 24 h. For AdV infection, serum was withdrawn 6 h beforehand and the infected myocytes were used within 48 h.
MST3 cloning and AdV vector preparation
The MST3 coding sequence was amplified from neonatal rat cardiac myocyte cDNA with Pfu polymerase using the forward primer 5′-GACGACAAGGGTACCATGGCCCACTCCCCGGTGCAG-3′ and the reverse primer 5′-CTTATCTAGAAGCTTTCAGTGCGCTGAGGCTCCTCCA-3′ to produce a MST3 product (containing the initiator methionine residue) flanked by the KpnI and HindIII sites (underlined). These restriction sites were then used for insertion into the corresponding sites in FLAG-pShuttle-CMV [31]. The product was sequenced and submitted to GenBank® (accession number EU371958.1). MST3 C-terminal deletion mutants were constructed similarly, but using the reverse primers 5′-CTTATCTAGAAGCTTTCAATCTGTCTCCACATCTGA-3′ [for MST3(Δ314–431)], 5′-CTTATCTAGAAGCTTTCATCCGTTCTCCAGATTCTTGG-3′ [for MST3(Δ341–431)] and 5′-CTTATCTAGAAGCTTTCATTTCAGCTCCGCAAACAGAG-3′ [for MST3(Δ377–431)]. Point mutations were introduced into MST3 by PCR using forward and reverse complementary primer pairs that contained the desired base change(s) flanked by 15 bases on either side. Two separate PCR amplifications were performed using the pShuttle-CMV forward primer (5′-GGTCTATATAAGCAGAGCTG-3′) in combination with the MST3 mutant reverse primer and the pShuttle-CMV reverse primer (5′-GTGGTATGGCTGATTATGATCAG-3′) in combination with the mutant MST3 forward primer, with wild-type MST3 as a template. Subsequently, a third PCR was performed using pShuttle-CMV forward and reverse primers and a mixture of the two purified mutant products from the first round of PCRs. The mutant PCR product generated was purified and cloned into the Kpn1 and HindIII sites of FLAG-pShuttle-CMV. Where multiple mutations were required, they were introduced together on the same primer pair if close together or, if further apart, were introduced sequentially. All constructs were sequenced to verify that the desired mutation(s) was present and that no further mutations had been introduced during the PCR procedure. AdVs were generated from the shuttle plasmids using the AdEasy™ XL Adenoviral Vector System (Stratagene), as described previously [30,31].
Immunoblotting
Following treatments, myocytes were washed twice with ice-cold PBS and scraped into Buffer A [20 mM 2-glycerophosphate (pH 7.5), 50 mM NaF, 5 mM dithiothreitol, 2 mM EDTA, 0.2 mM Na3VO4, 2 μM microcystin LR and 1% (v/v) Triton X-100] containing protease inhibitors {10 mM benzamidine, 300 μM PMSF, 200 μM leupeptin and 10 μM E-64 [trans-epoxysuccinyl-L-leucylamido-(4-guanidino)butane]} (100 μl per 35 mm dish and 150 μl per 60 mm dish). Samples were incubated on ice for 10 min and then centrifuged (10000 g for 5 min at 4°C).
Supernatants were denatured by heating at 100°C with 0.33 vol. of 4×SDS/PAGE sample buffer as described previously [30]. Non-specific binding sites were blocked with 5% (w/v) non-fat milk powder in TBST [Tris-buffered saline plus Tween 20; 20 mM Tris/HCl (pH 7.5), 137 mM NaCl and 0.1% Tween 20] for 30 min. Blots were incubated with primary antibodies in TBST containing 5% (w/v) BSA overnight at 4°C. The blots were washed three times with TBST [for 5 min at room temperature (21°C)], incubated with secondary antibodies [1:5000 dilution in TBST containing 1% (w/v) non-fat milk powder for 1 h at room temperature] and then washed again three times with TBST (for 5 min at room temperature). Bands were detected by ECL (enhanced chemiluminescence; Santa Cruz Biotechnology) using Hyperfilm MP and were quantified by scanning densitometry. Latterly, bands were detected with GE Healthcare ECL Plus and a GE ImageQuant 350 imager with one-dimensional gel analysis software. Unless divided by a vertical bar, all individual immunoblots shown are images of single gels, although the lane order has been altered by image manipulation in some cases to improve comprehensibility.
Immunoprecipitations and immunokinase assays
Myocytes were washed twice with ice-cold PBS and extracted into Buffer B [20 mM Tris/HCl (pH 7.5), 300 mM KCl, 5 mM MgCl2, 5 mM NaF, 1 mM EDTA, 0.2 mM Na3VO4, 2 μM microcystin LR, 10% (v/v) glycerol, 1% (v/v) Triton X-100 and 0.05% 2-mercaptoethanol containing the protease inhibitors as for Buffer A]. When myocytes had been pre-exposed to 100 nM calyculin A, calyculin A (100 nM) was also included in the extraction buffer. Samples were incubated on ice for 10 min and then centrifuged (10000 g for 5 min at 4°C). The supernatants were retained and samples were taken for immunoblotting to assess the input of the proteins subsequently immunoprecipitated. For immunokinase assays, supernatants (27.5 μl) were diluted to 150 μl with Buffer B and added to 15 μl of EZview™ red anti-FLAG M2 affinity gel (1:1 slurry in Buffer B), followed by overnight incubation at 4°C with rotation. The pellets were collected by centrifugation (10000 g for 1 min at 4°C) and washed three times in Buffer B (0.7 ml), once with kinase assay buffer [20 mM Tris/HCl (pH 7.5), 1 mM dithiothreitol, 1 μM microcystin LR, 4 μM leupeptin and 1 mM benzamidine; 150 μl) and finally resuspended in 45 μl of kinase assay buffer containing 0.5 mg/ml MBP and 1 μM PKA inhibitor (TTYADFIASGRTGRRNAIHD; Bachem).
FLAG–MST3 immunokinase assays were initiated by the addition of 5 μl of 1 mM [γ-32P]ATP (0.5 μCi/assay)/200 mM MgCl2 followed by incubation at 30°C for 30 min. Reactions were terminated by the addition of 5 μl of 0.5 M EDTA (pH 7.5), followed by cooling on ice. Samples were centrifuged (10000 g for 1 min at 4°C) and duplicate samples (20 μl) of supernatants were spotted on to Whatman P81 paper squares, which were then washed four times in 75 mM H3PO4 (for 15 min) on a rocking platform. Radioactivity was measured by Čerenkov radiation counting. Blanks (one with kinase assay buffer and [γ-32P]ATP/MgCl2, and one involving the assay of MBP kinase activity with immunoprecipitates from uninfected myocytes) were routinely included.
Dephosphorylations with PP2A
FLAG–MST3 in samples taken from pooled extracts of myocytes which had or had not been exposed to 100 nM calyculin A for 10 min was prepared essentially as described in the Immunoprecipitations and immunokinase assays section. Immunoprecipitates were washed three times with Buffer C [50mM Tris/HCl (pH 7.0), 0.15 mM EGTA, 0.1% 2-mercaptoethanol and 0.1% BSA] and resuspended in 25 μl of Buffer C. GST–MST3 was similarly precipitated using glutathione–Sepharose. Dephosphorylation was then initiated by the addition of PP2A (25 μl, 10 units/μl in Buffer C) in the absence or presence of 2 μM okadaic acid. Incubations were for up to 30 min at 30°C, and PP2A activity was terminated by the addition of 0.1 vol. of 50 μM okadaic acid/2 μM calyculin A. Immunoprecipitates were washed with Buffer C, centrifuged (10000 g for 5 min at 4°C) and the pellets extracted into 50 μl of 2×sample buffer for subsequent immunoblotting.
Gel filtration
Supernatants (prepared in Buffer B from 60 mm dishes of monocytes as described in the Immunoblotting section) of AdV–MST3-infected myocytes were applied to columns of Sephacryl S200HR (1.4 cm×85 cm, 131 ml inclusion volume measured using 3H2O) or Sephacryl S300HR (1.6 cm×95 cm, 185 ml inclusion volume) equilibrated with 50 mM Tris/HCl (pH 7.5), 250 mM NaCl, 1 mM EDTA, 1 mM dithiothreitol and 0.03% Brij-35 which had been standardized with proteins of known Stokes' radii. When extracts of calyculin A-treated myocytes were examined, the elution buffer additionally contained 50 nM calyculin A. Flow rates were 0.2–0.3 ml/min and fractions of 0.6–1.3 ml as required were collected. In some cases, fractions were centrifugally concentrated at 4°C by up to 10–15-fold in Amicon Ultra-2 10 kDa nominal molecular mass limit centrifugal filter devices (Millipore).
Immunocytochemistry
Myocytes (1.5×106 cells per 35 mm Primaria culture dish) were cultured on to glass coverslips precoated with 1% (w/v) gelatin followed by laminin (20 μg/ml in PBS). Myocytes were cultured as described in the Cardiac myocyte isolation and culture section, and were infected with AdV–FLAG–MST3 for 16 h in serum-free medium. After treatment, myocytes were washed with ice-cold PBS (3×0.5 ml), fixed in 4% (v/v) formaldehyde (10 min at room temperature), permeabilized with 0.1% Triton X-100 in PBS (10 min at room temperature) and incubated with 1% (w/v) BSA in PBS containing 0.1% Triton X-100 (10 min at room temperature) to block non-specific antibody binding. FLAG–MST3 was detected by incubation of cells with Santa Cruz Biotechnology mouse anti-FLAG mAbs (1:1000 dilution for 1 h at 37°C), then incubated with goat anti-(mouse IgG) coupled to Alexa Fluor® 488 (1:200 dilution for 1 h at 37°C) followed by Texas Red®-X phalloidin (Invitrogen; 1:200 dilution for 1 h at 37°C). Nuclei were stained with Hoechst 33258 (Sigma; 10 μg/ml for 15 min at 37°C). Coverslips were washed with PBS (3×0.5 ml) between each incubation, mounted with fluorescence mounting medium (Dako), viewed under a Zeiss Axioskop fluorescence microscope with a ×40 objective, and photographed using a digital camera.
Structural studies
The disorder probability plot was produced using the DISOclust method (http://www.reading.ac.uk/bioinf/DISOclust/) for the prediction of intrinsically unstructured regions in proteins [32]. Predicted secondary structure was obtained using the PSIPRED server (http://bioinf.cs.ucl.ac.uk/psipred/) [33]. The three-dimensional model was produced using the IntFOLD-TS method (http://www.reading.ac.uk/bioinf/IntFOLD/) [34,35] and was colour coded from blue (high accuracy) through green, yellow and orange to red (low accuracy) using the ModFOLDclust2 model quality assessment method (http://www.reading.ac.uk/bioinf/ModFOLD/) [36]. The image was rendered using PyMol (http://www.pymol.org). Protein sequences were taken from the NCBI (National Center for Biotechnology Information) protein database (http://www.ncbi.nlm.nih.gov/protein/). Alignments were carried out using ClustalW2 (http://www.ebi.ac.uk/Tools/msa/clustalw2/) with manual curation if necessary.
RESULTS AND DISCUSSION
Primary sequence of rat MST3
We cloned and sequenced rat MST3 (GenBank® accession number EU371958.1, submitted 23 January 2008). The predicted 431 residue sequence (Supplementary Figure S1) has 98.6% and 96.8% identity with the (431 residue) MST3 of mouse (GenBank® accession number NP_663440.1) and Homo sapiens (GenBank® accession number NP_001027467.2; cloned by Schinkmann and Blenis [6]) respectively, with mostly conservative or semi-conservative substitutions elsewhere. The ATP-binding site [GXGXXGX(14–23)K] in rat MST3 lies between residues 31 and 53, the activation loop serine/threonine is Thr178, and the ‘caspase cleavage’ site is V310ETD313 (AETD in H. sapiens). The residue in the 314 position is glycine in H. sapiens and mouse, but is serine in rat. Thus, although Gly314 has been identified as a possible N-myristoylation site that becomes available following caspase cleavage of MST3 [37,38], this cannot occur in the rat and N-myristoylation of MST3(314–431) may not be of universal importance.
Phosphorylation of MST3 in cardiac myocytes
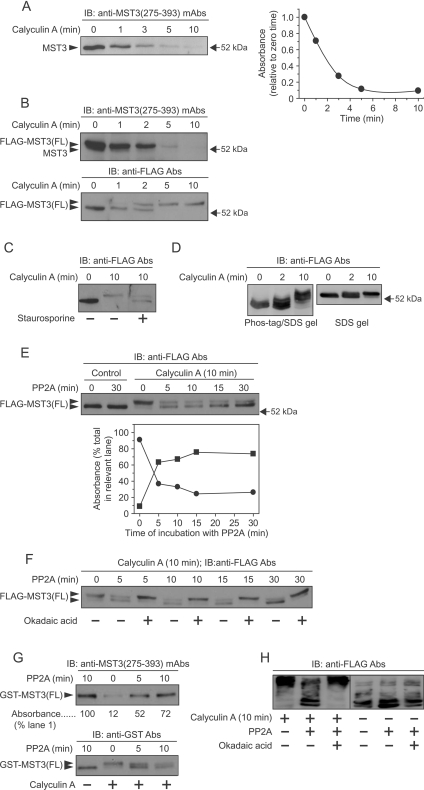
MST3(275–393) mAbs detected MST3 as a single band of ~52 kDa in cardiac myocyte extracts, and exposure of myocytes to calyculin A (100–200 nM) resulted in its rapid (complete in <10 min) loss (Figure 1A). To investigate this further, N-terminally FLAG-tagged rat full-length (431 residue) MST3 [FLAG–MST3(FL)] was expressed in cardiac myocytes using an AdV vector. In unstimulated myocytes, FLAG–MST3(FL) was detected as a band of ~54 kDa with anti-MST3(275–393) mAbs which predominated over endogenous MST3 (Figure 1B, upper panel). Exposure of myocytes to 100 nM calyculin A resulted in the loss of both endogenous MST3 and FLAG–MST3(FL) (Figure 1B, upper panel). Immunoblotting with anti-FLAG antibodies revealed that FLAG–MST3(FL) was not lost following calyculin A exposure, but instead migrated at ~58 kDa (Figure 1B, lower panel). Loss of the 54 kDa band and appearance of 58 kDa FLAG–MST3(FL) were essentially complete by 5–10 min (Figure 1B, lower panel), and there were no increases in bands of <58 kDa (Supplementary Figure S2 at http://www.BiochemJ.org/bj/442/bj4420595add.htm) indicating absence of detectable cleavage. The broad-spectrum serine/threonine protein kinase inhibitor staurosporine reduced the rate of the appearance of 58 kDa FLAG–MST3(FL) in calyculin A-treated myocytes (Figure 1C), suggesting that the band shift represents serine/threonine phosphorylation. Additional evidence that the 58 kDa band resulted from phosphorylation was obtained using phos-tag™ SDS/PAGE immunoblots [39]. Anti-FLAG antibodies detected multiple FLAG–MST3(FL) bands even in unstimulated myocytes (Figure 1D, left-hand panel). Following exposure of myocytes to calyculin A, the overall mobility of the bands was reduced, indicating that FLAG–MST3(FL) was increasingly phosphorylated. A ‘control’ SDS/PAGE immunoblot from the same experiment is shown for comparison (Figure 1D, right-hand panel).
Figure 1. Phosphorylation of MST3 and FLAG–MST3(FL) species.
Myocytes were uninfected (A), infected with AdV–FLAG–MST3(FL) (B–F and H) or infected with AdV–GST–MST3(FL) (G). They were exposed to 100 nM calyculin A for the times indicated and extracts were prepared. Proteins were separated by SDS/PAGE (A–C) or (E–G) phos-tag™ SDS/PAGE (D and H) followed by immunoblot (IB) analysis. (A) Extracts were immunoblotted with anti-MST3(275–393) mAbs and the bands (left-hand panel) were quantified (right-hand panel). (B) Extracts were immunoblotted with anti-MST3(275–393) mAbs (upper panel) to detect endogenous MST3 and FLAG–MST3(FL) or with anti–FLAG antibodies (Abs, lower panel) to detect FLAG-MST3(FL). (C) Myocytes were unstimulated, or exposed to calyculin A in the absence or presence of staurosporine (1 μM) and extracts were immunoblotted with anti-FLAG antibodies. (D) Extracts were analysed using phos-tag™ SDS/PAGE (left-hand panel) or SDS/PAGE (right-hand panel) and immunoblotted with anti-FLAG antibodies. (E–H) Myocytes expressing FLAG–MST3(FL) (E, F and H) or GST–MST3(FL) (G) were unstimulated or exposed to calyculin A for 10 min. FLAG–MST3(FL) was immunoprecipitated using anti-FLAG M2 affinity gel and GST–MST3(FL) was precipitated using glutathione–Sepharose. Precipitates were incubated with PP2A for the times indicated. In some instances (F and H), 2 μM okadaic acid was included in the PP2A incubations. Extracts were immunoblotted with anti-FLAG (E, F and H), anti–MST3(275–393) mAbs (G, upper panel), or anti-GST antibodies (G, lower panel). (E) The lanes showing extracts of myocytes exposed to calyculin A (upper panel) were quantified (lower panel). ●, upper band; ■, lower band. Experiments were repeated at least twice with similar results with different myocyte preparations.
We confirmed that serine/threonine phosphorylation of FLAG–MST3(FL) caused the shift from 54 kDa to 58 kDa (Figure 1B, lower panel) by treating FLAG–MST3(FL) immunoprecipitates from calyculin A-treated myocytes with recombinant PP2A. This resulted in the disappearance of the 58 kDa FLAG–MST3 band with the reappearance of the 54 kDa band (Figure 1E). This was prevented by inclusion of the PP1/PP2A inhibitor okadaic acid in the PP2A incubation (Figure 1F), indicating that proteolysis, for example, was not responsible. Furthermore, in glutathione–Sepharose precipitates of myocytes expressing GST–MST3(FL) which had been exposed to calyculin A, the band detected by anti-MST3(275–393) mAbs reappeared with PP2A treatment detected using anti-GST antibodies (Figure 1G, upper panel) with simultaneous reappearance of the more rapidly migrating dephosphorylated band (Figure 1G, lower panel). Analogous experiments using phos-tag™ SDS/PAGE gels and anti-FLAG antibodies confirmed that FLAG–MST3(FL) reverted to the more rapidly migrating forms and this was prevented by okadaic acid (Figure 1H). However, even in PP2A-treated immunoprecipitates, multiple FLAG–MST3 bands remained clearly visible in phos-tag™ SDS/PAGE gels (Figure 1H). We interpret this as indicating that either residues other than serine/threonine (e.g. tyrosine) are phosphorylated or that MST3 contains phosphorylated serine/threonine residues that are resistant to PP2A. There are at least two precedents for the latter interpretation [40,41].
The only other agent that resulted in the appearance of the 58 kDa FLAG–MST3 in cardiac myocytes was okadaic acid (results not shown). A very recent paper [15] has also shown that okadaic acid reduced the mobility of endogenous MST3 in HEK (human embryonic kidney)-293 cells [detected using mAbs to an epitope in the C-terminus of MST3(FL)], which was reversed by incubation with exogenous purified PP2A. The list of agonists/stimuli that do not activate (i.e. reduce the mobility to 58 kDa, as explained below) MST3 includes Gq-protein-coupled receptor agonists (e.g. endothelin-1 and α-adrenergic agonists), Gs-protein-coupled receptor agonists (e.g. isoprenaline), peptide growth factors (e.g. epidermal growth factor and insulin) and stress stimuli such as tunicamycin (to induce endoplasmic reticulum stress), hyperosmotic shock and oxidative stress (results not shown).
In summary, we conclude that exposure of myocytes to calyculin A induces serine/threonine phosphorylation of MST3(FL). Because anti-MST3(275–393) mAbs fail to recognize FLAG–MST3(FL) following calyculin A exposure (Figure 1B, upper panel), the phosphorylation site(s) probably lies between residues 275 and 393 and is distinct from the previously identified phosphorylation of MST3 on Thr178 of its activation loop [19].
Identification of a novel phosphorylation site in MST3
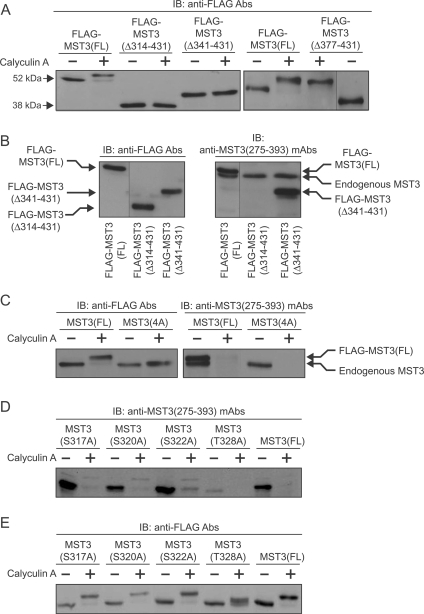
To identify the potential phosphorylation site(s) additional to MST3(Thr178) deletion mutants were generated and expressed in myocytes: FLAG–MST3(Δ314–431) with deletion of Ser314–His431, FLAG–MST3(Δ341–431) with deletion of Thr341–His431 and FLAG–MST3(Δ377–431) with deletion of Glu377–His431. Incubation of myocytes with calyculin A reduced the mobility of FLAG–MST3(FL) or FLAG–MST3(Δ377–431) on SDS/PAGE, but not that of FLAG–MST3(Δ314–431) or MST3(Δ341–431) (Figure 2A). Thus the site(s) of phosphorylation responsible for reduced migration probably lies between Ser314 and Lys376. In addition, although the anti-MST3(275–393) mAbs recognized FLAG–MST3(FL) and MST3(Δ341–431), they did not recognize FLAG–MST3(Δ314–431) (Figure 2B). This suggests that the epitope recognized by the MST3(275–393) mAbs probably lies between Ser314 and Gly340.
Figure 2. Identification of a novel phosphorylation site (Thr328) in MST3.
Myocytes were infected with the appropriate AdV vectors. Myocytes expressing FLAG–MST3(FL), MST3 truncation mutants [FLAG–MST3(Δ314–431), FLAG–MST3(Δ341–431) or FLAG–MST3(Δ377–431)] or MST3 with alanine substitutions [FLAG–MST3(4A) with S317A, S320A, S322A and T328A mutations or FLAG–MST3(S317A), FLAG–MST3(S320A), FLAG–MST3(S322A); FLAG–MST3(T328A)] were unstimulated (A–E) or exposed to calyculin A (100 nM for 10 min) (A and C–E) and extracts prepared. Proteins were separated by SDS/PAGE followed by immunoblot (IB) analysis with anti-FLAG antibodies (Abs) (A) (B and C, left-hand panels) and (E) or anti-MST3(275–393) mAbs (B and C, right-hand panels) and (D). Experiments were repeated at least twice with different myocyte preparations with similar results.
We used alanine scanning to identify the phosphorylation site on MST3, focusing first on the Thr341–Lys376 region because of the differing behaviour of FLAG–MST3(Δ314–431) and FLAG–MST3(Δ341–431) to FLAG–MST3(Δ377–431) following calyculin A exposure (Figure 2A). In this regard, a proteomics study had shown that MST3(Thr341) was phosphorylated in a lymphoma cell line [42]. All six serine/threonine residues in the Thr341–Lys376 region of FLAG–MST3(FL) were mutated to alanine either individually or all together, and the proteins expressed in myocytes. On SDS/PAGE, the mobilities of all seven mutants were still reduced to 58 kDa by calyculin A (Supplementary Figure S3A at http://www.BiochemJ.org/bj/442/bj4420595add.htm). Thus, although the MST3(341–376) region is required for the phosphorylation(s) resulting in reduced mobility on SDS/PAGE (Figure 2A), it does not contain the phosphorylation site(s) and presumably represents a docking domain. We next examined the Ser314–Gly340 region. We had separately examined FLAG–MST3(S314G) earlier because of the difference at residue 314 between rat (serine) and H. sapiens or mouse (glycine; Supplementary Figure S1), and found that its migration was reduced to the same extent by exposure to calyculin A as that of FLAG–MST3(FL) (results not shown). The appearance of 58 kDa FLAG–MST3 when Ser317, Ser320, Ser322 and Thr328 were all mutated to alanine [FLAG–MST3(4A)] was not reduced to the same extent as FLAG–MST(FL), although a slight reduction was apparent (Figure 2C, left-hand panel). Furthermore, anti–MST3(275–393) mAbs failed to recognize FLAG–MST3(4A) with or without exposure of myocytes to calyculin A (Figure 2C, right-hand panel). With individual single alanine substitutions, although FLAG–MST3(T328A) remained detectable using FLAG antibodies, this mutation alone caused the loss of the anti–MST3(275–393) mAb epitope even in the absence of exposure to calyculin A (Figure 2D), and the finding that the mobility of FLAG–MST3(T328A) was not reduced to the same extent as FLAG–MST3(FL) by calyculin A indicated that phosphorylation of FLAG–MST3(Thr328) was responsible for the major reduction in FLAG–MST3(FL) mobility (Figure 2E). The mobility of FLAG–MST3(T328A) was still slightly reduced in myocytes exposed to calyculin A, presumably resulting from phosphorylation of another residue [potentially FLAG-MST3(Thr178)]. It is noteworthy that endogenous MST3 was still detectable by anti-MST3(275–393) mAbs in FLAG–MST3(T328A)-infected myocytes that had not been exposed to calyculin A (fourth lane from the right-hand side of Figure 2D), showing that the failure of these mAbs to recognize FLAG–MST3(T328A) did not result from technical problems.
PhosphSitePlus® (http://www.phosphosite.org) lists a further four potentially phosphorylatable serine residues in the MST3(275–393) region (A296EQpSHDDpSpSpSEDSDAETDGQASGGSDSGDWIFTIR330; where pS represents potentially phosphorylatable serine residues) identified by proteomics approaches [43,44]. (Readers should rely on sequence information rather than numerical annotation because of the confusion in MST3(a)/MST3b terminology.) However, serine-to-alanine mutation of each of these serine residues (Ser299, Ser303, Ser304 and Ser305) and Ser308 in addition, or mutation of all residues in FLAG–MST3 produced proteins that were indistinguishable from FLAG—MST3 (FL) in response to calyculin A (Supplementary Figure S3B). Our overall conclusions from the experiments shown in Figure 2 are as follows. First, that the MST3(341–376) region is required for the reduced migration of FLAG–MST3(FL) resulting from its phosphorylation, but that this region does not contain the phosphorylation site responsible for the reduced migration. Secondly, cis-autophosphorylation of MST3(Thr328) is responsible for the greater proportion of the calyculin A-induced reduction in FLAG–MST3(FL) migration. Thirdly, the identity and the phosphorylation of MST3(Thr328) is crucial for the recognition of MST3 by anti-MST3(275–393) mAbs.
Regulation of MST3 kinase activity
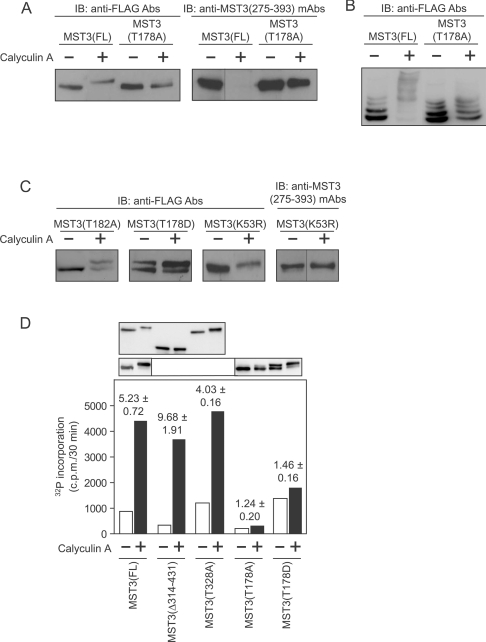
We next examined the relative mobilities of FLAG–MST3(FL) containing mutations in or close to the activation loop by SDS/PAGE. Mutation of the activation loop Thr178 phosphorylation site [FLAG–MST3(T178A)] prevented the appearance of any 58 kDa FLAG–MST3 band following exposure of myocytes to calyculin A (Figure 3A, left-hand panel) and the anti-MST3(275–393) mAb epitope was also preserved (Figure 3A, right panel). Phos-tag™/SDS gels (Figure 3B) confirmed the result shown in the left-hand panel of Figure 3(A). Thus phosphorylation of MST3(Thr328) requires MST3 kinase activity and the data suggest that this autophosphorylation occurs in cis rather than in trans (since endogenous MST3 would have been activated by calyculin A in these experiments). GCKs reportedly possess a ‘secondary’ phosphorylation site lying in the conserved GTP(C,F,Y)WMAPE sequence in the P+1 loop (Thr182 in MST3), phosphorylation of which may be required for activation [2]. Phosphorylation of this residue has been detected experimentally by proteomics approaches [44,45], but care must be taken in annotation because of sequence similarities between GCKIII members in this region. Compared with FLAG–MST3(FL) (Figure 3A), there was only a partial shift of FLAG–MST3(T182A) to a lower mobility species in myocytes exposed to calyculin A (Figure 3C), possibly indicating a requirement of MST3(Thr182) phosphorylation for MST3 activation as assessed by MST3(Thr328) cis-autophosphorylation. However, the analogous threonine residue in the catalytic subunit of PKA is important in the kinase–protein substrate interaction [46], and this is also true for other protein serine/threonine kinases. Thus it is equally likely that the FLAG–MST3(T182A) mutation reduces the affinity of MST3 for its substrates, in this case, FLAG–MST3(Thr328). Gordon et al. [15] recently obtained similar results to those of the present study using FLAG–MST(T178A) and FLAG–MST3(T182A). They also studied the FLAG–MST3(T172A) mutation since MST3(phosphoThr172) has also been detected by proteomics approaches [44,47,48]. The mobility of FLAG–MST3(T172A) was still reduced by okadaic acid [15], and the conclusion was that phosphorylation of this site is unimportant in the appearance of 58 kDa FLAG–MST3(FL) [15].
Figure 3. Phosphorylation of MST3(Thr178), but not MST3(Thr328), is required for MST3 activity.
Myocytes were infected with the appropriate AdV vectors. Myocytes expressing FLAG–MST3(FL), FLAG–MST3(T178A), FLAG–MST3(T178D), FLAG–MST3(K53R), FLAG–MST3(Δ314–431) or FLAG–MST3(T328A) were unstimulated or exposed to calyculin A (100 nM for 10 min) and extracts were prepared. Proteins were separated by SDS/PAGE (A, C and D) or phos-tag™ SDS/PAGE (B) followed by immunoblot (IB) analysis with anti-FLAG antibodies (Abs) (A, left-hand panel), (B and C, left-hand three panels) and (D), or anti–MST3(275–393) mAbs (A and C, right-hand panels). Immunoblots are representative of experiments repeated at least twice with different myocyte preparations. (D) Immunokinase assays of FLAG–MST3(FL) and FLAG–MST3 mutants. Myocytes were unstimulated (open columns) or exposed to calyculin A (100 nM for 10 min; filled columns). Incorporation of 32P from [γ-32P]ATP into MBP was measured and the results from a single experiment are shown. Values above the columns (means±S.E.M.; n=3 separate preparations of myocytes) are the ratios of kinase activities (c.p.m. incorporated/30 min of incubation) in myocytes in the absence or presence of calyculin A. The panels above the histogram are immunoblots with anti-FLAG antibodies of extracts of myocytes expressing the various FLAG–MST3 species demonstrating that similar quantities of FLAG–MST3 were used in the immunokinase assays.
The phosphomimetic mutation, FLAG–MST3(T178D), resulted in the constitutive appearance of 58 kDa FLAG–MST3(T178D), suggesting stimulation of cis-autokinase activity, though 54 kDa FLAG–MST3(T178D) was never completely lost (Figure 3C). Calyculin A did not cause a complete shift of FLAG–MST3(T178D) to the 58 kDa band (Figure 3C), although there may have been an increase in the relative abundance of 58 kDa FLAG–MST3(T178D) species following exposure of myocytes to calyculin A compared with untreated myocytes (Figure 3C), which presumably results from increased preservation of FLAG–MST3(Thr328) phosphorylation. Mutation of the essential Lys53 in the MST3 ATP-binding site to an arginine residue [FLAG–MST3(K53R)] prevented the appearance of any 58 kDa FLAG–MST3 on exposure of myocytes to calyculin A and the MST3(275–393) mAb epitope was preserved (Figure 3C).
In vitro immunokinase activities of the various MST3 mutants were assessed by phosphorylation of MBP. Myocytes were infected with AdV–FLAG–MST3 constructs titrated to produce essentially equal expression of FLAG–MST3 species (Figure 3D). MST3 is reportedly more active in such assays with Mn2+ rather than with Mg2+ as the metal ion cofactor, although the relative magnitude varies with metal ion concentration [6,19,49]. In our assays, FLAG–MST3(FL) was only ~30% as active with 20 mM Mn2+ in untreated or calyculin A-treated myocytes as with 20 mM Mg2+ (results not shown), thus Mg2+ was routinely used. Activities of FLAG–MST3(FL), FLAG–MST3(Δ314–431) and FLAG-MST3(T328A) were stimulated to a similar overall level by calyculin A (Figure 3D), suggesting that FLAG–MST3(Thr328) phosphorylation does not play any direct role in the regulation of MST3 activity. Furthermore, the fact that activity of FLAG–MST3(1–313) was indistinguishable from FLAG–MST3(FL) (Figure 3D) confirms that phosphorylation of MST3(Thr178) in the catalytic domain did not contribute significantly to the reduced migration of FLAG–MST3(FL). Because FLAG–MST3(Δ314–431) was not constitutively active, the concept that caspase cleavage of MST3 results in constitutive activation [24,28] could not be confirmed in our system. FLAG–MST3(Δ341–431) and FLAG–MST3(Δ377–431) were activated similarly to FLAG–MST3(Δ314–431) and FLAG–MST3(FL) (results not shown). As expected, FLAG–MST3(T178A) exhibited a very low level of relative activation by calyculin A (Figure 3D) and FLAG–MST3(K53R) was completely inactive (results not shown), consistent with the findings of Lu et al. [19]. FLAG–MST3(T178D) may be slightly more active in the absence of calyculin A exposure than FLAG-MST3(FL) and, as expected, was only very slightly activated by calyculin A. Our general conclusion is that the activation of FLAG–MST3(FL) and the appearance of the FLAG–MST3 [i.e. FLAG–MST3(phosphoThr178/phosphoThr328)] band at 58 kDa are different aspects of the same process.
Although we have concluded that the phosphorylation of MST3(Thr328) represents a cis-autophosphorylation, the kinase(s) responsible for phosphorylation of MST3(Thr178) is unclear. Previous work suggests that the MST3(Thr178) phosphorylation is also an autophosphorylation [6,19]. FLAG–MST3(T178A) expresses a very low, but detectable, MBP kinase activity (Figure 3D), whereas in contrast FLAG–MST3(K53R) is completely inactive (results not shown). Hence autophosphorylation of MST3(Thr178) could lead to rapid autocatalytic activation following PP1/PP2A inhibition. In addition, we examined the rat MST3 primary sequence (Supplementary Figure S1) with Scansite (http://scansite.mit.edu). At medium stringency, Scansite identified PKA as a putative MST3(Thr178) kinase. However, isoprenaline did not cause any reduction in mobility in FLAG–MST3(FL) on SDS/PAGE that should result from MST3(Thr178) phosphorylation and subsequent MST3(Thr328) cis-autophosphorylation (results not shown). At high stringency, MST3(Ser305) was identified as a putative casein kinase 2 site. Although it is possible that this site is phosphorylated, its phosphorylation does not contribute to the reduction in mobility seen with calyculin A (Supplementary Figure S3B).
Lu et al. [19] identified two tryptic phosphopeptides in autophosphorylated recombinant MST3(FL), one of which contained phosphoThr178 that was responsible for autoactivation. The second tryptic phosphopeptide was not identified and could contain MST3(phosphoThr328). We did not detect ‘constitutive’ autophosphorylation (Figure 1B, lower panel) or autoactivation (Figure 3D) of FLAG–MT3(FL) in myocytes in the absence of calyculin A. However, immunoblots in the study by Lu et al. [19] of immunoprecipitated HA–MST3(FL) expressed in COS-1 cells [with antibodies against residues 310–432 (sic)] detected two bands. These were identified as ‘unphosphorylated’ HA–MST3(FL) and autophosphorylated HA–MST3(FL). but, from the extent of the reduction in mobility, the latter may represent HA–MST3(phosphoThr178/phosphoThr328) rather than HA–MST3(phosphoThr178).
Canonical pathway of activation of MST3 in cardiac myocytes
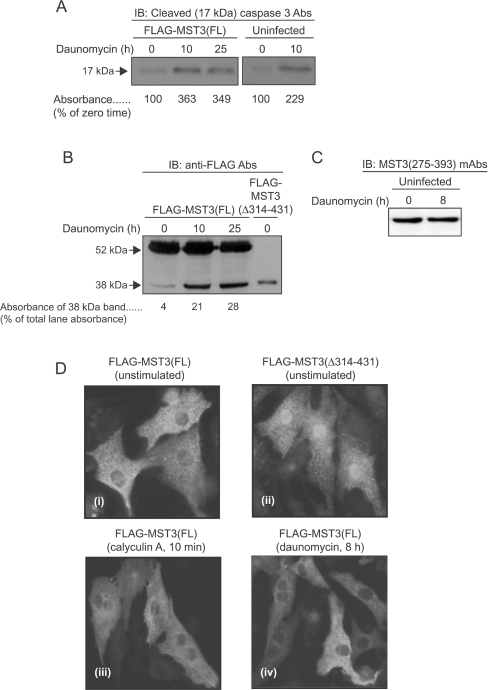
Others have shown that MST3 is cleaved immediately C-terminal to Asp313 by caspases during apoptosis, generating a species that translocates to the nucleus [24,28]. The anthracycline daunomycin stimulates apoptosis and caspase activation in cardiac myocytes [50–52], thus we examined its effects on cleavage of FLAG–MST3(FL). As expected, daunomycin increased the appearance of the active 17 kDa fragment of caspase 3 in uninfected cardiac myocytes or myocytes expressing FLAG–MST3(FL), confirming stimulation of apoptosis (Figure 4A). There was detectable cleavage of FLAG–MST3(FL) to a form that co-migrated with FLAG–MST3(Δ314–431) (Figure 4B), indicating that induction of apoptosis resulted in MST3 cleavage. However, there was no detectable loss of the endogenous immunosignal detected by MST3(275–393) mAbs [which do not recognize FLAG–MST3(Δ314–431); Figure 2B] in uninfected myocytes (Figure 4C). We also examined the subcellular localization of FLAG–MST3(FL) following activation by calyculin A and of FLAG–MST3(Δ314–431). In unstimulated cells, FLAG–MST3(FL) was mainly cytoplasmic [Figure 4D(i)], whereas FLAG–MST3(Δ314–431) constitutively accumulated in the nucleus [Figure 4D(ii)] even though it cannot be enzymically active (Figure 3D). No significant accumulation of FLAG–MST3(FL) in the nucleus was detected following exposure to calyculin A [Figure 4D(iii)]. We could not detect any nuclear translocation of cleaved FLAG–MST3 into the nucleus in myocytes expressing FLAG–MST3(FL) following exposure to daunomycin [Figure 4D(iv)], presumably because it represents only a comparatively minor species (Figure 4B). Thus aspects of the canonical pathway for activation of MST3 operate in cardiac myocytes, in that MST3 is cleaved during apoptosis (Figure 4B) and the inactive cleaved form can potentially migrate to the nucleus [Figure 4D(ii)].
Figure 4. Activation of caspase 3 and cleavage of FLAG–MST3(FL) in myocytes exposed to daunomycin.
Myocytes were uninfected or were infected with AdV–FLAG–MST3(FL) or AdV–FLAG–MST3(Δ314–431) (to identify the location of the ‘caspase-cleaved’ form of FLAG-MST3) and (A–C) extracts were prepared. The numbers below the various immunoblot lanes represent relative band absorbances. (A and B) Myocytes were exposed to 0.5 μM daunomycin for the times indicated. Extracts were immunoblotted (IB) with antibodies (Abs) to cleaved (17 kDa) caspase 3 (A), anti-FLAG antibodies (B) or MST3(275–393) (C). (D) Immunocytochemical localization of FLAG–MST3(FL) or FLAG–MST3(Δ314–431) in unstimulated [(Di) and (Dii)] myocytes, and in myocytes exposed to calyculin A [100 nM for 10 min; (Diii)] or daunomycin [0.5 μM for 8 h; (Div)]. Experiments were repeated at least twice with different myocyte preparations with similar results.
Structural features of MST3 and changes associated with activation by calyculin A
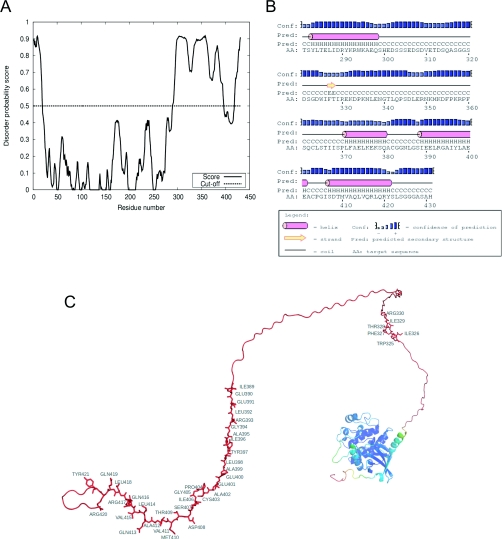
Since the principal mechanism of activation of MST3 by calyculin A does not appear to involve caspase cleavage, we took a computational and biochemical approach to investigate the structure of MST3. The X-ray structure of the catalytic domain (residues 1–303) of MST3 has been determined [53,54], but little is known about the regulatory domain. We first assessed the degree of disorder of rat MST3 using DISOclust [32]. The initial ~20 amino acids at the N-terminus constitute a disordered region that precedes a highly ordered structure (~270 residues) associated with the (conserved) kinase catalytic domain (Figure 5A). In contrast, the regulatory domain [MST3(314–431)] is highly disordered apart from a relatively low-probability-ordered region (residues 397–418) towards the C-terminus. Residues 281–431 were examined using PSIPRED [33]. This detected a region of low-probability strand structure [W325IFTIR330; note that this contains MST3(Thr328)] and a helical region between residues 389 and 421 (Figure 5B) corresponding to the dip in disorder probability between residues 397 and 418 in Figure 5(A). A representation of the three-dimensional model for MST3(FL) using the IntFOLD-TS method [34,35] shows the disordered N-terminal domain, the ordered catalytic domain and the largely disordered regulatory domain (Figure 5C). The strand W325IFTIR330 and the C-terminal predicted helical region sequences are marked.
Figure 5. Modelling of MST3 structure.
(A) Disorder probability plot for MST3(FL) produced with the DISOclust method for the prediction of intrinsically unstructured regions in proteins. (B) Graphical view of predicted secondary structure of MST3(281–431) from the PSIPRED server. (C) Representation of the three-dimensional model for MST3(FL) full-length sequence, produced using the IntFOLD-TS method. Colouring represents predicted local errors, from blue (high accuracy) through green, yellow and orange to red (low accuracy) using the ModFOLDclust2 model quality assessment method. The intrinsically unstructured residues that have a high predicted error are shown in red. Residue labelling and the stick representation indicates the WIFTIR sequence and the C-terminal predicted helical region. The image was rendered using PyMol.
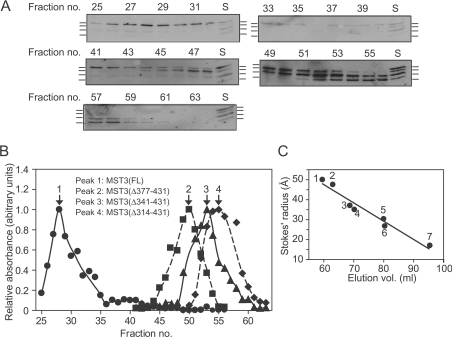
Stokes radii (a) of the various FLAG–MST3 species were determined by gel filtration. The Stokes radius of FLAG–MST3(FL) by Sephacryl S200HR (Figures 6A and 6B) or S300HR (Figures 7A, 7C and 7E) chromatography was 47.3–49.6 Å (where 1 Å=0.1 nm; Table 1), considerably greater than the value of 29.2 Å calculated for a globular FLAG–MST3 monomer of ~49 kDa molecular mass. Either FLAG–MST3(FL) is highly asymmetric (axial ratio ~20 for an unhydrated prolate ellipsoid as calculated from the frictional coefficient [55]) which could result from the highly disordered structure of its regulatory domain (Figure 5) or it is complexed with other proteins (including possibly itself). Using myocytes infected variously with combinations of FLAG–MST3(FL), HA–MST3(FL) and GST–MST3(FL) followed by immunoprecipitation or glutathione–Sepharose precipitation then immunoblotting, we were unable to find any evidence for homomultimer formation (results not shown). The Stokes radius of FLAG–MST3(Δ377–431) was significantly less than that of FLAG–MST3(FL) and, although still highly asymmetric, was closer to the Stokes radius calculated for a globular protein (Figures 6A and 6B, and Table 1), suggesting that the C-terminal MST3(377–431) sequence may be involved in a heterologous interaction with another protein. As would be predicted, FLAG–MST3(Δ341–431) and FLAG–MST3(Δ314–431) also eluted at volumes closer to their calculated Stokes radii for corresponding globular proteins (Figure 6A and 6B, and Table 1).
Figure 6. Sephacryl S200HR chromatography of FLAG–MST3.
Extracts of unstimulated AdV-infected myocytes individually expressing FLAG–MST3(FL), FLAG–MST3(Δ314–431), FLAG–MST3(Δ341–431) and FLAG–MST3(Δ377–431) were combined and subjected to gel-filtration chromatography using Sephacryl S200HR. (A) Immunoblotting of column fractions with anti-FLAG antibodies. The lowest mobility band is FLAG–MST3(FL) and the highest is FLAG–MST3(Δ314–431). Lane S is a standard containing combined unfractionated extracts from myocytes expressing the four MST3 species. (B) Elution profile showing quantification of band intensities. ●, FLAG–MST3(FL); ■, FLAG–MST3(Δ377–431); ▲, FLAG–MST3(Δ341–431); and ◆, FLAG–MST3(Δ314–431). (C) The Sephacryl S200HR column was standardized with proteins of known Stokes radii (taken from the literature). 1: IgG, 50.0 Å; 2: BSA dimer, 47.5 Å; 3: rabbit muscle phosphorylase b monomer, 37.0 Å; 4: BSA monomer, 35.0 Å; 5: ovalbumin, 30.5 Å; 6: β-lactoglobin, 26.7 Å; 7: cytochrome c, 17.0 Å. Experiments were repeated three times with different myocyte preparations with similar results.
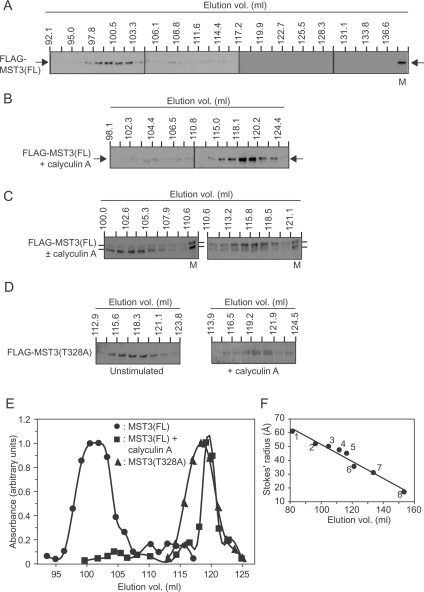
Figure 7. Sephacryl S300HR chromatography of FLAG–MST3 in myocyte extracts.
AdV-infected myocytes expressing (A–C) FLAG–MST3(FL) or (D) FLAG–MST3(T328A) were unstimulated or exposed to calyculin A (100 nM for 10 min) and the extracts were subjected to gel-filtration chromatography using Sephacryl S300HR. (A–D) Column fractions were immunoblotted with anti-FLAG antibodies: (A) fractions from unstimulated myocytes; (B) fractions from myocytes exposed to calyculin A; (C) fractions from mixed extracts from unstimulated myocytes and myocytes exposed to calyculin A; and (D) fractions from unstimulated myocytes (left-hand panel) or myocytes exposed to calyculin A (right-hand panel). In (A), M is a marker lane of FLAG–MST3(FL) in whole myocyte extracts. In (C), M is a marker lane of mixed whole extracts of FLAG–MST3(FL) from myocytes unexposed or exposed to calyculin A. (E) Elution profiles for the quantified imunoblots. ●, FLAG–MST3(FL); ■, FLAG–MST3(FL) from myocytes exposed to calyculin A; and ▲, FLAG–MST3(T328A). (F) The Sephacryl S300HR column was standardized with proteins of known Stokes radii (taken from the literature). 1: ferritin, 61.0 Å; 2: catalase tetramer, 52.0 Å; 3: IgG, 50.0 Å; 4: BSA dimer, 47.5 Å; 5: catalase dimer, 45.0 Å; 6: BSA monomer, 35.0 Å; 7: Hb, 31.0 Å; 8: cytochrome c, 17.0 Å. Experiments were repeated at least twice with different myocyte preparations with similar results.
Table 1. Stokes radii of FLAG–MST3.
Stokes radii were measured using calibrated Sephacryl S200HR or S300HR columns (see Figures 6 and 7). Molecular mass (M in Da) was calculated from amino acid sequences. Frictional ratios (f/f0) were calculated from the formula: f/fo=a/(3vM/4π N)1/3 where a in the Stokes radius, v is the partial specific volume (assumed to be 0.74 ml/g) and N is Avagadro's number. The theoretical values of Stokes radii for globular proteins were calculated assuming a value of 1.2 for f/f0 [75]. Axial ratios for prolate, unhydrated ellipsoids of revolution were derived from published Tables [55].
| FLAG–MST3 species | M (Da) | a (Å), S300HR | a (Å), S200HR | Theoretical monomeric a (Å) | (f/f0) | Axial ratio |
|---|---|---|---|---|---|---|
| FLAG–MST3(FL) | 48984 | 49.6±0.6 (8) | 47.3±0.3 (3) | 29.2 | 1.95–2.04 | 19–21 |
| FLAG–MST3(FL) in calyculin A-treated (100 nM, 10 min) myocytes | 48984 | 40.1±0.2 (7) | – | 29.2 | 1.65 | 12 |
| FLAG–MST3(T328A) | 48954 | 39.0±0.7 (3) | – | 29.2 | 1.61 | 11 |
| FLAG–MST3(Δ377–431) | 43180 | – | 35.2±0.6 (3) | 28.0 | 1.51 | 9.4 |
| FLAG–MST3(Δ341–431) | 39089 | – | 32.9±0.6 (3) | 27.1 | 1.46 | 8.5 |
| FLAG–MST3(Δ314–431) | 36255 | – | 31.3±0.3 (3) | 26.4 | 1.42 | 7.8 |
Exposure of myocytes expressing FLAG–MST3(FL) to calyculin A caused a dramatic reduction in the Stokes radius, suggesting dissociation of a macromolecular complex involving FLAG–MST3(FL) or a dramatic shape change (Figures 7A–7C and 7E, and Table 1). Mixing of FLAG–MST3(FL) extracts from myocytes exposed or not exposed to calyculin A clearly showed the separation of inactive FLAG–MST3(FL) (Figure 7C, lower band) from activated FLAG-MST3(FL) (Figure 7C, upper band). Furthermore, the Stokes radius of FLAG–MST3(T328A) is significantly less (P<0.001) than FLAG–MST3(FL) (Figures 7D and 7E, and Table 1) and its elution volume was not significantly altered by exposure of myocytes to calyculin A (Figure 7D), suggesting that it does not interact with other proteins in the same manner as FLAG–MST3(FL).
Interaction of MST3 with GOLGA2/gm130 and MO25, and the function of MST3(Thr328) phosphorylation
YSK1/SOK1 (another member of the GCKIII subgroup) interacts with the Golgi protein GOLGA2/gm130 [56–59] with residues 20–302 of YSK1/SOK1 (this represents the entire catalytic domain) being important in this interaction [56]. Fidalgo et al. [58] have interpreted the data from [56] as indicating that residues 270–302 are critical in the interaction of YSK1/SOK1 with GOLGA2/gm130. However, an interaction between MST3 and GOLGA2/gm130 could not be identified [56]. This is surprising, given that the residues in rat MST3 {MST3(274–306), which also contain the bipartite NLS [28]} are homologous to YSK1/SOK1(270–302) (24/33 identity with 4/33 conservative replacements). We examined the interaction between FLAG–MST3(FL) and GOLGA2/gm130 in myocytes with and without calyculin A exposure. GOLGA2/gm130 was detected in immunoprecipitates of FLAG–MST3(FL) in unstimulated myocytes indicating an association (Figure 8A). Incubation of myocytes with calyculin A resulted in a decrease in this association (Figures 8A and 8B). Interactions (not necessarily direct and sometimes mediated by GOLGA2/gm130 [58]) between GCKIII members and CCM3/PDCD10, one of the proteins important in cerebral cavernous malformations [11], have been demonstrated [12,15,58,60,61]. We therefore also attempted to detect interactions between FLAG–MST3 and CCM3/PDCD10, but were unsuccessful (results not shown). However, further investigation of interactions between MST3 and heterologous proteins is warranted especially in view of recent work on striatins. Striatins (most abundant in the brain [62,63] although splice variants are expressed in the heart [64]) are PP2A B-type regulatory subunit scaffolding proteins which form complexes [STRIPAKs (striatin-interacting phosphatase and kinases)] with PP2A and additionally contain GCKIII members, CCM3/PDCD10 and phocein/Mob3 (which is present in the Golgi and other membranous structures) [15,60,65,66].
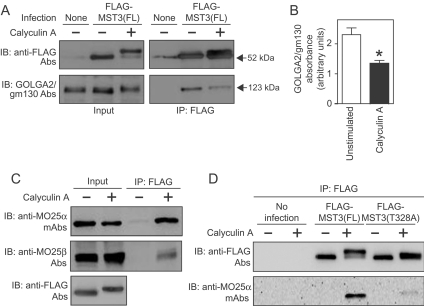
Figure 8. Co-immunoprecipitation of FLAG–MST3 with GOLGA2/gm130 or MO25.
Uninfected or AdV-infected myocytes expressing FLAG–MST3(FL) or FLAG–MST3(T328A) were unstimulated or exposed to calyculin A (100 nM for 10 min) and extracts were prepared. FLAG–MST3(FL) was immunoprecipitated (IP) with anti-FLAG M2 affinity gel and subjected to immunoblot (IB) analysis. (A) Co-immunoprecipitation of GOLGA2/gm130 with FLAG–MST3(FL). Inputs (left-hand panels) or immunoprecipitates (right-hand panels) of myocyte extracts were immunoblotted with anti-FLAG antibodies (Abs, upper panels) or antibodies against GOLGA2/gm130 (lower panels). (B) Quantification of GOLGA2/gm130 co-immunoprecipitated with FLAG–MST3(FL). Results are means±S.E.M. for three separate preparations of myocytes; *P<0.001 compared with unstimulated myocytes. (C) Co-immunoprecipitation of MO25α and MO25β with FLAG–MST3(FL). All myocytes were infected with AdV–FLAG–MST3(FL). The two lanes to the left-hand side show inputs of myocyte extracts with immunoprecipitates in the two lanes to the right-hand side. Extracts were immunoblotted with anti-MO25α mAbs (top panel), or antibodies to MO25β (middle panel) or FLAG (bottom panel). (D) FLAG–MST3(T328A) does not interact with MO25α. Anti-FLAG M2 affinity gel was used to immunoprecipitate FLAG–MST3 from myocytes expressing FLAG–MST3(FL) (middle lanes) or myocytes expressing FLAG–MST3(T328A) (right-hand lanes). Control immunoprecipitations from uninfected myocytes are shown in the left lanes. The upper panel shows FLAG immunoreactivity and the lower panel shows MO25α immunoreactivity in the immunoprecipitates. Experiments were repeated at least twice with different myocyte preparations with similar results.
MO25 (of which there are two isoforms, MO25α and MO25β) is an evolutionarily conserved protein first identified by its expression during early murine embryogenesis [67,68]. It is best characterized as a scaffolding protein in a heterotrimer with the STRAD pseudokinases and the LKB1 (liver kinase B1) tumour suppressor kinase [3]. Binding of STRADα to MO25 involves four interaction sites (sites A–D) in STRADα: Glu105 (site A), Ile138 (site B), Tyr185 (site C) and the C-terminal Trp429–(Glu430)–Phe431 [W(E)F] motif (site D) [69,70]. A recent analysis of human Ste20-related kinases revealed that all four sites are conserved in the GCKIII and GCKVI subgroups, with sites A–C lying in the catalytic domain and site D in the C-terminal regulatory domain [4]. In rat MST3 (Supplementary Figure S1), at Glu58, Leu90 and Tyr134 constitute sites A–C respectively, and the STRADα site DW(E)F motif is replaced by a WIF motif at residues 325–327, immediately N-terminal to MST3(Thr328). Surface plasmon resonance showed that MO25 isoforms (MO25α and MO25β) bind to the two GCKVI members [OXSR1 (oxidative-stress responsive 1)/OSR1/KIAA1101 and SPAK (SPS1-related proline-alanine-rich protein kinase)/PASK/Stk39 (serine threonine kinase 39) and the GCKIII member MST4/MASK with Kd1 values in the 3–10 μM range (as opposed to about 10–50 nM with STRADα) [3,4]. MST3 and YSK1/SOK1 were not studied. The activities of the wild-type GCKIII kinases are stimulated 2–5-fold by MO25α [4]. Stimulation of the activities of the GCKVI members is greater (70–100-fold), but the threonine residue of their activation loops had been mutated phosphomimetically to glutamic acid, so the situation is not necessarily comparable.
Because the sites involved in interaction with MO25 are conserved in all five GCKIII/GCKVI members, we examined the interaction between MO25 and FLAG–MST3(FL). Neither MO25α nor MO25β co-immunoprecipitated with FLAG–MST3(FL) in myocyte extracts in the absence of exposure to calyculin A (Figure 8C, top and middle panels). However, following exposure of myocytes to calyculin A and associated phosphorylation of FLAG–MST3(Thr328) (Figure 8C, bottom panel)], MO25α and MO25β each co-immunoprecipitated with FLAG–MST3(FL) (Figure 8C, top and middle panels). In an analogous experiment with myocytes expressing FLAG–MST3(FL) or FLAG–MST3(T328A) at approximately equal levels (Figure 8D, upper panel), MO25α did not co-immunoprecipitate with FLAG–MST3(T328A) to any significant extent following exposure of myocytes to calyculin A (Figure 8D, lower panel), indicative of an absolute requirement for phosphorylation of this site for association. Thus phosphorylation of MST3(Thr328) leads to decreased association with GOLGA2/gm130, together with increased association with MO25.
Filippi et al. [4] were unable to demonstrate co-immunoprecipitation of MO25 with any GCKIII or GCKVI kinase and attributed this to the high Kd values for the interactions. Our co-immunoprecipitation experiments show that calyculin A-activated FLAG–MST3(FL) does interact with MO25α (and MO25β) (Figures 8C and 8D), an interaction that does not occur with calyculin A-activated FLAG–MST3(T328A) (Figure 8D). Interestingly, the W325IFTIR330 sequence of MST3 corresponding to a low-probability strand region (Figures 5B and 5C) is conserved in both YSK1/SOK1 (W319TFPPTIR326) and MST4/MASK (W324SFTTVR330) (Table 2). In the GCKVI members, where the WIF motif is replaced by WEW motif, the residue in the +1 position relative to the C-terminal tryptophan is a serine which lies in a highly acidic region in these kinases (Table 2). Although phosphorylation of a serine in the regulatory domain of SPAK/PASK/Stk39 or OXSR1/OSR1/KIAA1101 has been identified, the relevant residue lies in the −14 position relative to the serine residue of the WEWS motif [71–73]. Given that all of the GCKIII and GCKVI members except YSK1/SOK1 contain a serine/threonine residue +1 to the W(I/S/E)(F/W) motif (and, even in YSK1/SOK1, there is a threonine residue at +3), we suggest that the scheme of activation that we are proposing for MST3 might be more widely applicable to other GCKIII/VI members. Furthermore, we can speculate that the relatively low affinity of GCKIII/GCKVI members for MO25 [4] might be attributable to the serine/threonine residue immediately C-terminal to the STRADα-like site D being unphosphorylated.
Table 2. Sequences of the WX(F/W)(S/T)-like sequence in GCKIII and GCKVI subgroups in H. sapiens and mouse.
The WX(F/W)(S/T)-like sequence corresponds to site D in STRADα [4]. In Xenopus laevis MST3 (GenBank® acession number NP_001085728), the sequence is W323NFTRR328, and in Danio rerio MST3 (GenBank® acession number NP_998642), it is W315IFTIR320. It is also conserved in MST3 of many other vertebrates. For the GCKVI subgroup, the STRADα site D-like ‘WEW’ sequence has been extended on either side to show the acidic nature of the region.
| GCK subgroup | GenBank® accession number (H. sapiens), with residue numbers of the WX(F/W)(S/T)-like sequence in parentheses | GenBank® accession number (mouse), with residue numbers of the WX(F/W)(S/T)-like sequence in parentheses | WX(F/W)(S/T)-like sequence |
|---|---|---|---|
| GCKIII subgroup | |||
| MST3 (Stk24) | NP_001027467 (325–330) | NP_663440 (325–330) | WIF–TIR |
| YSK1/SOK1 (Stk25) | NP_006365 (319–326) | NP_067512 (319–326) | WTFPPTIR |
| MST4/MASK | NP_057626 (324–330) | NP_598490 (324–330) | WSFT-TVR |
| GCKVI subgroup | |||
| SPAK/PASK (Stk39) | NP_037365 (378–395) | NP_058562 (390–407) | EDGDWEWSDDEMDEKSEE |
| OXSR1/OSR/KIAA1101 | NP_005100 (332–349) | NP_598746 (332–349) | EDGGWEWSDDEFDEESEE |
A model for non-canonical regulation of MST3
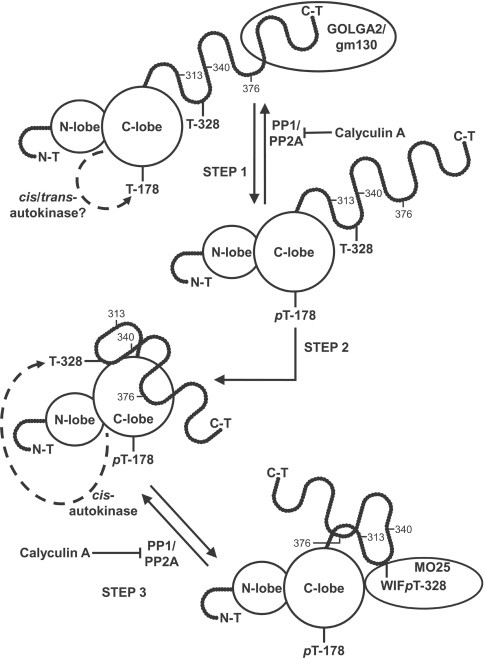
From our experiments, we have developed the following model (Figure 9). MST3 is bound to the Golgi apparatus protein GOLGA2/gm130 [59] in the basal state, probably through the MST3(377–431) region. GOLGA2/gm130 might retain MST3 in the cytoplasm which may be why FLAG–MST3(Δ314–431) (which gel-filtration experiments suggest is unable to interact with GOLGA2/gm130) constitutively localizes to the nucleus [Figure 4D(ii)]. Calyculin A activates MST3 by promoting phosphorylation of MST3(Thr178), and this leads to cis-autophosphorylation of MST3(Thr328) and dissociation of the MST3–GOLGA2/gm130 complex. The MST3(Thr328) cis-autophosphorylation requires the ‘docking domain’ encompassed by MST3 residues 341–376 and does not directly affect MST3 activity. MST3(Thr328) lies within a conserved WIFTIR sequence that may represent a structured region in the regulatory domain which is, overall, highly disordered. Cis-autophosphorylation of MST3(Thr328) simultaneously allows MO25 to bind to MST3. The function of the MST3–MO25 interaction is unknown as yet. In the MO25–STRAD–LKB1 complex, the GCKVI homologue STRAD is involved in nucleocytoplasmic shuttling of LKB1 and the interaction between LKB1 and STRAD (stabilized by MO25) localizes LKB1 to the cytoplasm [69,74]. It is possible that the various complexes that MST3 forms are related to the determination of subcellular localization.
Figure 9. A model of the non-canonical pathway of MST3 regulation.
For details, see the Results and Discussion section. N-T, N-terminus; C-T, C-terminus; N-lobe and C-lobe, lobes of catalytic domain. The numbers associated with the disordered regulatory domain denote the relevant amino acid residues (Asp313, Gly340 and Lys376) at the C-termini of FLAG–MST3(Δ314–431), FLAG–MST3(Δ341–431) and FLAG–MST3(Δ377–431) respectively.
Online data
AUTHOR CONTRIBUTION
Stephen Fuller performed the majority of the experimental work including the preparation of the many AdV constructs. Liam McGuffin was responsible for the modelling of the MST3 structure. Andrew Marshall performed some of the immunoblots. Alejandro Giraldo performed the immunofluorescence staining. Sampsa Pikkarainen made the initial observation shown in Figure 1(A) and assisted with the molecular cloning of MST3. Angela Clerk prepared the cardiac myocytes. Peter Sugden performed the hydrodynamic characterization of MST3. The experiments were conceived by Angela Clerk and Peter Sugden in conjunction with Stephen Fuller. The paper was written primarily by Peter Sugden with the assistance of Angela Clerk, and Stephen Fuller and Liam McGuffin also contributed.
ACKNOWLEDGEMENTS
We also thank Dario Alessi and Paola de los Heros (MRC Protein Phosphorylation Unit, University of Dundee, Dundee, U.K.) for providing us with the MO25β antibody.
FUNDING
This work was supported by the British Heart Foundation [grant number PG/07/060/23276] and Fondation Leducq [grant number CV05-02].
References
- 1.Dan I., Watanabe N. M., Kusumi A. The Ste20 group kinases as regulators of MAP kinase cascades. Trends Cell Biol. 2001;11:220–230. doi: 10.1016/s0962-8924(01)01980-8. [DOI] [PubMed] [Google Scholar]
- 2.Delpire E. The mammalian family of sterile 20p-like protein kinases. Pflügers Arch. 2009;458:953–967. doi: 10.1007/s00424-009-0674-y. [DOI] [PubMed] [Google Scholar]
- 3.Alessi D. R., Sakamoto K., Bayascas J. R. LKB1-dependent signaling pathways. Annu. Rev. Biochem. 2006;75:137–163. doi: 10.1146/annurev.biochem.75.103004.142702. [DOI] [PubMed] [Google Scholar]
- 4.Filippi B. M., de los Heros P., Mehellou Y., Navratilova I., Gourlay R., Deak M., Plater L., Toth R., Zeqiraj E., Alessi D. R. MO25 is a master regulator of SPAK/OSR1 and MST3/MST4/YSK1 protein kinases. EMBO J. 2011;30:1730–1741. doi: 10.1038/emboj.2011.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pombo C. M., Force T., Kyriakis J., Nogueira E., Fidalgo M., Zalvide J. The GCK II and III subfamilies of the STE20 group kinases. Front. Biosci. 2007;12:850–859. doi: 10.2741/2107. [DOI] [PubMed] [Google Scholar]
- 6.Schinkmann K., Blenis J. Cloning and characterization of a human STE20-like protein kinase with unusual cofactor requirements. J. Biol. Chem. 1997;272:28695–28703. doi: 10.1074/jbc.272.45.28695. [DOI] [PubMed] [Google Scholar]
- 7.Pombo C. M., Bonventre J. V., Molnar A., Kyriakis J., Force T. Activation of a human Ste20-like kinase by oxidant stress defines a novel stress response pathway. EMBO J. 1996;15:4537–4546. [PMC free article] [PubMed] [Google Scholar]
- 8.Osada S., Izawa M., Saito R., Mizuno K., Suzuki A., Hirai S., Ohno S. YSK1, a novel mammalian protein kinase structurally related to Ste20 and SPS1, but is not involved in the known MAPK pathways. Oncogene. 1997;14:2047–2057. doi: 10.1038/sj.onc.1201043. [DOI] [PubMed] [Google Scholar]
- 9.Qian Z., Lin C., Espinosa R., LeBeau M., Rosner M. R. Cloning and characterization of MST4, a novel Ste20-like kinase. J. Biol. Chem. 2001;276:22439–22445. doi: 10.1074/jbc.M009323200. [DOI] [PubMed] [Google Scholar]
- 10.Dan I., Ong S.-E., Watanabe N. M., Blagoev B., Nielsen M. M., Kajikawa E., Kristiansen T. Z., Mann M., Pandey A. Cloning of MASK, a novel member of the mammalian germinal center kinase III subfamily, with apoptosis-inducing properties. J. Biol. Chem. 2002;277:5929–5939. doi: 10.1074/jbc.M110882200. [DOI] [PubMed] [Google Scholar]
- 11.Riant F., Bergametti F., Ayrignac X., Boulday G., Tournier-Lasserve E. Recent insights into cerebral cavernous malformations: the molecular genetics of CCM. FEBS J. 2010;277:1070–1075. doi: 10.1111/j.1742-4658.2009.07535.x. [DOI] [PubMed] [Google Scholar]
- 12.Zheng X., Xu C., Di Lorenzo A., Kleaveland B., Zou Z., Seiler C., Chen M., Cheng L., Xiao J., He J., et al. CCM3 signaling through sterile 20-like kinases plays an essential role during zebrafish cardiovascular development and cerebral cavernous malformations. J. Clin. Invest. 2010;120:2795–2804. doi: 10.1172/JCI39679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Irwin N., Li Y.-M., O'Toole J. E., Benowitz L. I. Mst3b, a purine-sensitive Ste20-like protein kinase, regulates axon outgrowth. Proc. Natl. Acad. Sci. U.S.A. 2006;103:18320–18325. doi: 10.1073/pnas.0605135103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhou T.-H., Ling K., Guo J., Zhou H., Wu Y.-L., Jing Q., Ma L., Pei G. Identification of a human brain-specific isoform of mammalian STE20-like kinase 3 that is regulated by cAMP-dependent protein kinase. J. Biol. Chem. 2000;275:2513–2519. doi: 10.1074/jbc.275.4.2513. [DOI] [PubMed] [Google Scholar]
- 15.Gordon J., Hwang J., Carrier K. J., Jones C. A., Kern Q. L., Moreno C. S., Karas R. H., Pallas D. C. Protein phosphatase 2a (PP2A) binds within the oligomerization domain of striatin and regulates the phosphorylation and activation of the mammalian Ste20-like kinase Mst3. BMC Biochem. 2011;12:54. doi: 10.1186/1471-2091-12-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rowland E. A., Müller T. H., Goldstein M., Greene L. A. Cell-free detection and characterization of a novel nerve growth factor-activated protein kinase in PC12 cells. J. Biol. Chem. 1987;262:7504–7513. [PubMed] [Google Scholar]
- 17.Volonté C., Rukenstein A., Loeb D. M., Greene L. A. Differential inhibition of nerve growth factor responses by purine analogues: correlation with inhibition of a nerve growth factor-activated protein kinase. J. Cell Biol. 1989;109:2395–2403. doi: 10.1083/jcb.109.5.2395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Volonté C., Greene L. A. Nerve growth factor-activated protein kinase N. Characterization and rapid near homogeneity purification by nucleotide affinity-exchange chromatography. J. Biol. Chem. 1992;267:21663–21670. [PubMed] [Google Scholar]
- 19.Lu T.-J., Lai W.-Y., Huang C.-Y. F., Hsieh W.-J., Yu J.-S., Hsieh Y.-J., Chang W.-T., Leu T.-H., Chang W.-C., Chuang W.-J., et al. Inhibition of cell migration by autophosphorylated mammalian sterile 20-like kinase 3 (MST3) involves paxillin and protein-tyrosine phosphatase-PEST. J. Biol. Chem. 2006;281:38405–38417. doi: 10.1074/jbc.M605035200. [DOI] [PubMed] [Google Scholar]
- 20.Suganuma M., Fujiki H., Okabe S., Nishiwaki S., Brautigan D., Ingebritsen T. S., Rosner M. R. Structurally different members of the okadaic acid class selectively inhibit protein serine/threonine but not tyrosine phosphatase activity. Toxicon. 1992;30:873–878. doi: 10.1016/0041-0101(92)90385-i. [DOI] [PubMed] [Google Scholar]
- 21.Stegert M. R., Hergovich A., Tamaskovic R., Bichsel S. J., Hemmings B. A. Regulation of NDR protein kinase by hydrophobic motif phosphorylation mediated by the mammalian Ste20-like kinase MST3. Mol. Cell. Biol. 2005;25:11019–11029. doi: 10.1128/MCB.25.24.11019-11029.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wu H.-Y., Lin C.-Y., Lin T.-Y., Chen T.-C., Yuan C.-J. Mammalian Ste20-like protein kinase 3 mediates trophoblast apoptosis in spontaneous delivery. Apoptosis. 2008;13:283–294. doi: 10.1007/s10495-007-0161-x. [DOI] [PubMed] [Google Scholar]
- 23.Chen C.-B., Ng J. K.W, Choo P.-H., Wu W., Porter A. G. Mammalian sterile 20-like kinase 3 (MST3) mediates oxidative stress-induced cell death by modulation JNK activation. Biosci. Rep. 2009;29:405–415. doi: 10.1042/BSR20090096. [DOI] [PubMed] [Google Scholar]
- 24.Huang C.-Y. F., Wu Y.-M., Hsu C.-Y., Lee W.-S., Lai M-D., Lu T.-J., Huang C.-L., Leu T.-H., Shih H.-M., Fang H.-I., et al. Caspase activation of mammalian Sterile 20-like kinase 3 (Mst3). Nuclear translocation and induction of apoptosis. J. Biol. Chem. 2002;277:34367–34374. doi: 10.1074/jbc.M202468200. [DOI] [PubMed] [Google Scholar]
- 25.Lin C.-Y., Wu H.-Y., Wang P.-L., Yuan C.-J. Mammalian Ste20-like protein kinase 3 induces a caspase-independent apoptotic pathway. Int. J. Biochem. Cell Biol. 2010;42:98–105. doi: 10.1016/j.biocel.2009.09.012. [DOI] [PubMed] [Google Scholar]
- 26.Lorber B., Howe M. L., Benowitz L. I., Irwin N. Mst3b, an Ste20-like kinase, regulates axon regeneration in mature CNS and PNS pathways. Nat. Neurosci. 2009;12:1407–1414. doi: 10.1038/nn.2414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zai L., Ferrari C., Dice C., Subbaiah S., Havton L. A., Coppola G., Geschwind D., Irwin N., Huebner E., Strittmatter S. M., Benowitz L. I. Inosine augments the effects of a Nogo receptor blocker and of environmental enrichment to restore skilled forelimb use after stroke. J. Neurosci. 2011;31:5977–5988. doi: 10.1523/JNEUROSCI.4498-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee W.-S., Hsu C.-Y., Wang P-L., Huang C.-Y. F., Chang C.-H., Yuan C.-J. Identification and characterization of the nuclear import and export signals of the mammalian Ste20-like protein kinase 3. FEBS Lett. 2004;572:41–45. doi: 10.1016/j.febslet.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 29.Iwaki K., Sukhatme V. P., Shubeita H. E., Chien K. R. α- and β-Adrenergic stimulation induces distinct patterns of immediate early gene expression in neonatal rat myocardial cells. fos/jun expression is associated with sarcomere assembly; Egr-1 induction is primarily an α1-mediated response. J. Biol. Chem. 1990;265:13809–13817. [PubMed] [Google Scholar]
- 30.Sugden P. H., Markou T., Fuller S. J., Tham E. L., Molkentin J. D., Paterson H. F., Clerk A. Monophosphothreonyl extracellular signal-regulated kinases 1 and 2 (ERK1/2) are formed endogenously in intact cardiac myocytes and are enzymically active. Cell. Signalling. 2011;23:468–477. doi: 10.1016/j.cellsig.2010.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fuller S. J., Pikkarainen S., Tham E. L., Cullingford T. E., Molkentin J. D., Cornils H., Hergovich A., Hemmings B. A., Clerk A., Sugden P. H. Nuclear Dbf2-related protein kinases (NDRs) in isolated cardiac myocytes and the myocardium: activation by cellular stresses and by phosphoprotein serine/threonine phosphatase inhibitors. Cell. Signalling. 2008;20:1564–1577. doi: 10.1016/j.cellsig.2008.04.013. [DOI] [PubMed] [Google Scholar]
- 32.McGuffin L. J. Intrinsic disorder prediction from the analysis of multiple protein fold recognition models. Bioinformatics. 2008;24:1798–1804. doi: 10.1093/bioinformatics/btn326. [DOI] [PubMed] [Google Scholar]
- 33.McGuffin L. J., Bryson K., Jones D. T. The PSIPRED protein structure prediction server. Bioinformatics. 2000;16:404–405. doi: 10.1093/bioinformatics/16.4.404. [DOI] [PubMed] [Google Scholar]
- 34.Roche D. B., Tetchner S. J., McGuffin L. J. FunFOLD: an improved automated method for the prediction of ligand binding residues using 3D models of proteins. BMC Bioinformatics. 2011;12:160. doi: 10.1186/1471-2105-12-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Roche D. B., Buenavista M. T., Tetchner S. J., McGuffin L. J. The IntFOLD server: an integrated web resource for protein fold recognition, 3D model quality assessment, intrinsic disorder prediction, domain prediction and ligand binding site prediction. Nucleic Acids Res. 2011;39:W171–W176. doi: 10.1093/nar/gkr184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McGuffin L. J., Roche D. B. Rapid model quality assessment for protein structure predictions using the comparison of multiple models without structural alignments. Bioinformatics. 2010;26:182–188. doi: 10.1093/bioinformatics/btp629. [DOI] [PubMed] [Google Scholar]
- 37.Martin D. D. O., Vilas G. L., Prescher J. A., Rajaiah G., Falck J. R., Bertozzi C. R., Berthiaume L. G. Rapid detection, discovery, and identification of post-translationally myristoylated proteins during apoptosis using a bio-orthogonal azidomyristate analog. FASEB J. 2008;22:797–806. doi: 10.1096/fj.07-9198com. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Martin D. D.O., Beauchamp E., Berthiaume L. G. Post-translational myristoylation: fat matters in cellular life and death. Biochimie. 2011;93:18–31. doi: 10.1016/j.biochi.2010.10.018. [DOI] [PubMed] [Google Scholar]
- 39.Kinoshita E., Kinoshita-Kikuta E., Takiyama K., Koike T. Phosphate-binding tag, a new tool to visualize phosphorylated proteins. Mol. Cell Proteomics. 2006;5:749–757. doi: 10.1074/mcp.T500024-MCP200. [DOI] [PubMed] [Google Scholar]
- 40.Humphries K. M., Deal M. S., Taylor S. S. Enhanced dephosphorylation of cAMP-dependent protein kinase by oxidation and thiol modification. J. Biol. Chem. 2005;280:2750–2758. doi: 10.1074/jbc.M410242200. [DOI] [PubMed] [Google Scholar]
- 41.Keshwani M. M., von Daake S., Newton A. C., Harris T. K., Taylor S. S. Hydrophobic motif phosphorylation is not required for activation loop phosphorylation of p70 ribosomal protein S6 kinase 1 (S6K1) J. Biol. Chem. 2011;286:23552–23558. doi: 10.1074/jbc.M111.258004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shu H., Chen S., Bi Q., Mumby M., Brekken D. L. Identification of phosphoproteins and their phosphorylation sites in the WEHI-231 B lymphoma cell line. Mol. Cell Proteomics. 2004;3:279–286. doi: 10.1074/mcp.D300003-MCP200. [DOI] [PubMed] [Google Scholar]
- 43.Daub H., Olsen J. V., Bairlein M., Gnad F., Oppermann F. S., Körner R., Greff Z., Kéri G., Stemmann O., Mann M. Kinase-selective enrichment enables quantitative phosphoproteomics of the kinome across the cell cycle. Mol. Cell. 2008;31:438–448. doi: 10.1016/j.molcel.2008.07.007. [DOI] [PubMed] [Google Scholar]
- 44.Huttlin E. L., Jedrychowski M. P., Elias J. E., Goswami T., Rad R., Beausoleil S. A., Villén J., Haas W., Sowa M. E., Gygi S. P. A tissue-specific atlas of mouse protein phosphorylation and expression. Cell. 2010;143:1174–1189. doi: 10.1016/j.cell.2010.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dephoure N., Zhou C., Villén J., Beausoleil S. A., Bakalarski C. E., Elledge S. J., Gygi S. P. A quantitative atlas of mitotic phosphorylation. Proc. Natl. Acad. Sci. U.S.A. 2008;105:10762–10767. doi: 10.1073/pnas.0805139105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yang J., Ten Eyck L. F., Xuong N.-H., Taylor S. S. Crystal structure of a cAMP-dependent protein kinase mutant at 1.26Å: new insights into the catalytic mechanism. J. Mol. Biol. 2004;336:473–487. doi: 10.1016/j.jmb.2003.11.044. [DOI] [PubMed] [Google Scholar]
- 47.Thingholm T. E., Larsen M. R., Ingrell C. R., Kassem M., Jensen O. N. TiO2-based phosphoproteomic analysis of the plasma membrane and the effects of phosphatase inhibitor treatment. J. Proteome. Res. 2008;7:3304–3313. doi: 10.1021/pr800099y. [DOI] [PubMed] [Google Scholar]
- 48.Oppermann F. S., Gnad F., Olsen J. V., Hornberger R., Greff Z., Kéri G., Mann M., Daub H. Large-scale proteomics analysis of the human kinome. Mol. Cell Proteomics. 2009;8:1751–1764. doi: 10.1074/mcp.M800588-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lu T.-J., Huang C.-Y. F., Yuan C.-J., Lee Y.-C., Leu T.-H., Chang W.-C., Lu T.-L., Jeng W. Y., Lai M. D. Zinc ion acts as a cofactor for serine/threonine kinase MST3 and has a distinct role in autophosphorylation of MST3. J. Inorg. Biochem. 2005;99:1306–1313. doi: 10.1016/j.jinorgbio.2005.03.003. [DOI] [PubMed] [Google Scholar]
- 50.Poizat C., Sartorelli V., Chung G., Kloner R. A., Kedes L. Proteasomemediated degradation of the coactivator p300 impairs cardiac transcription. Mol. Cell. Biol. 2000;20:8643–8654. doi: 10.1128/mcb.20.23.8643-8654.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fukazawa R., Miller T. A., Kuramochi Y., Frantz S., Kim Y.-D., Marchionni M. A., Kelly R. A., Sawyer D. B. Neuregulin-1 protects ventricular myocytes from anthracycline-induced apoptosis via erbB4-dependent activation of PI3-kinase/Akt. J. Mol. Cell. Cardiol. 2003;35:1473–1479. doi: 10.1016/j.yjmcc.2003.09.012. [DOI] [PubMed] [Google Scholar]
- 52.Pikkarainen S., Kennedy R. A., Marshall A. K., Tham E. L., Lay K., Kriz T. A., Handa B. S., Clerk A., Sugden P. H. Regulation of expression of the rat orthologue of mouse double minute 2 (MDM2) by H2O2-induced oxidative stress in neonatal rat cardiac myocytes. J. Biol. Chem. 2009;284:27195–27210. doi: 10.1074/jbc.M109.037887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ko T.-P., Jeng W.-Y., Liu C.-I., Lai M.-D., Wu C.-L., Chang W.-J., Shr H.-L., Lu T.-J., Wang A. H.-J. Structures of human MST3 kinase in complex with adenine, ADP and Mn2+. Acta Crystallogr. D Biol. Crystallogr. 2010;66:145–154. doi: 10.1107/S0907444909047507. [DOI] [PubMed] [Google Scholar]
- 54.Record C. J., Chaikuad A., Rellos P., Das S., Pike A. C., Fedorov O., Marsden B. D., Knapp S., Lee W. H. Structural comparison of human mammalian ste20-like kinases. PLoS ONE. 2010;5:e11905. doi: 10.1371/journal.pone.0011905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Westley F., Cohen I. Tables of values relating the axial ratio to the frictional ratio of an ellipsoid of revolution. Biopolymers. 1966;4:201–204. [Google Scholar]
- 56.Preisinger C., Short B., De Corte V., Bruyneel E., Haas A., Kopajtich R., Gettemans J., Barr F. A. YSK1 is activated by the Golgi matrix protein GM130 and plays a role in cell migration through its substrate 14-3-3ζ. J. Cell Biol. 2004;164:1009–1020. doi: 10.1083/jcb.200310061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nogueira A., Fidalgo M., Molnar A., Kyriakis J., Force T., Zalvide J., Pombo C. M. SOK1 translocates from the Golgi to the nucleus upon chemical anoxia and induces apoptotic cell death. J. Biol. Chem. 2008;283:16248–16258. doi: 10.1074/jbc.M709724200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fidalgo M., Fraile M., Pires A., Force T., Pombo C., Zalvide J. CCM3/PDCD10 stabilizes GCKIII proteins to promote Golgi assembly and cell orientation. J. Cell Sci. 2010;123:1274–1284. doi: 10.1242/jcs.061341. [DOI] [PubMed] [Google Scholar]
- 59.Nakamura N. Emerging new roles of GM130, a cis-Golgi matrix protein, in higher order cell functions. J. Pharmacol. Sci. 2010;112:255–264. doi: 10.1254/jphs.09r03cr. [DOI] [PubMed] [Google Scholar]
- 60.Goudreault M., D'Ambrosio L. M., Kean M. J., Mullin M. J., Larsen B. G., Sanchez A., Chaudhry S., Chen G. I., Sicheri F., Nesvizhskii A. I., et al. A PP2A phosphatase high density interaction network identifies a novel striatin-interacting phosphatase and kinase complex linked to the cerebral cavernous malformation 3 (CCM3) protein. Mol. Cell Proteomics. 2009;8:157–171. doi: 10.1074/mcp.M800266-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ceccarelli D. F., Laister R. C., Mulligan V. K., Kean M. J., Goudreault M., Scott I. C., Derry W. B., Chakrabartty A., Gingras A.-C., Sicheri F. CCM3/PDCD10 heterodimerizes with germinal center kinase III (GCKIII) proteins using a mechanism analogous to CCM3 homodimerization. J. Biol. Chem. 2011;286:25056–25064. doi: 10.1074/jbc.M110.213777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Castets F., Bartoli M., Barnier J. V., Baillat G., Salin P., Moqrich A., Bourgeois J.-P., Denizot F., Rougon G., Calothy G., Monneron A. A novel calmodulinbinding protein, belonging to the WD-repeat family, is localized in dendrites of a subset of CNS neurons. J. Cell Biol. 1996;134:1051–1062. doi: 10.1083/jcb.134.4.1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Benoist M., Gaillard S., Castets F. The striatin family: a new signaling platform in dendritic spines. J. Physiol. (Paris) 2006;99:146–153. doi: 10.1016/j.jphysparis.2005.12.006. [DOI] [PubMed] [Google Scholar]
- 64.Sanghamitra M., Talukder I., Singarapu N., Sindhu K. V., Kateriya S., Goswami S. K. WD-40 repeat protein SG2NA has multiple splice variants with tissue restricted and growth responsive properties. Gene. 2008;420:48–56. doi: 10.1016/j.gene.2008.04.016. [DOI] [PubMed] [Google Scholar]
- 65.Moreno C. S., Lane W. S., Pallas D. C. A mammalian homolog of yeast MOB1 is both a member and a putative substrate of striatin family-protein phosphatase 2A complexes. J. Biol. Chem. 2001;276:24253–24260. doi: 10.1074/jbc.M102398200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Baillat G., Moqrich A., Castets F., Baude A., Bailly Y., Benmerah A., Monneron A. Molecular cloning and characterization of phocein, a protein found from the Golgi complex to dendritic spines. Mol. Biol. Cell. 2001;12:663–673. doi: 10.1091/mbc.12.3.663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Miyamoto H., Matsushiro A., Nozaki M. Molecular cloning of a novel mRNA sequence expressed in cleavage stage mouse embryos. Mol. Reprod. Dev. 1993;34:1–7. doi: 10.1002/mrd.1080340102. [DOI] [PubMed] [Google Scholar]
- 68.Nozaki M., Onishi Y., Togashi S., Miyamoto H. Molecular characterization of the Drosophila Mo25 gene, which is conserved among Drosophila, mouse, and yeast. DNA Cell Biol. 1996;15:505–509. doi: 10.1089/dna.1996.15.505. [DOI] [PubMed] [Google Scholar]
- 69.Boudeau J., Baas A. F., Deak M., Morrice N. A., Kieloch A., Schutkowski M., Prescott A. R., Clevers H. C., Alessi D. R. MO25α/β interact with STRADα/β enhancing their ability to bind, activate and localize LKB1 in the cytoplasm. EMBO J. 2003;22:5102–5114. doi: 10.1093/emboj/cdg490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zeqiraj E., Filippi B. M., Goldie S., Navratilova I., Boudeau J., Deak M., Alessi D. R., van Aalten D. M.F. ATP and MO25α regulate the conformational state of the STRADα pseudokinase and activation of the LKB1 tumour suppressor. PLoS Biol. 2009;7:e1000126. doi: 10.1371/journal.pbio.1000126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Vitari A. C., Deak M., Morrice N. A., Alessi D. R. The WNK1 and WNK4 protein kinases that are mutated in Gordon's hypertension syndrome phosphorylate and activate SPAK and OSR1 protein kinases. Biochem. J. 2005;391:17–24. doi: 10.1042/BJ20051180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Moriguchi T., Urushiyama S., Hisamoto N., Iemura S., Uchida S., Natsume T., Matsumoto K., Shibuya H. WNK1 regulates phosphorylation of cationchloride-coupled cotransporters via the STE20-related kinases, SPAK and OSR1. J. Biol. Chem. 2005;280:42685–42693. doi: 10.1074/jbc.M510042200. [DOI] [PubMed] [Google Scholar]
- 73.Gagnon K. B., Delpire E. On the substrate recognition and negative regulation of SPAK, a kinase modulating Na+-K+-2Cl− cotransport activity. Am. J. Physiol. Cell Physiol. 2010;299:C614–C620. doi: 10.1152/ajpcell.00074.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Dorfman J., Macara I. G. STRADα regulates LKB1 localization by blocking access to importin-α, and by association with Crm1 and exportin-7. Mol. Biol. Cell. 2008;19:1614–1626. doi: 10.1091/mbc.E07-05-0454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Siegel LM., Monty K. J. Determination of molecular weights and frictional ratios of proteins in impure systems by the use of gel filtration and density gradient centrigation. Application to crude preparations of sulfite and hydroxylamine reductases. Biochim. Biophys. Acta. 1966;112:346–362. doi: 10.1016/0926-6585(66)90333-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.