Abstract
The interaction between β-catenin and B-cell CLL/lymphoma 9 (BCL9), critical for the transcriptional activity of β-catenin, is mediated by a helical segment from BCL9 and a large binding groove in β-catenin. Design of potent, metabolically stable BCL9 peptides represents an attractive approach to inhibit the activity of β-catenin. In this study, we report the use of the Huisgen 1,3-dipolar cycloaddition reaction to generate triazole-stapled BCL9 α-helical peptides. The high efficiency and mild conditions of this “click” reaction combined with the ease of synthesis of the necessary unnatural amino acids allows for facile synthesis of triazole-stapled peptides. We have performed extensive optimization of this approach and identified the optimal combinations of azido and alkynyl linkers necessary for stapling BCL9 helices. The unsymmetrical nature of the triazole staple also allowed the synthesis of double-stapled BCL9 peptides, which show a marked increase in helical character and an improvement in binding affinity and metabolic stability relative to wild-type and linear BCL9 peptides. This study lays the foundation for further optimization of these triazole-stapled BCL9 peptides as potent, metabolically stable and cell-permeable inhibitors to target the β-catenin and BCL9 interaction.
Introduction
Upregulation of β-catenin, the primary mediator of the Wnt signaling pathway, plays an important role in the tumorigenesis of several types of human cancer, including colon cancer, prostate cancer and melanoma 1. Functioning as a transcriptional activator, β-catenin controls the expression of a number of key genes that regulate the cell cycle and apoptosis. Transcriptional activation mediated by β-catenin requires the formation of a β-catenin super-complex, consisting of B-cell lymphoma 9 (BCL9), BCL9-like (B9L), a T-cell factor/lymphoid enhancer factor (TCF/LEF), the cAMP response element-binding protein (CBP) and other cofactors 2. It has been proposed that small-molecule inhibitors capable of disrupting the protein-protein interactions of β-catenin and its key cofactors may be a promising strategy to block β-catenin activity 2. Previous efforts have indeed led to the successful identification of several classes of small-molecule inhibitors that are capable of disrupting the interactions of β-catenin with TCF or CBP, thereby inhibiting β-catenin-mediated transcriptional activation 2. Some of these inhibitors also cause inhibition of cell growth in cancer cells with constitutively activated β-catenin 2.
Analysis of the recently reported crystal structure of β-catenin in a complex with BCL9 and TCF showed that the interaction between β-catenin and BCL9 is distinct from that of other β-catenin binding partners 3 and is mediated by a well-defined binding groove in β-catenin and several hydrophobic and charged residues from the α-helix of BCL9. This binding interaction, with a Kd value of 0.5 μM, suggests that the β–catenin-BCL9 interaction may be an attractive site for the design of potent and specific small-molecule inhibitors capable of blocking β-catenin activity.
The interaction between BCL9 and β–catenin proteins is mediated by an approximately 25-residue helical segment from BCL9 and a large binding groove in β–catenin, and design of non-peptide, potent small-molecule inhibitors to block this protein-protein interaction (PPI) would be expected to be difficult. Indeed, no such small-molecule inhibitors have been reported to date. An alternative strategy to target this PPI is to design and synthesize stapled helical peptides based upon BCL9. In the last few years, it has been demonstrated that stapled helical peptides, designed to induce conformational constraint in the side chains of the helical peptide residues, may lead to compounds with enhanced cell-permeability and metabolic stability.4
One of the most notable methods of helix stabilization has been the use of ring-closing olefin metathesis (RCM) to link the side chains of the i and i+4 or i+7 positions (Figure 1A)5. This method of producing hydrocarbon “stapled” α-helical peptides, introduced first by Blackwell and Grubbs5a, and further refined by Verdine et al.5b has been shown to increase helical propensity and peptide binding proclivity, while decreasing the rate of peptide cleavage by proteases. Furthermore, these hydrocarbon stapled peptides have been shown to have cellular activity in several biological settings.6
Figure 1. Comparison of helix stapling techniques.
(A) Hydrocarbon stapling. Peptides are synthesized on resin and the S-olefin-containing amino acids are incorporated at the i and i+4 positions. The olefins are then connected by means of an on-resin RCM reaction and then the peptide is cleaved and purified. For clarity, only i/i+4 stapling is depicted. (B) Triazole stapling. Peptides are synthesized on resin and the azido and alkynyl amino acids are incorporated at the i and i+4 positions. The peptide is then cleaved and cyclized in solution using the Huisgen Cu(I)-mediated 1,3-dipolar cycloaddition reaction.
More recently Cantel et al. demonstrated that the Cu(I)-mediated Huisgen 1,3-dipolar cycloaddition reaction 7 (a.k.a. a “click” reaction) can be used to generate a 1,4-substituted 1,2,3-triazole between side chain azido and alkynyl moieties at the i and i+4 positions of a peptide chain (Figure 1B) that effectively mimics the use of the analogous lactam bridge8. This cycloaddition reaction has been popularized by Sharpless and co-workers in the past decade and has been called a “click” reaction due to its simplicity, high yields, mild reaction conditions and biocompatibility 9. The use of cheaper, less toxic copper reagents also makes click-mediated stapling an attractive alternative to ruthenium-based RCM-type cyclization methods. Unlike hydrocarbon stapling, however, this method of triazole stapling has not been rigorously tested and its influence on peptide helicity and protease resistance has not been examined to date.
In the present study, we employed the triazole stapling method to stabilize BCL9 α-helical peptides for targeting the BCL9/β-catenin protein-protein interaction. Furthermore, in the process of developing triazole-stapled BCL9 peptide inhibitors, we optimized the linker lengths, staple orientation and amino acid stereochemistry required for generation of the optimal triazole staples in our peptides. We also investigated various stapling sites and successfully generated double triazole-stapled peptides that exhibit over 90% helicity and improved protease resistance.
Results
In contrast to hydrocarbon stapling, which has been extensively optimized and utilized in a number of different model systems5b, 6a–c, 10, the triazole stapling method has not been well studied and there has been only limited investigation into the optimal length of the linker, the order of the azide and alkynyl residues, or the use of L versus D enantiomers. Cantel et al. demonstrated that cyclization of L-norleucine(εN3) [i.e. Nle(εN3)] and L-propargylglycine (Pra) substituted at the i and i+4 positions, respectively, mimicked the amide-stapled peptide inhibitor of parathyroid hormone-related peptide (PTHrP), albeit with minor differences in the peptide backbone 8. Using this combination as a starting point, we systematically examined the effect of linker length, position of the triazole ring within the staple, and use of different stereoisomers for the staple.
Analysis of the crystal structure of BCL9 in a complex with β-catenin indicated that E360 and Q364 were suitable residues for use in triazole stapling (Figure 2). These residues were both exposed to solvent and located in the central region of our optimized BCL9 24-mer peptide11. Thus, a triazole staple between these positions would be predicted to have a significant stabilizing effect on both the N- and C-terminal portions of the peptide. Other solvent exposed residues were also examined but arginine residues were omitted from consideration so as to maintain an overall positive charge for our triazole-stapled peptides. Verdine et al. have demonstrated that the likelihood that stapled peptides will be cell permeable are increased when the stapled peptide has a net neutral or positive charge6a.
Figure 2. Solvent-exposed residues in BCL9.
The crystal structure (PDB code: 2GL7) of BCL9 (grey) in complex with β-catenin (cyan) shows that L356, E360, Q364, and D368 (colored magenta) are exposed to solvent with Q364 having the only fully resolved side chain. In order to maintain a high net positive charge for the stapled peptide, solvent-exposed arginine residues were not considered for stapling sites. The BCL9 sequence is shown at the top with the corresponding solvent-exposed residues colored magenta. E360 and Q364 are numbered.
Before introducing a triazole staple, we used computational modeling to examine the different combinations of linker lengths and isomers for use in the staple. Various combinations of L-Nle(εN3) or L-norvaline(δN3) [Nva(δN3)] and L- or D-Pra linkers were examined and the locally minimized structures of the triazole-stapled peptides were compared with the BCL9 peptide from the co-crystal structure (Figure 3). As shown in Figure 3A–C, using only L-amino acids or shortening the length of the staple by using Nva(δN3) caused noticeable changes in the backbone conformation. On the other hand, the L-Nle(εN3)/D-Pra combination of residues at the E360 and Q364 (i and i+4, respectively) positions showed the least amount of distortion in the peptide backbone (Figure 3D). Thus, our first triazole stapled peptides incorporated L-Nle(εN3) in place of E360 and either L- or D-Pra in place of Q364.
Figure 3. Molecular dynamic simulations of possible triazole-stapled BCL9 peptides.
Top: The BCL9 24-mer. Highlighted in red are the residues replaced by azido and alkynyl amino acids (magenta *). (A)–(D) Locally minimized structures of triazole-stapled BCL9 peptides using various combinations of L-Nle(εN3) or L-Nva(δN3) and L- or D-Pra. The combinations of un-natural amino acids used are given above each structure. The BCL9 peptide from the reported co-crystal structure with β-catenin (PDB code: 2GL7) is shown in yellow. Locally minimized BCL9 peptides are shown in green with the triazole staple highlighted in magenta/blue.
To synthesize the requisite Fmoc-protected L-Nle(εN3)-OH residue, we converted Fmoc-L-Lys-OH to its azide derivative in one step according to the reported method12. Cyclization of the peptides was monitored by analytical RP-HPLC, with the cyclized product typically having a retention time differing from the linear precursor peptide by approximately 0.1–0.6 min (Figure S1, Supplementary Information). We found that the conditions of Cantel et al. 8, which employed 4.4 equivalents of CuSO4 and ascorbic acid, and a peptide concentration of 1 mg/mL in 2:1 H2O/t-BuOH worked well for our system. As evidence of the ease of this click reaction, we observed that reaction overnight as previously reported was unnecessary as the reaction appeared to be complete within 20–60 min at room temperature. We did observe partial oxidation of our BCL9 peptides, probably at M372. Mutation of M372 to Leu (the corresponding residue in the B9L homolog; peptide 2 in Table 1) in subsequent experiments eliminated this problem and did not affect the binding affinity of our peptides.
Table 1.
Optimization of linkers for triazole stapling of BCL9 peptides.
| Peptide | Sequence | Linear Ki ± SD (μM)a | % Helicity |
|---|---|---|---|
| 1 | Ac-351LSQEQLEHRERSLQTLRDIQRMLF-NH2 | 0.60 ± 0.14 | 44 |
| 2 | Ac-351LSQEQLEHRERSLQTLRDIQRLLF-NH2 | 0.94 ± 0.17 | 47 |
| 3 |
|
1.4 ± 0.2 | 44 |
| 4 |
|
4.9 ± 0.9 | 45 |
| 5 |
|
2.4 ± 0.2 | 42 |
| 6 |
|
1.4 ± 0.1 | 49 |
| 7 |
|
1.9 ± 0.4 | 50 |
| 8 |
|
3.6 ±1.0 | 49 |
| 9 |
|
11.0 ±1.0 | 32 |
| 10 |
|
2.2 ± 0.2 | 34 |
| 11 |
|
0.33 ± 0.06 | 66 |
| 12 |
|
0.13 ± 0.05 | 90 |
| 13 |
|
NB | 26 |
| 14 |
|
3.0 ± 0.3 | 48 |
| 15 |
|
~30 ± 5b | 30 |
| 16 |
|
0.77 ± 0.18 | NDc |
| 17 |
|
1.17 ± 0.51 | 57 |
| 18 |
|
4.3 ± 0.4 | 67 |
indicates L-amino acid;
indicates D-amino acid; NB: no binding; ND: not determined
Ki values were determined using a BCL9-competitive FP binding assay
Ki value was extrapolated from an incomplete binding curve due to poor solubility
Peptide too insoluble for accurate measurement
Using these reaction conditions, we synthesized two sets of triazole-stapled peptides bearing either L- or D-Pra at position 364 (11 (3T) and 12 (4T), respectively in Table 1). The L-Nle(εN3) and L-Pra combination gave a linear precursor peptide 3 with a binding affinity to β-catenin half that of the wild type BCL9 24-mer (Table 1). In contrast, the triazole-stapled peptide 11 was twice as potent as the wild type peptide 1 and 4 times more potent than its linear precursor 3. The linear precursor peptide 4 with the L-Nle(εN3) and D-Pra combination, was 8-fold weaker than the wild type, which was expected as a result of the introduction of a D-amino acid into the sequence. However, the triazole-stapled peptide 12 was 4-fold more potent than the wild type BCL9 24-mer and 38-fold more potent than its linear precursor. These results supported our computational modeling which suggested that the use of D-Pra may cause less distortion of the α-helical peptide backbone in the triazole-stapled peptide.
Circular dichroism (CD) spectroscopy confirmed that the triazole stapled peptides were indeed more helical than the linear wild-type peptide 1 or their linear precursors 3 and 4. Peptide 11, which utilizes all L-amino acids showed an improvement to 66% helical content in PBS compared to 44–45% for the wild-type peptide 1 and the uncyclized peptides 3 and 4 (Figure 4). Peptide 12, which incorporates the D-Pra at the i+4 position of the staple, was determined to be 90% helical. Although the helicity for 12 seemed unusually high, it was correlated with its higher binding affinity than 11.
Figure 4. CD spectra of triazole-stapled BCL9 peptides.
Peptides were dissolved in PBS, pH 7.4 at a final concentration of approximately 50 μM. Exact peptide concentrations were determined using the BCA Protein Assay Kit. The spectra were averaged over 10 scans. Percent helicity was calculated from the mean residue ellipticity, θ, at 222 nm.
Baldwin and co-workers showed that far-UV CD spectrum of a peptide can be affected by an aromatic group within the peptide, thus yielding a false enhancement of helical propensity of the peptide13. To study the effect of 1, 2, 3-triazole aromatic ring to CD spectrum and helical propensity, we synthesized unstapled peptides 31 and 32, which contain a triazole ring based upon peptides 11 and 12 and analyzed their CD spectra (Figure S2, Supporting Information). Our data showed that the CD spectra for peptides 31 and 32 are very similar to those of peptides 3 and 4 without the triazole ring. The calculated helical propensities for peptides 3, 4, 31 and 32 are also very close (Figure S2, Supporting Information). These data thus further confirmed that the triazole-staple strategy via cyclization is successful in enhancing the helical conformations for BCL9 peptides.
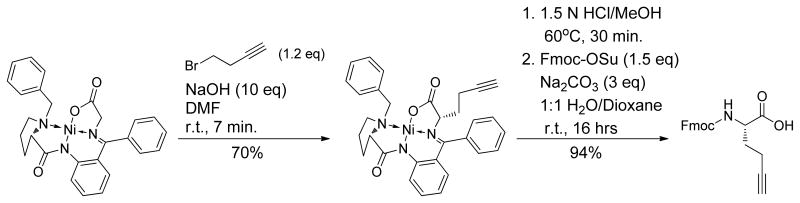
We next sought to determine the optimal length of the triazole staple and the position of the triazole within the staple and to address these questions, synthesized the azido derivative of Fmoc-L-ornithine [Fmoc-L-Nva(δN3)], which is one carbon shorter than the lysine used in the synthesis of Fmoc-Nle(εN3). In addition, we used a Ni(II) complex of a glycine Schiff’s base with (S)-2-[N,(N′-benzylprolyl)amino]benzophenone (BPB; Scheme 1) to synthesize propargylalanine (Paa), which is one carbon longer than Pra 14. Issad et al. recently reported on the use this complex for accessing a range of amino acids that can be used for triazole stapling8. The starting complex is relatively simple to synthesize on large scale and its use for the synthesis of mono-substituted amino acids is straightforward and gives acceptable yields.
Scheme 1.
Synthesis of Fmoc-L-propargylalanine (Paa)
With several different azido and alkynyl amino acids in hand, we systematically examined the effect of shortening or lengthening the staple, moving the triazole within the staple, or reversing the direction of the triazole staple (Table 1). All linear uncyclized peptides had low micromolar binding affinities (peptides 5–10), but when the peptides were cyclized, significant differences became apparent. Shortening the linker by one carbon (13) resulted in complete loss of binding affinity for β-catenin and a reduction in helical structure. Lengthening the linker by one carbon (14) also resulted in weaker binding affinity (2-fold weaker than uncyclized, 5-fold weaker than BCL9 24-mer 1) and showed no helix stabilizing effect. In addition, shifting the position of the triazole by one atom (15) markedly reduced the binding affinity (50-fold weaker than BCL9 24-mer) and also resulted in significantly worse peptide solubility. Shifting of the triazole to a more central position in the staple also perturbed the helical structure of the peptide, indicating that the dipole of the triazole itself may have an influence on the peptide structure. Reversing the linker did not significantly affect the binding affinity when using both L-Pra and L-Nle(εN3) (16), but the solubility of the resulting peptide was decreased. The reversed staple could also tolerate use of the D-form of the Nle(N3) at the i+4 position (17), but using D-Pra at position 360 (18) resulted in 6-fold weaker binding affinity.
We thus concluded that for triazole stapling of one turn of an α-helix, the optimal staple consists of an 8 atom linker consisting of L-Nle(εN3) and D-Pra at the i and i+4 positions, respectively. Although the use of the D-Pra at the i+4 position was only marginally better than use of L-Pra, the incorporation of the D-amino acid served as a negative control for peptide cyclization. Unlike the analogous RCM reaction in which the linear and stapled peptides differ in mass by 28 (C2H4), the triazole-stapled peptides have molecular weights that are identical to their linear uncyclized precursors. Without NMR or IR spectra to confirm the formation of the triazole ring, the successful triazole stapling was deduced from both the shift in retention time in the analytical RP-HPLC chromatogram (Figure S1) and the 10–38 fold difference in binding affinities between the linear and triazole stapled peptides (compare 4 and 12; 9 and 17).
We then investigated different stapling sites within our BCL9 24mer peptides (Table 2). Since this 24mer peptide is fully helical with six complete helical turns, there were a number of positions that could be used for stapling, several of which are illustrated in Figure 2. Using the optimal L-Nle(εN3)/D-Pra combination of linkers, we positioned the triazole staple closer to the N-terminus (19 and 20), or closer to the C-terminus (21 and 22), but found little difference in binding affinity. The peptide could also tolerate the reverse staple towards the C-terminus, again with little change in binding affinity (24). Although the C-terminal stapled peptide 22 retained a high degree of helical character, there was a decrease in the helical content of 20 and 24. In the case of 20, we hypothesized that the Thr residue, which is known to disfavor helical structure15, might be the reason for this decrease in helicity and that positioning a single staple away from this Thr residue was insufficient to maintain helical structure throughout the peptide. For 24, we speculated that the drop in helicity is associated with the reversed orientation and triazole staple and possibly a different effect of the triazole dipole with the peptide itself. This same modest, but noteworthy drop in binding affinity and helical content was also seen in the reversed staples at the original position (compare peptides 16 and 17 to 11 and 12). Nevertheless, we confirmed three potential stapling sites in our BCL9 peptide and demonstrated that either orientation of the triazole staple could be used, albeit with a preference for the azide/alkyne orientation at the i/i+4 positions over the reverse orientation.
Table 2.
Optimization of triazole stapling location and number of staples for BCL9 peptides.
| Peptide | Sequence | Linear Ki ± SD (μM)a | % Helicity |
|---|---|---|---|
| 1 | Ac-351LSQEQLEHRERSLQTLRDIQRMLF-NH2 | 0.60 ± 0.14 | 44 |
| 4 |
|
4.9 ± 0.9 | 45 |
| 12 |
|
0.13 ± 0.05 | 90 |
| 19 |
|
1.97 ± 0.20 | 21 |
| 20 |
|
0.41 ± 0.05 | 60 |
| 21 |
|
2.01 ± 0.10 | 31 |
| 22 |
|
0.26 ± 0.04 | 81 |
| 23 |
|
6.66 ± 0.39 | 16 |
| 24 |
|
0.56 ± 0.05 | 42 |
| 25 |
|
13.2 ± 0.9 | 12 |
| 26 |
|
0.41 ± 0.07 | 95 |
| 27 |
|
14.5 ± 1.1 | 20 |
| 28 |
|
0.61 ± 0.01 | 96 |
| 29 |
|
3.6 ± 0.1 | NDc |
| 30 |
|
0.19 ± 0.03 | 99 |
indicates L-amino acid;
indicates D-amino acid
Ki values were determined using a BCL9-competitive FP binding assay
Peptide too insoluble for accurate measurement
Based on the data obtained from the peptides stapled at different positions, we proposed that a double stapled peptide may further improve binding affinity and helical content and possibly metabolic stability. In contrast to hydrocarbon stapling, we hypothesized that the unsymmetrical nature of the triazole staple would allow for two staples to be placed adjacent to one another on consecutive turns of the helix, simply by reversing the orientation of the second staple. Having the two staples in opposite orientations would effectively put incompatible linkers (i.e. both alkynes or both azides) from each staple adjacent to each other and reduce the chance of “mixed staples” (Figure 5). It should be noted, however, that Verdine et al. recently demonstrated that hydrocarbon stapling between the i and i+3 or i+4 positions could be used to generate double hydrocarbon staples in a polyalanine model peptide, when the staples were separated by a two residue spacing 10a. This short spacing was shown to place the two adjacent olefins (one from each staple) in an unfavorable position to generate mixed staples. However, in our peptide the majority of staple locations that fail to interfere with binding interactions or reduce the number of positively charged residues occur exactly at the i and i+4 positions relative to each other. These positions may thus lead to significant amounts of mixed stapling when using the hydrocarbon stapling procedure.
Figure 5. Double triazole-stapled peptides.
Reversing the order of the azido and alkynyl linkers favors the generation of a double triazole staple (left) and discourages the formation of mixed staples (right).
To generate the first double triazole-stapled BCL9 peptide, we combined the staple combinations and positions used in 20 and 24 to synthesize 26 (Table 2). Reversing both staples allowed us to synthesize 28. Both 26 and 28 showed reasonable binding affinity, and their helical percentages were increased to >90% (Table 2). Since we suspected that the reverse triazole staples [i.e. L-Pra/D-Nle(εN3)] might be causing a slight decrease in the potency of our peptides, we synthesized 30 in which the triazole staples were spaced out by one full turn of the helix. This additional spacing allowed for both staples to utilize the optimal L-Nle(εN3)/D-Pra linker combination and resulted in a 2–3 fold improvement in the binding affinity of 30 which also had a helical content >95%.
Finally we tested whether these double triazole-stapled peptides with a very high helicity would be resistant to proteolytic degradation. Using cell media taken from actively cultured SW480 colon cancer cells, we examined the rate of proteolysis of the wild-type peptide 2 and of two double triazole-stapled BCL9 peptides 28 and 30. The double triazole-stapled peptides 28 and 30 both demonstrated a markedly improved stability over the linear peptide 2 (Figure 6).
Figure 6. Stability of linear and double triazole-stapled BCL9 peptides.
Peptides were diluted in cell culture media (Leibovitz-15, 10% FBS, 50 U/mL penicillin and streptomycin) taken from actively cultured SW480 cells, and incubated at 37°C. Aliquots were taken at various time points, quenched with 1% TFA/MeOH and analyzed by LC-MS.
Discussion
Design of non-peptide, small molecules to inhibit protein-protein interactions (PPIs) is a major challenge in modern drug discovery due to large surface area of the interactions between proteins. For this reason, peptide-based compounds can serve as an alternative for targeting PPIs. In the past, the use of peptides has been limited to cell-free in vitro experiments due to their poor stability and cell permeability. To overcome these major issues, researchers have focused on the design of conformationally restricted peptides that stabilize the secondary structure of peptides and improve their metabolic stability. To stabilize α-helical peptides, a number of approaches have been investigated including the use of helix caps, disulfide linkages, amide linkages and hydrocarbon linkages4. Verdine’s group demonstrated that RCM between side chain olefins at the i and i+4 or i+7 positions could be used to generate hydrocarbon staples that significantly improve the helical character of peptides in aqueous solution5b. This method of hydrocarbon stapling has been shown to increase peptide binding affinity, cell permeability and protease resistance5b, 6a, 6c, d, 16 but it requires the multi-step synthesis of the requisite olefin amino acids and uses the fairly toxic ruthenium catalysts.
Triazole stapling was recently introduced by Cantel et al. and shown to be a good replacement for a lactam bridge between the i and i+4 positions of a peptide helix8. In addition, the Huisgen 1,3-dipolar cycloaddition reaction employed to generate the triazole staple uses inexpensive and non-toxic reagents and the requisite azido and alkynyl amino acids are easily obtained from commercial sources or can be synthesized in one step. However, this method of triazole stapling has not been extensively studied and its effects on peptide binding affinities, helical propensity, metabolic stability and cell permeability have not been addressed.
In the present study, we employed the use of triazole stapling to generate stabilized α-helical BCL9 peptides as inhibitors of the BCL9/β-catenin binding interaction. In the process, we performed a detailed study of the optimal combinations of azido and alkynyl linkers. After examining a range of linkers, we found that for stabilization of one turn of the helix the optimal triazole-containing staple was eight atoms in length. Furthermore, the helical propensity of the peptide was extremely dependent on the position of the triazole within the staple, with the preferred staple having the triazole ring off-center. In addition, there appeared to be a slight preference for the azido and alkynyl amino acids to be at the i and i+4 positions, respectively, instead of in the reverse arrangement. Our computational modeling, binding studies and CD studies also indicated a preference for the use of D-Pra rather than the L-Pra at the i+4 position. In addition, use of D-Pra also provided an important negative control for the cyclization reaction, since completion of the reaction could not be determined by mass spectrometry due to the same molecular weight before and after the reaction. Introducing D-Pra into the peptide caused a significant decrease in the activity of the linear precursor peptide and the peptides regained their potency only after the successful completion of the triazole staple.
As with any type of “stapling” method, the optimal location of the staple within the peptide sequence needs to be determined experimentally. Examination of the crystal structure of BCL9 in a complex with β-catenin (PDB code: 2GL7) allowed identification of several combinations of solvent exposed residues that could be replaced in order to generate the triazole staple. As additional considerations for selection of a stapling site, we attempted to maximize the substitution of acidic residues and avoided the replacement of basic residues so as to increase the overall positive charge of the peptide. Bernal et al. demonstrated that the probability of cell permeability of hydrocarbon-stapled peptides was improved when the peptide had an overall neutral or positive charge 6a. Accordingly, we examined three different locations and found that either the central position (residues 360 and 364) or the C-terminal position (residues 364 and 368) were effective in stabilizing the helical structure of the peptide and in maintaining potent binding affinity. We hypothesize that these two locations work well due to their close proximity to T365. Threonine residues are known to disfavor helical structure 15, and this residue may possibly cause the “unraveling” of the peptide helix when the staple is located some distance away.
We next explored the design of double triazole-stapled peptides. The synthesis of a double hydrocarbon-stapled polyalanine peptide was recently demonstrated, but it requires that the two staples be separated by only two residues 10a. In our case, the only suitable stapling sites that did not require mutation of an arginine residue were spaced all at exactly the i and i+4 positions relative to each other, making hydrocarbon stapling problematic. In contrast, triazole stapling uses unsymmetrical linkers so by reversing the orientation of one staple relative to the other, we were able to generate double triazole-stapled peptides with a minimal amount of mixed stapling. These double triazole-stapled peptides showed binding affinities comparable to most of the single stapled peptides; however, their helical content was increased to over 90% in aqueous solution. We also found that separating the staples by one and a half turns of the helix also allowed us to generate a double triazole-stapled peptide with increased potency and over 90% helicity. We hypothesized that in longer peptides, those having more than four helical turns, a single triazole staple may be inadequate to prevent the unraveling of the ends of the peptide. However, by utilizing two strategically placed staples these longer peptides can be stabilized much more effectively, which will increase their proteolytic stability and potentially also their cell permeability. Indeed, we found that in addition to one of the single stapled peptides, two of our double-triazole stapled peptides (28 and 30) showed a marked improvement in its stability in cell media. Studies are currently in progress to evaluate the activity of these triazole-stapled BCL9 peptides in cell-based assays. If these peptides prove to be cell permeable, they will serve as useful biological tools with which to evaluate the true effects of inhibiting the interaction of BCL9 with β-catenin.
Conclusion
In this study, we performed extensive optimization of the triazole stapling strategy to generate single and double triazole-stapled BCL9 peptides. Our data showed that the combination of L-Nle(εN3) and D-Pra substituted at the i and i+4 positions, respectively, produced the best results for generation of single triazole-stapled peptides. These peptides demonstrated increased potency and improved helical structure. In addition, double-stapled peptides were over 90% in helical propencities and demonstrated potent binding to β-catenin. Several of the designed single and double triazole-stapled peptides also showed improved resistance to proteolytic degradation. Finally, we showed that triazole stapling can be used to generate double-stapled peptides in scenarios in which hydrocarbon stapling could lead to significant amounts of mixed stapling. Hence, triazole stapling can be an attractive approach for the design of stabled α-helical peptides with improved binding affinities toward their molecular targets, as well as improved metabolic stabilities.
Experimental Section
General synthesis of Fmoc-azido amino acids
Fmoc-Nle(εN3)-OH and Fmoc-L-Nva(δN3)-OH were synthesized from Fmoc-Lys-OH and Fmoc-L-Orn(Boc)-OH, respectively, according to the previously reported protocol 12. The L- and D- enantiomers of Fmoc-Nle(εN3)-OH along with Fmoc-L-Orn(Boc)-OH were purchased from Advanced Chemtech or ChemImpex International, Inc.
Propargylalanine (Paa)-Ni-BPB
(S)-Gly-Ni-BPB14b (2.78 mmol) and crushed NaOH (27.8 mmol) were suspended in anhydrous DMF under vacuum. The flask was purged with nitrogen and the reaction was stirred at room temperature for 7 min. 4-Bromo-1-butyne (3.83 mmol) was added via syringe and the reaction was stirred for an additional 6 min, then poured into 5% acetic acid (120 mL) and extracted with toluene (3 x 40 mL). The combined organic extracts were washed with brine (20 mL), dried over MgSO4, filtered, concentrated in vacuo and purified by flash column chromatography (4:1 ethyl acetate/acetone) to afford the product as a red solid (1.94 mmol; 70%). Rf 0.3 (4:1 EtOAc/Acetone); [a]D20 = +2390° (c = 0.024, CHCl3); 1H NMR (300 MHz, CDCl3) δ 8.13 (d, J=8.4 Hz, 1H), 8.05 (d, J=6.9 Hz, 2H), 7.56-7.46 (m, 3H), 7.36 (t, J=7.5 Hz, 2H), 7.28-7.12 (m, 3H), 7.01-6.99 (m, 1H), 6.70-6.61 (m, 2H), 4.45 (d, J=12.6 Hz, 1H), 3.99 (dd, J= 8.7, 3.6 Hz, 1H), 3.70-3.60 (m, 1H), 3.59 (d, J=12.6 Hz, 1H), 3.54-3.44 (m, 2H), 2.81-2.75 (m, 2H), 2.60-2.49 (m, 1H), 2.46-2.02 (m, 4H), 1.90-1.83 (m, 1H), 1.80 (t, J=2.7 Hz, 1H); 13C NMR (75.5 MHz, CDCl3) δ 180.34, 178.80, 171.04, 142.26, 133.71, 133.33, 133.2, 132.26, 131.55, 130.07, 129.74, 129.11, 128.96, 128.94, 128.89, 128.50, 127.85, 127.61, 127.18, 126.35, 123.77, 120.76, 82.65, 70.18, 69.58, 69.38, 63.13, 57.07, 54.04, 34.09, 30.67, 23.90, 15.03; ESI-MS [M+Na]+ calcd. for C31H29N3NaNiO3 572.15, found 572.27.
Fmoc-Paa-OH
(S)-Paa-Ni-BPB (4.8 mmol) was dissolved in 2:1 MeOH/CH2Cl2 (15 mL) and added dropwise over 10 min to a refluxing solution of 3N HCl/MeOH at 60°C. The reaction was stirred for an additional 20 min and then evaporated to dryness, redissolved in H2O (10 mL), and evaporated to dryness again. The crude residue was dissolved in H2O (30 mL), washed with CHCl3 (6 x 20 mL). The combined organic washes were extracted with H2O (15 mL) and the combined aqueous extracts were neutralized with Na2CO3. Additional Na2CO3 (15 mmol) was added and the solution was cooled to 0°C. Fmoc-N-hydroxysuccinimide ester (7.5 mmol) in dioxane (45 mL) was added dropwise and the reaction was then allowed to warm slowly to room temperature and were stirred overnight. The reaction was diluted with H2O (100 mL), acidified with concentrated HCl and extracted with EtOAc (3 x 40 mL). The combined organic extracts were dried (MgSO4), filtered, concentrated in vacuo, and purified by flash column chromatography (1:1 EtOAc/hexane, 1% AcOH) to afford the product as a colorless solid. The product was redissolved in an ether/hexane mixture, concentrated in vacuo and dried under high vacuum to remove all traces of AcOH leaving the product as a solid (4.5 mmol; 94%). [a]D20 = −14.64° (c = 3.35, MeOH); 1H NMR (300 MHz, CDCl3) δ 7.76 (d, J = 7.5 Hz, 2H), 7.59 (d, J = 6.9 Hz, 2H), 7.40 (t, J = 7.5 Hz, 2H), 7.31 (t, J = 7.2 Hz, 2H), 5.40 (d, J = 8.1 Hz, 1H), 4.52-4.39 (m, 3H), 4.23 (t, J = 6.6 Hz, 1H), 2.31-2.28 (m, 2H), 2.20-2.17 (m, 1H), 2.01-1.92 (m, 2H); 13C NMR (75.5 MHz, CDCl3) δ 176.08, 156.11, 143.77, 143.60, 141.34, 127.78, 127.11, 125.05, 120.04, 82.41, 69.87, 67.16, 53.07, 47.14, 30.78, 14.97. ESI-MS [M+Na]+ calcd. for C21H19NNaO4 372.12, found 372.13.
General Peptide Synthesis
Automated solid-phase peptide synthesis was performed on an Applied Biosciences 433A peptide synthesizer unless otherwise indicated. L- and D- enantiomers of Fmoc-Pra-OH were purchased from Advanced Chemtech or Chem-Impex International, Inc. All peptides were cleaved and deprotected with 87.5% TFA, 5% dithiothreitol, 5% H2O, 2.5% triisopropylsilane, and then purified by semi-preparative RP-HPLC using a Waters Sunfire™ 19x150 C18 column followed by lyophilization for 16–24 hours. Mass spectrometry was performed with a Thermo-Finnigan LCQ Deca or LCQ Fleet electrospray ionization mass spectrometer. Purity of all the synthesized peptides was determined by HPLC to be >95% and such information is provided in Table S1 in the Supplementary Information.
Triazole-stapled peptides. General procedure
Pure, lyophilized linear peptide and CuSO4·5H2O (4.4 equivalents) were dissolved in 2:1 H2O/t-BuOH to give a final peptide concentration of 1 mg/mL. Alternatively, hydrophobic peptides can be dissolved in a minimum amount of 6M guanidine hydrochloride solution and then added dropwise to the CuSO4/H2O/t-BuOH solution to prevent peptide precipitation. Sodium L-ascorbate (4.4 equivalents) dissolved in H2O (2 mL) was added slowly. The reaction was stirred at room temperature for 30–90 min. The reaction was then concentrated in vacuo and purified by semi-preparative RP-HPLC. Typical yields of cyclized product were 40–60%. Triazole-stapled peptides typically had retention times approximately 0–0.6 min longer than their linear, unstapled precursor.
Modeling of triazole staples
A helical peptide segment of BCL9 (residues S352 to F374) taken from the crystal structure of BCL9 in a complex with β-catenin (PDB code 2GL7) was used as a template with which to construct stapled peptides. Triazole staples of varying lengths, orientations and stereochemistries (at the peptide backbone) were constructed. After the staple was formed, structures were locally minimized using Sybyl (Tripos, Inc., St. Louis, MO). The locally minimized structures were then superimposed on the original BCL9 peptide segment obtained from the crystal structure.
Fluorescence Polarization (FP) Competitive Binding Assays
Peptide binding affinities for β-catenin were determined using our optimized BCL9-competitive FP binding assay and the BCL9-derived fluorescent probe 8-F described previously11.
CD Experiments
CD measurements were performed at room temperature (20°C) using a Jasco J-715 and a quartz flow cell with a 1 mm path length. Peptides were dissolved in phosphate buffered saline (PBS, pH 7.4) at a concentration of approximately 0.17 mg/mL. Precise peptide concentrations were then determined using the BCA Protein Assay Kit (Pierce) according to the manufacturer’s protocol. Spectra were collected at 50 nm/min, using a band width of 1 nm, averaged over 10 scans, and the baseline (PBS only) was subtracted from each spectrum. The mean residue ellipticity [θ]222 at 222 nm was calculated using equation 1,
| (1) |
where θ222 is the measured ellipticity at 222 nm, C is the molar concentration of the peptide, and n is the number of amino acid residues in the peptide. The maximum mean ellipticity [θ]max was calculated using equation 2 as reported previously 17,
| (2) |
where T is the temperature in degrees Celsius and k is the number of non-hydrogen-bonded peptide carbonyls18. For N-terminal acetylated peptides k = 3. Percent helicity was then calculated by equation 3,
| (3) |
Peptide Stability Assays
Leibovitz-15 cell culture media supplemented with 10% fetal bovine serum and 50 units/mL of penicillin/streptomycin was used to culture SW480 colon cancer cells. Cells were cultured for 3 days at 37°C, in 5% CO2 in a humidified incubator and then the cell media was collected and centrifuged at 4,000 rpm (900 rcf) for 5 min. The cleared cell culture media was then chilled on ice. Tryptophan was dissolved in PBS at a concentration of 1 mM and then added to the cell culture media to give a final tryptophan concentration of 200 μM. Peptides (20 mM in DMSO) were then added to the cell culture media to give a final peptide concentration of 200 μM. The peptides were incubated in cell culture media at 37°C. Aliquots of the reaction mixture were removed at specific time intervals and quenched with three volumes of 1% TFA in methanol, incubated on ice for 10 min and then centrifuged at 13,000 rpm for a further 10 min. The supernatant was then analyzed by RP-HPLC-MS using a Surveyor Plus HPLC equipped with a 2.1x150 mm Waters C18 Sunfire® column and a Surveyor Plus PDA detector and interfaced with a Thermo Finnigan LCQ Fleet electrospray ionization mass spectrometer (ESI-MS). Chromatographic separation was achieved with a flow rate of 0.4 mL/min and a 4.5%/min gradient of solvent B (acetonitrile, 0.05% TFA, 0.05% AcOH) in solvent A (H2O, 0.05% TFA, 0.05% AcOH). The LC-MS chromatogram was analyzed by integrating the chromatographic peak for the triply-charged parent peptide and dividing it by the area under the chromatographic peak of the tryptophan internal control (at 280 nm). The percentage of peptide remaining at each time point was normalized to the value calculated for the 0 min time point. LC-MS analysis of each time point was performed in triplicate.
Supplementary Material
Acknowledgments
We thank Dr. Ari Gafni of the Department of Biophysics at the University of Michigan for the use of his CD spectropolarimeter. This work is supported in part by the Department of Homeland Security, University of Michigan Regents, the American Foundation for Pharmaceutical Education, and the National Institutes of Health (R21NS066494).
Non-standard abbreviations
- BCL9
B-cell CLL/lymphoma 9
- B9L
BCL9-like
- TCF/LEF
T-cell factor/lymphoid enhancer factor
- CBP
cAMP response element-binding protein
- PPI
protein-protein interaction
- RCM
ring-closing olefin metathesis
- BPB
(S)-2-[N,(N′-benzylprolyl)amino]benzophenone
- Paa
propargylalanine
- Pra
L-propargylglycine
- PTHrP
parathyroid hormone-related peptide
- CD
Circular dichroism
- FP
Fluorescence Polarization
Footnotes
Supporting Information Available
Complete listing of peptide sequences, characterization and additional proteolytic stability assays. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Polakis P. Wnt signaling and cancer. Genes & Development. 2000;14(15):1837–1851. [PubMed] [Google Scholar]
- 2.Takemaru KI, Ohmitsu M, Li FQ. An oncogenic hub: beta-catenin as a molecular target for cancer therapeutics. Handbook of Experimental Pharmacology. 2008;(186):261–284. doi: 10.1007/978-3-540-72843-6_11. [DOI] [PubMed] [Google Scholar]
- 3.Sampietro J, Dahlberg CL, Cho US, Hinds TR, Kimelman D, Xu WQ. Crystal structure of a beta-catenin/BCL9/Tcf4 complex. Molecular Cell. 2006;24(2):293–300. doi: 10.1016/j.molcel.2006.09.001. [DOI] [PubMed] [Google Scholar]
- 4.(a) Haridas V. From Peptides to Non-Peptide Alpha-Helix Inducers and Mimetics. European Journal of Organic Chemistry. 2009;(30):5112–5128. [Google Scholar]; (b) Henchey LK, Jochim AL, Arora PS. Contemporary strategies for the stabilization of peptides in the alpha-helical conformation. Current Opinion in Chemical Biology. 2008;12(6):692–697. doi: 10.1016/j.cbpa.2008.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.(a) Blackwell HE, Grubbs RH. Highly efficient synthesis of covalently cross-linked peptide helices by ring-closing metathesis. Angew Chem Int Edit. 1998;37(23):3281–3284. doi: 10.1002/(SICI)1521-3773(19981217)37:23<3281::AID-ANIE3281>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]; (b) Schafmeister CE, Po J, Verdine GL. An all-hydrocarbon cross-linking system for enhancing the helicity and metabolic stability of peptides. Journal of the American Chemical Society. 2000;122(24):5891–5892. [Google Scholar]
- 6.(a) Bernal F, Tyler AF, Korsmeyer SJ, Walensky LD, Verdine GL. Reactivation of the p53 tumor suppressor pathway by a stapled p53 peptide. Journal of the American Chemical Society. 2007;129(9):2456–2457. doi: 10.1021/ja0693587. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Moellering RE, Cornejo M, Davis TN, Del Bianco C, Aster JC, Blacklow SC, Kung AL, Gilliland DG, Verdine GL, Bradner JE. Direct inhibition of the NOTCH transcription factor complex. Nature. 2009;462(7270):182–188. doi: 10.1038/nature08543. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Walensky LD, Kung AL, Escher I, Malia TJ, Barbuto S, Wright RD, Wagner G, Verdine GL, Korsmeyer SJ. Activation of apoptosis in vivo by a hydrocarbon-stapled BH3 helix. Science. 2004;305(5689):1466–1470. doi: 10.1126/science.1099191. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Walensky LD, Pitter K, Morash J, Oh KJ, Barbuto S, Fisher J, Smith E, Verdine GL, Korsmeyer SJ. A stapled BID BH3 helix directly binds and activates BAX. Molecular Cell. 2006;24(2):199–210. doi: 10.1016/j.molcel.2006.08.020. [DOI] [PubMed] [Google Scholar]
- 7.Huisgen R, Szeimies G, Mobius L. 1.3-Dipolare Cycloadditionen. 32. Kinetik der Additionen organischer Azide an CC-Mehrfachbindungen. Chemische Berichte. 1967;100(8):2494–2507. [Google Scholar]
- 8.Cantel S, Isaad Ale C, Scrima M, Levy JJ, DiMarchi RD, Rovero P, Halperin JA, D’Ursi AM, Papini AM, Chorev M. Synthesis and conformational analysis of a cyclic peptide obtained via i to i+4 intramolecular side-chain to side-chain azide-alkyne 1,3-dipolar cycloaddition. The Journal of Organic Chemistry. 2008;73(15):5663–5674. doi: 10.1021/jo800142s. [DOI] [PubMed] [Google Scholar]
- 9.Kolb HC, Sharpless KB. The growing impact of click chemistry on drug discovery. Drug Discov Today. 2003;8(24):1128–1137. doi: 10.1016/s1359-6446(03)02933-7. [DOI] [PubMed] [Google Scholar]
- 10.(a) Kim YW, Kutchukian PS, Verdine GL. Introduction of all-hydrocarbon i, i+3 staples into alpha-helices via ring-closing olefin metathesis. Organic Letters. 2010;12(13):3046–2049. doi: 10.1021/ol1010449. [DOI] [PubMed] [Google Scholar]; (b) Kim YW, Verdine GL. Stereochemical effects of all-hydrocarbon tethers in i, i+4 stapled peptides. Bioorganic & Medicinal Chemistry Letters. 2009;19(9):2533–2536. doi: 10.1016/j.bmcl.2009.03.022. [DOI] [PubMed] [Google Scholar]
- 11.Kawamoto SA, Thompson AD, Coleska A, Nikolovska-Coleska Z, Yi H, Wang S. Analysis of the interaction of BCL9 with beta-catenin and development of fluorescence polarization and surface plasmon resonance binding assays for this interaction. Biochemistry. 2009;48(40):9534–41. doi: 10.1021/bi900770z. [DOI] [PubMed] [Google Scholar]
- 12.Katayama H, Hojo H, Ohira T, Nakahara Y. An efficient peptide ligation using azido-protected peptides via the thioester method. Tetrahedron Lett. 2008;49(38):5492–5494. [Google Scholar]
- 13.Chakrabartty A, Kortemme T, Padmanabhan S, Baldwin RL. Aromatic side-chain contribution to far-ultraviolet circular dichroism of helical peptides and its effect on measurement of helix propensities. Biochemistry. 1993;32(21):5560–5565. doi: 10.1021/bi00072a010. [DOI] [PubMed] [Google Scholar]
- 14.(a) Belokon YN, Bespalova NB, Churkina TD, Cisarova I, Ezernitskaya MG, Harutyunyan SR, Hrdina R, Kagan HB, Kocovsky P, Kochetkov KA, Larionov OV, Lyssenko KA, North M, Polasek M, Peregudov AS, Prisyazhnyuk VV, Vyskocil S. Synthesis of alpha-amino acids via asymmetric phase transfer-catalyzed alkylation of achiral nickel(II) complexes of glycine-derived Schiff bases. Journal of the American Chemical Society. 2003;125(42):12860–12871. doi: 10.1021/ja035465e. [DOI] [PubMed] [Google Scholar]; (b) Belokon YN, Tararov VI, Maleev VI, Savel’eva TF, Ryzhov MG. Improved procedures for the synthesis of (S)-2-[N-(N′-benzylprolyl)amino]benzophenone (BPB) and Ni(II) complexes of Schiff’s bases derived from BPB and amino acids. Tetrahedron-Asymmetry. 1998;9(23):4249–4252. [Google Scholar]
- 15.Rohl CA, Chakrabartty A, Baldwin RL. Helix propagation and N-cap propensities of the amino acids measured in alanine-based peptides in 40 volume percent trifluoroethanol. Protein Science. 1996;5(12):2623–37. doi: 10.1002/pro.5560051225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.(a) Chapman RN, Dimartino G, Arora PS. A highly stable short alpha-helix constrained by a main-chain hydrogen-bond surrogate. Journal of the American Chemical Society. 2004;126(39):12252–12253. doi: 10.1021/ja0466659. [DOI] [PubMed] [Google Scholar]; (b) Nikolovska-Coleska Z, Wang RX, Fang XL, Pan HG, Tomita Y, Li P, Roller PP, Krajewski K, Saito NG, Stuckey JA, Wang SM. Development and optimization of a binding assay for the XIAP BIR3 domain using fluorescence polarization. Analytical Biochemistry. 2004;332(2):261–273. doi: 10.1016/j.ab.2004.05.055. [DOI] [PubMed] [Google Scholar]
- 17.(a) Wang D, Chen K, Kulp JL, Iii, Arora PS. Evaluation of biologically relevant short alpha-helices stabilized by a main-chain hydrogen-bond surrogate. Journal of the American Chemical Society. 2006;128(28):9248–9256. doi: 10.1021/ja062710w. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Shepherd NE, Hoang HN, Abbenante G, Fairlie DP. Single turn peptide alpha helices with exceptional stability in water. Journal of the American Chemical Society. 2005;127(9):2974–2983. doi: 10.1021/ja0456003. [DOI] [PubMed] [Google Scholar]; (c) Luján-Upton H, Okamoto Y, Walser AD. Fluorescence lifetime of terbium(III) as a probe for the ion-binding properties of tactic poly(methacrylic acids) Journal of Polymer Science Part a-Polymer Chemistry. 1997;35(3):393–398. [Google Scholar]
- 18.Rohl CA, Baldwin RL. Deciphering rules of helix stability in peptides. Methods in Enzymology. 1998;295:1–26. doi: 10.1016/s0076-6879(98)95032-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.