Abstract
Gelsolin is the most widely expressed member of the actin capping and severing family of proteins. There are two isoforms of gelsolin: isoform 1, a secretory (plasma) protein that is 51 amino acids longer than isoform 2, a cytosolic protein, at the N-terminus; the first 27 amino acids is a signal sequence. Both isoforms are coded by a single gene and differ as a result of alternative initiation site/ splicing. The level of gelsolin in the blood and cerebrospinal fluid (CSF) is altered in many diseases including amyloidoses and other neurodegenerative disorders. Although quantitative analysis of gelsolin has been reported, lack of suitable antibodies makes it impossible to differentiate these two isoforms by immunodetection techniques and no other technique is available. Therefore, ambiguity exists whether gelsolin present in circulation is isoform 1 or also isoform 2 released from lysed cells. We report in this communication a mass spectrometry approach to identify isoform 1 of gelsolin immunopurified from human plasma and CSF. Recombinant isoform 1 was used as reference.
Keywords: gelsolin, HIV, CSF, plasma, biomarker
Introduction
Gelsolin, an actin-binding 82-kD protein, is the most widely expressed member of the actin severing family of proteins. Isoforms 1 and 2 of gelsolin are coded by a single gene, but differ as a result of alternative initiation and/or splicing [1, 2]. Isoform 1, a secretory protein found in plasma and cerebrospinal fluid (CSF) is longer than isoform 2 by 51 amino acids at the N-terminal end. The first 27 amino acids is a signal sequence that engages gelsolin into the secretion pathway in the endoplasmic reticulum and is subsequently removed prior to the release. The remaining 24 amino acids long peptide differentiates cytosolic and secreted isoforms [3]. A third form of gelsolin produced by oligodendrocytes has been reported by Vouyiouklis et al. [4]. The N-terminal sequence of this isoform is different than both the cytoplasmic and the plasma isoforms; however, this form is less documented and has not been characterized yet. Figure 1A presents a model of the two isoforms of the gelsolin molecule.
Figure 1.
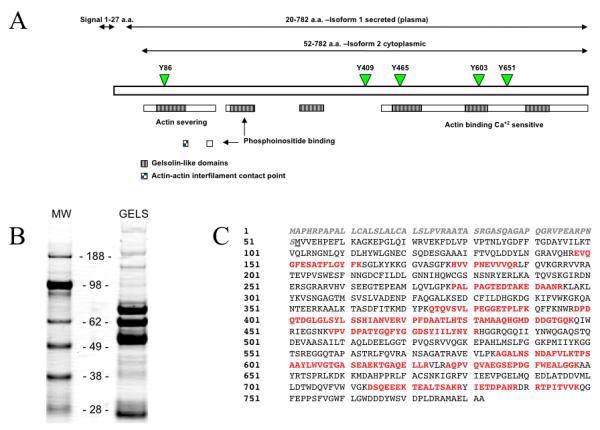
(A) Schematic representation of gelsolin and its molecular features. Green arrowheads and numbers indicate five tyrosine residues and their locations that can be potentially phosphorylated. The dotted blue line indicates a disulfide bridge. Adapted from UniPortKB (http://www.uniprot.org/uniprot/P06396, database entry P06396). (B) 1DE of 25ug immunopurified Gelsolin (GELS, right) and SeeBlue® Plus2 Pre-Stained Standard (MW, left) on a NuPAGE® Novex® 4-12% Bis-Tris Gel. (C) Amino acid sequence of gelsolin. Peptides from tryptic digest that were identified by nano-LC-MS/MS analysis are in red. The secretory (plasma) isoform of gelsolin is longer than the cytoplasmic intracellular isoform by 51 a.a. and contains both a secretory signal sequence and an additional N-terminal fragment. These 51 a.a. precede the methionine translation start site indicator of isoform 1 and are shown in grey, bold italics.
Gelsolin has five potential sites of phosphorylation [5], however there is no consensus about which sites, if any, are phosphorylated in vivo. Yuan et al. [6] and Marcus et al. [7] postulated that gelsolin circulating in the CSF is phosphorylated, however, the authors did not provide further experimental details.
Functions of plasma gelsolin are largely unknown [8-10]. It is distributed throughout body via body fluids and has a half-life time of 2 to 3 days in plasma [11]. Gelsolin is not synthesized in the liver and very little is known about regulation of its expression. It has been postulated that neurons is a major source of gelsolin in the central nervous system (CNS) [6, 12] however oligodendrocytes [4] have been shown to produce extracellular gelsolin as well. Eventually, how much of a total pool of gelsolin originates from neuron and how much from oligodendrocytes has not been determined yet. Gelsolin -/- mice were used to show roles of gelsolin in a number of pathological conditions such as inflammation, cancer and amyloidosis [13]. Although gelsolin -/- mice provide great information about the functions of this protein, it is impossible to make a straightforward conclusion related to each isoform unless alternative spicing is perturbed.
Reports from our laboratory show that in HIV-infected individuals, the level of gelsolin is increased in plasma and decreased in CSF [14, 15]. Quantitation of each isoform of gelsolin circulating in body fluids is impossible due to lack of appropriate antibody. Therefore we attempted to use mass spectrometry to discriminate between these two isoforms in plasma and CSF.
Materials and methods
Plasma samples
Sera samples were obtained from the National NeuroAIDS Tissue Consortium (NNTC, https://web.emmes.com/study/hbb/) under Request# R101. The UNMC Institutional Review Board approved use of clinical samples in this study (#196-05-EX). Besides patient classification [non-demented (ND) and HIV-associated dementia (HAD) as provided by NNTC], no other criteria (age, race, gender, T-cell count, viral load, etc.) were applied for sample selection. Gelsolin samples were pooled based on clinical classification provided by NNTC prior to affinity purification.
Reagents
NaHCO3, NaCl, Ethanolamine, acetic acid, Trizma base, HCl, sodium acetate, CHAPS, urea and bromophenol blue were obtained from Sigma Aldrich (St. Louis, MO, USA). Gelsolin protein (human recombinant) was from Cytoskeleton inc. (Denver, CO). HPLC water was obtained from Fisher Scientific (Pittsburg, PA, USA). NuPAGE Bis-Tris 4-12% gels were obtained from Invitrogen Corp. (Carlsbad, CA, USA). Pierce® Chromatography cartridges Protein G were obtained from Pierce (Rockford, IL, USA). HiTrap™ NHS-activated HP column was obatined from GE healthcare (Pittsburg, PA, USA).
Gelsolin immunoaffinity purification
A polyclonal goat anti-gelsolin antibody recognizing both isoforms of gelsolin was obtained from Santa Cruz (Santa Cruz, CA, USA) and purified by Protein-G affinity chromatography according to the manufacturer protocol using HiTrap column. The HiTrap column coupled to the goat anti-gelsolin antibody was washed with 3 ml of PBS containing 2% of Tween 20 (binding buffer) and 3 ml of 0.1 M glycine, pH 2 (eluting buffer). The column was then equilibrated using 10 ml of binding buffer. Plasma samples, diluted at a 1:10 ratio in binding buffer, were passed twice through the column. Resin was rinsed with 20 ml of binding buffer. Gelsolin was eluted using 4 ml of eluting buffer and 500 μl fractions were collected and neutralized with 50 μl of 1 M Tris-HCl, pH 7.5. The column was equilibrated using 10 ml of binding buffer. Eluted fractions were analyzed by NanoDrop; fractions containing proteins were pooled, dialyzed and stored at −80°C until use.
Mass spectrometry analysis
Samples of immunopurifed gelsolin (25 μg) and recombinant gelsolin (2 μg) were dried and solubilized with 20 μl NuPAGE sample buffer prior to gel loading. 1DE was performed using the NuPAGE gel system (Invitrogen Corp.) in 4–12% gradient Bis-Tris gels under reducing conditions.
After electrophoresis, gels were stained with brilliant blue G-colloidal concentrate (Sigma Aldrich) and protein bands were excised using a razor blade. A two-step destaining technique was performed using 200 μL of 20 mM NH4HCO3/ 50% acetonitrile (ACN) followed by 200 μL 100% ACN. After destaining, the gel slices were dried and incubated with 0.1 μg/μL trypsin (Promega, Madison, WI) overnight at 37°C. Peptides were extracted with 0.1% TFA/60% ACN, dried and re-suspended in 0.1% TFA. Next, the samples were purified using reverse phase C18 Zip Tips® (Millipore, Billerica, MA) according to manufacturer’s procedure and samples were re-suspended in 0.1% formic acid in water prior to MS analysis.
Mass spectrometric analysis was performed using ESI-LC-MS/MS system in a nano-spray configuration using a microcapillary RP-C18 column (New Objectives, Woburn, MA). Tandem mass spectrometry analyses were performed with an ion trap mass spectrometer LCQ-Deca XP Plus (Thermo Scientific Inc.). The spectra were searched using Sequest™ algorithm (BioWorks 3.2 software, Thermo Scientific Inc.) using the following parameters: threshold for Dta generation = 10000, precursor ion mass tolerance = 1.4, peptide tolerance = 2.00 and fragment ions tolerance = 1.00. The NCBI database in FASTA format (http://ftp.ncbi.nih.gov) was used for analyses of sequence alignment with the criteria of two missed cleavage sites allowed and at least two peptides were required for protein identification.
The MALDI-TOF-TOF 4800 mass spectrometer (Applied Biosystems) was used for MS and MS/MS analysis. One microliter of in-gel digested gelsolin was spotted directly onto a MALDI plate (Applied Biosystems, Foster City, CA) and co-crystallized with α-cyano-4-hydroxycinnamic acid (CHCA) matrix (5 mg/mL in 50% ACN-0.1% TFA). Droplets were dried at room temperature. Spectra acquisition and processing was performed with the 4000 series Explorer software (Applied Biosystems) version 3.5.1 in positive reflectron mode at fixed laser fluency with low mass gate and delayed extraction. External calibration was performed with a mixture of five external standards (Applied Biosystem). Peptide masses were acquired by steps of 50 spectra for masses between 800 to 3000 Da. MS spectra were summed from 1000 laser shots by an Nd-YAG laser operating at 355 nm and 200 Hz. MS/MS spectra was acquired in 1-kV positive mode, and 1000 shots were summed in increments of 50. Database searching was realized using the MatrixScience website (www.matrixscience.com) for MS and MS/MS interrogations on human proteins from Swiss-Prot databank (www.expasy.org). The search parameters were as follows: carbamidomethylation for cysteins and oxidation for methionines as variable modifications, one missed tryptic cleavage was permitted, and mass accuracy tolerance was set at 100 ppm for precursors and 0.5 Da for fragments.
Results
1D SDS-PAGE revealed that immunoaffinity purification of gelsolin isolated multiple forms/fragments. Eight bands ranging in molecular weight between 19 and 86 kDa show the presence of gelsolin confirmed by mass spectrometric analysis, using nano-LC-LCQDecaPlus (Thermo Electron). The predominant band corresponded to 86 kDa, which is the expected size of full-length form of this protein (Figure 1B). However, to discriminate between plasma and cytosolic gelsolin isoforms any peptide derived from first 27 amino acids ought to be identified. We used recombinant plasma gelsolin as a reference sample for mass spectrometric analysis. We used recombinant plasma gelsolin as a reference for mass spectrometric method development and to determine the feasibility of the approach. Second, use this method on immunopurified gelsolin.
Electrospray Ionization (ESI) nano-LC-MS/MS using high sensitivity LTQOrbitrap mass spectrometer, with recombinant gelsolin, identified peptide PNSMVVEHPEFLK belonging to isoform 1 of gelsolin (Figure 2A). Surprisingly the N-terminal amino acid of this peptide is proline. Arg-Pro bond is cleaved by trypsin at very low rate. While the MS peak of this peptide was triply charged and the intensity of the peak was low, the sequence coverage of this peptide, obtain by MS/MS (Figures 2B and 2C) shows efficient fragmentation. The low intensity of the MS peak implies that this method might not be fully effective for characterization of low amounts of immunopurified gelsolin.
Figure 2. Tandem mass spectrometry identification of peptide belonging to isoform 1 of gelsolin.
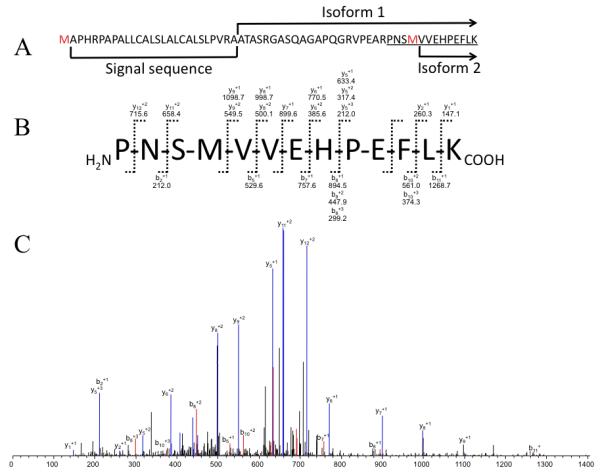
(A) Amino acid sequence of the N-terminal fragment of gelsolin showing the signal peptide and start (N-terminus) of isoforms 1 and 2. Underlined is sequence of peptide derived by trypsin digest of isoform 1 and identified in nano-LC-LTQOrbitrap analysis. (B) Fragment ions of identified peptide PNSMVVEHPEFLK detected by ESI mass spectrometric analysis. (C) Corresponding MS/MS spectrum of PNSMVVEHPEFLK peptide.
MALDI-TOF/TOF analysis of recombinant gelsolin tryptic digest was able to highlight, by peptide mass fingerprinting (PMF) one peptide with 2079.06 m/z corresponding to a peptide specific of gelsolin isoform 1 underlined on figure 3A. The MS/MS analysis of this peptide presents sufficient fragmentation rate to cover the sequence of the peptide (Figure 3B). The same strategy used for immunopurified gelsolin provides a lower intensity of fragments (Figure 3 C and D). Nevertheless, the sequence coverage allows the identification of gelsolin isoform 1 and both MS/MS spectra, from recombinant plasma gelsolin and purified are comparable. Finally, contaminant peak with 2095,06 m/z (Figure 3C) corresponds, according to the PMF, to the same peptide (VPNEARPNSMVVEHPEFLK) with oxidized methionine. We were not able to separate those two peptides for the fragmentation but the contamination does not seem to interfere with the fragmentation with unreliable transitions. Also, as of now, our studies focusing on the forms of gelsolin circulating in the CSF and blood have failed to document phosphorylation.
Figure. 3. MALDI-MS/MS identification of peptide differentiating isoform 1 (plasma) and 2 (cytoplasmic) of gelsolin.
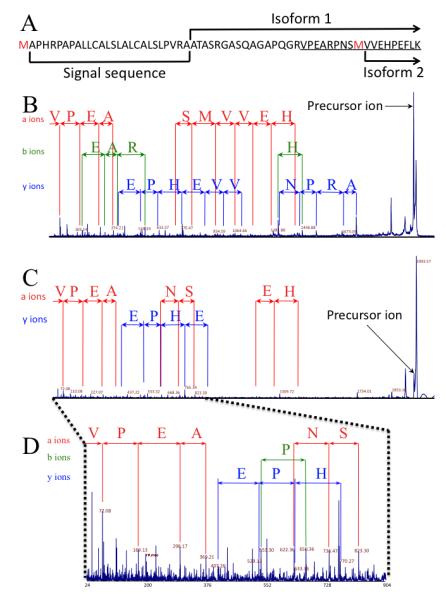
(A) Underlined is the sequence of peptide derived by trypsin digest of isoform 1 and identified in MALDI-TOF/TOF analysis. Red methionines are marked for reference and indicate the translation start sites for the different isoforms prior to post-translational modifications. (B) MALDI-TOF/TOF spectrum of VPEARPNSMVVEHPEFLK peptide derived by tryptic digest of recombinant isoform 1. (C) MALDI-TOF/TOF sequencing of VPEARPNSMVVEHPEFLK peptide derived from trypsin digestion of plasma gelsolin immunopurified from plasma of HIV-1 infected individuals. (D) Expanded region, 24 to 904 m/z of spectrum shown in C.
For ease of understanding, a-ions have been color-coded in red, b-ions have been color-coded in green, and y-ions have been color-coded in blue.
Discussion
Changes in gelsolin expression in plasma has been observed during several diseases such as liver failure, myocardial infarction, septic shock, myonecrosis [16], rheumathoid arthritis [17] and trauma, [18, 19]. In CSF, gelsolin expression is linked to multiple sclerosis [20] and other neurological diseases such as idiopathic cephalgia, ischialgia due to discopathy or subarachoid haemorhage [10]. The exact role of gelsolin in these diseases is not known, though several functions have been postulated. Nevertheless, increasing experimental evidence shows that plasma gelsolin (isoform 1) may play a wide range of roles in biological processes during disease and/or development than previously thought, thus its functions are not limited to actin capping and severing. For example, secreted gelsolin was shown to enhance the binding of beta(2) glycoprotein 1 (Apolipoprotein H, APOH) to α5β1 integrin [21] and alter signaling via p38 MAPK, present lysophosphatidic acid and other inflammatory mediators to their receptors [8], act as a scaffolding protein that links β2GPI (beta 2 glycoprotein 1, ApoH) and integrin/fibronectin in induction of Tissue Factor via integrin activation of the p38 MAPK and NF-κB pathways [22], block TLR-dependent NF-κB nuclear translocation in astrocytes [9] etc. These - as well as other functions - are just beginning to be investigated in in-depth studies.
Although it is assumed that gelsolin present in circulation is isoform 1, the mechanism of secretion and exact source have not been characterized experimentally. Moreover, there is not direct experimental evidence of which isoform is present in the circulation due to lack of antibodies distinguishing these two isoforms. It cannot be excluded that isoform 2 is released from cytoplasm due to cell lysis under various pathological conditions.
In this study, we used tandem mass spectrometry for identification of isoforms of gelsolin affinity purified from plasma and CSF of HIV-infected patients. One reason for using this particular source of gelsolin is our consistent observation that levels of gelsolin in plasma and CSF are altered during HIV infection. While the level of circulating gelsolin in CSF is decreased in patients with HIV-associated dementia, the level of gelsolin in blood is increased. This counterintuitive observation can be attributed to the fact that CNS has its own cells producing gelsolin (neurons and oligodendrocytes) while source of gelsolin in periphery is not certain. Skeletal, smooth and cardiac muscles are postulated to be major source and liver produces very little amounts of this protein [1, 23].
As expected, ESI and MALDI types of ionization provided different but complementary results. Both approaches showed the presence of peptides unique for secretory isoform 1. One unexpected result was identification of peptide H2N-PNSMVVEHPEFLK-COOH, which was derived from recombinant isoform 1. This suggests that trypsin cleaved Arg-Pro bond at position –R(48)-P(49)- . This particular peptide bond is cleaved by trypsin at very low level comparing to other -R-X- bonds. Our interpretation is that trypsin digest of recombinant form of gelsolin produced very low amounts of this peptide, however ionization by ESI was sufficient that precursor ion of this peptide was selected for fragmentation in data dependent mode. This was observed using nano-LC-LTQ Orbitrap method of data acquisition with the 5 top intensity precursor ions selected for data dependent (MS/MS fragmentation) scans. Trypsin digest of immunopurified gelsolin did not yield enough of H2N-PNSMVVEHPEFLK-COOH peptide for identification using MALDI type of ionization as well as ESI-nano-LC-LCQDecaPlus was not sensitive enough.
In conclusion, for the first time we present experimental evidence that gelsolin isolated from blood or CSF contains secretory isoform 1. Moreover, we have also shwon that peptides characteristic for isoform 1 can be identified by both modes of ionization, ESI and MALDI. While, MALDI-TOF/TOF analysis seems to be the most adapted method to characterize isoform 1 of gelsolin, our data also suggests that this approach may not work when gelsolin is in complex with a mixture of proteins because of either low levels of precursor ions due inefficient ionization or low level of specific peptides. iTRAQ or 18O labeling of trypsin derived peptides originating from recombinant gelsolin will provide quantitative measures whether immunopurified gelsolin contains only isoform 1 or is a mixture of both isoforms.
Acknowledgments
We would like to thank Ms. Jayme Wiederin and Ms. Robin Taylor for help in preparation of this manuscript. This work was partially supported by the National Institutes of Health 1 P20DA026146-01 and 2 P01 NS043985-05.
References
- [1].Kwiatkowski DJ, Stossel TP, Orkin SH, Mole JE, Colten HR, Yin HL. Nature. 1986;323:455. doi: 10.1038/323455a0. [DOI] [PubMed] [Google Scholar]
- [2].Stella MC, Schauerte H, Straub KL, Leptin M. J Cell Biol. 1994;125:607. doi: 10.1083/jcb.125.3.607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Yin HL, Kwiatkowski DJ, Mole JE, Cole FS. J Biol Chem. 1984;259:5271. [PubMed] [Google Scholar]
- [4].Vouyiouklis DA, Brophy PJ. J Neurochem. 1997;69:995. doi: 10.1046/j.1471-4159.1997.69030995.x. [DOI] [PubMed] [Google Scholar]
- [5].De Corte V, Demol H, Goethals M, Van Damme J, Gettemans J, Vandekerckhove J. Protein Sci. 1999;8:234. doi: 10.1110/ps.8.1.234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Yuan X, Desiderio DM. J Proteome Res. 2003;2:476. doi: 10.1021/pr025589a. [DOI] [PubMed] [Google Scholar]
- [7].Marcus K, Immler D, Sternberger J, Meyer HE. Electrophoresis. 2000;21:2622. doi: 10.1002/1522-2683(20000701)21:13<2622::AID-ELPS2622>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- [8].Bucki R, Byfield FJ, Kulakowska A, McCormick ME, Drozdowski W, Namiot Z, Hartung T, Janmey PA. J Immunol. 2008;181:4936. doi: 10.4049/jimmunol.181.7.4936. [DOI] [PubMed] [Google Scholar]
- [9].Bucki R, Levental I, Kulakowska A, Janmey PA. Curr Protein Pept Sci. 2008;9:541. doi: 10.2174/138920308786733912. [DOI] [PubMed] [Google Scholar]
- [10].Kulakowska A, Drozdowski W, Sadzynski A, Bucki R, Janmey PA. Eur J Neurol. 2008;15:584. doi: 10.1111/j.1468-1331.2008.02133.x. [DOI] [PubMed] [Google Scholar]
- [11].Smith DB, Janmey PA, Herbert TJ, Lind SE. J Lab Clin Med. 1987;110:189. [PubMed] [Google Scholar]
- [12].Paunio T, Kangas H, Heinonen O, Buc-Caron MH, Robert JJ, Kaasinen S, Julkunen I, Mallet J, Peltonen L. J Biol Chem. 1998;273:16319. doi: 10.1074/jbc.273.26.16319. [DOI] [PubMed] [Google Scholar]
- [13].Spinardi L, Witke W. Subcell Biochem. 2007;45:55. doi: 10.1007/978-1-4020-6191-2_3. [DOI] [PubMed] [Google Scholar]
- [14].Wiederin J, Rozek W, Duan F, Ciborowski P. Proteome Sci. 2009;7:8. doi: 10.1186/1477-5956-7-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Rozek W, Ricardo-Dukelow M, Holloway S, Gendelman HE, Wojna V, Melendez LM, Ciborowski P. J Proteome Res. 2007;6:4189. doi: 10.1021/pr070220c. [DOI] [PubMed] [Google Scholar]
- [16].Suhler E, Lin W, Yin HL, Lee WM. Crit Care Med. 1997;25:594. doi: 10.1097/00003246-199704000-00007. [DOI] [PubMed] [Google Scholar]
- [17].Osborn TM, Verdrengh M, Stossel TP, Tarkowski A, Bokarewa M. Arthritis Res Ther. 2008;10:R117. doi: 10.1186/ar2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Dahl B, Schiodt FV, Ott P, Gvozdenovic R, Yin HL, Lee WM. Shock. 1999;12:102. doi: 10.1097/00024382-199908000-00002. [DOI] [PubMed] [Google Scholar]
- [19].Mounzer KC, Moncure M, Smith YR, Dinubile MJ. Am J Respir Crit Care Med. 1999;160:1673. doi: 10.1164/ajrccm.160.5.9807137. [DOI] [PubMed] [Google Scholar]
- [20].Rithidech KN, Honikel L, Milazzo M, Madigan D, Troxell R, Krupp LB. Mult Scler. 2009;15:455. doi: 10.1177/1352458508100047. [DOI] [PubMed] [Google Scholar]
- [21].Bohgaki M, Matsumoto M, Atsumi T, Kondo T, Yasuda S, Horita T, Nakayama KI, Okumura F, Hatakeyama S, Koike T. J Cell Mol Med. 2009 doi: 10.1111/j.1582-4934.2009.00940.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Bohgaki M, Atsumi T, Yamashita Y, Yasuda S, Sakai Y, Furusaki A, Bohgaki T, Amengual O, Amasaki Y, Koike T. Int Immunol. 2004;16:1633. doi: 10.1093/intimm/dxh166. [DOI] [PubMed] [Google Scholar]
- [23].Kwiatkowski DJ. Curr Opin Cell Biol. 1999;11:103. doi: 10.1016/s0955-0674(99)80012-x. [DOI] [PubMed] [Google Scholar]


