Abstract
The tropoelastin monomer undergoes stages of association by coacervation, deposition onto microfibrils, and cross-linking to form elastic fibers. Tropoelastin consists of an elastic N-terminal coil region and a cell-interactive C-terminal foot region linked together by a highly exposed bridge region. The bridge region is conveniently positioned to modulate elastic fiber assembly through association by coacervation and its proximity to dominant cross-linking domains. Tropoelastin constructs that either modify or remove the entire bridge and downstream regions were assessed for elastogenesis. These constructs focused on a single alanine substitution (R515A) and a truncation (M155n) at the highly conserved arginine 515 site that borders the bridge. Each form displayed less efficient coacervation, impaired hydrogel formation, and decreased dermal fibroblast attachment compared to wild-type tropoelastin. The R515A mutant protein additionally showed reduced elastic fiber formation upon addition to human retinal pigmented epithelium cells and dermal fibroblasts. The small-angle X-ray scattering nanostructure of the R515A mutant protein revealed greater conformational flexibility around the bridge and C-terminal regions. This increased flexibility of the R515A mutant suggests that the tropoelastin R515 residue stabilizes the structure of the bridge region, which is critical for elastic fiber assembly.
Keywords: tropoelastin assembly, protease resistance
Elastic fibers confer the elastic and recoil properties required for repetitive and reversible deformation of elastic tissues during normal function (1–3). The assembly of elastic fibers from tropoelastin, the soluble precursor of elastin, is classically defined by stages of tropoelastin synthesis, coacervation, microfibrillar deposition, and cross-linking. Tropoelastin is secreted by elastogenic cells such as smooth muscle cells, endothelial cells, and fibroblasts (1, 2). At the cell surface, the monomers cluster through hydrophobic domain interactions in an aqueous environment (3–6) by the entropically driven process of coacervation (7). These tropoelastin assemblies remain attached through the C terminus to cell-surface integrin αvβ3 and glycosaminoglycans (8–10) until deposition on microfibrillar scaffolds (11), which direct the shape and orientation of elastic fibers (10, 12, 13). Microfibrillar proteins recruit lysyl oxidase (14, 15), which reacts with specific tropoelastin lysine residues to form cross-links (1, 11, 16). These cross-links occur at multiple sites in the molecule (17) and are enriched in domains 19–25 (1, 16). Cross-linking imposes expansional constraints on elastin and renders elastic fibers resilient under repetitive mechanical stretching (1, 12, 18).
The recently solved nanostructure of tropoelastin has allowed the main functional regions of tropoelastin to be placed within a structural context (19). Most of the elasticity of the molecule is conferred by a coiled region (20) that extends from domain 2 to domain 18. A protrusion from the coil region around domains 20–24 corresponds to a predicted hinge region (21). The bridge region is located around domains 25 and 26, which connects to a foot-like region containing the cell-interactive C terminus. Unlike the coil and foot regions, little is known about the functional role of the bridge region. One model is that the bridge is involved in elastic fiber assembly, because it is likely to encompasses the essential domain 26 and resides near other large hydrophobic domains that are important for coacervation (3, 5). The bridge is also proximal to the central domains that dominate intermolecular cross-linking to form elastin. The bridge region is exposed and contains a protease-susceptible arginine 515 (R515) site at the junction of domains 25 and 26 (3, 22). This site is highly conserved in confirmed mammalian tropoelastin sequences (Fig. S1).
We describe here the role of the bridge region in tropoelastin function. We assessed the elastogenic capacity of two tropoelastin constructs with perturbations in this region: R515A, a recombinant human tropoelastin isoform where the R515 residue is mutated to an alanine; and M155n, which corresponds to the larger N-terminal segment of tropoelastin after cleavage at R515 (23) (Fig. 1). The self-assembly and cell interaction functionalities of these constructs were compared against mature, full-length human tropoelastin (WT). The nanostructure of R515A, which was solved using small-angle X-ray scattering and compared with that of WT, accounts for its altered physical and biological properties.
Fig. 1.
Domain structures of the tropoelastin species used in this study (residue 515 indicated). Hydrophobic and hydrophilic domains are represented by white and gray blocks, respectively.
Results and Discussion
R515A Tropoelastin Is Resistant to Proteolytic Cleavage.
Conversion of the protease-susceptible R515 site to A515 (Fig. S2) conferred resistance to digestion with human plasma kallikrein (Fig. S3). A 2 h incubation with the serine protease resulted in cleavage of WT tropoelastin into 45 and 15 kDa fragments (3) over time. In contrast, the R515A mutant resisted kallikrein cleavage. Evolutionary conservation of this arginine in all confirmed mammalian tropoelastin sequences is consistent with a functional role in the protein.
Mutant Tropoelastin Species Display Impaired Coacervation.
The association of tropoelastin by coacervation represents the first essential stage of elastogenesis and crucially permits the subsequent incorporation of tropoelastin into the elastic fiber (24). WT, R515A, and M155n all exhibited temperature-dependent coacervation (3, 23, 25–27) (Fig. 2A). As each tropoelastin solution reached the critical temperature, there was a sharp rise in turbidity. No increase in turbidity was observed beyond this threshold temperature, which was distinct for each tropoelastin species. Full coacervation of WT was observed at 35 °C; R515A at 40 °C; and M155n at 45 °C.
Fig. 2.
(A) Temperature-dependent coacervation of 10 mg/mL WT (●), R515A (□), and M155n (▴) tropoelastin. (B) Coacervation time of 10 mg/mL WT (●), R515A (□), and M155n (▴) tropoelastin at a range of temperatures.
In addition to these thermodynamic differences, the temporal requirement for coacervation was likewise specific to each tropoelastin species (Fig. 2B). A similar trend was observed where WT tropoelastin coacervated most rapidly and M155n most slowly. However, the differences in coacervation time decreased with increasing temperatures and converged above 50 °C.
Differences in the coacervation profiles of WT and R515A tropoelastin were assessed by solution particle size analysis (Fig. 3). At 25 °C, all R515A and most WT species were 8–18 nm (i.e., monomers). At 30 °C, R515A remained as monomers, whereas WT associated to give 1.0- to 2.0-μm particles. At 35 °C, R515A formed similar associates but WT aggregated into larger 2.0- to 6.4-μm droplets consistent with full-sized tropoelastin coacervates (28). At 40 °C, over 90% of the R515A coacervate comprised particles that were similarly sized to the WT. The initial association of WT at 30 °C and R515A at 35 °C was consistent with the distinct temperatures at which each species first exhibited coacervation.
Fig. 3.
Particle sizes of 10 mg/mL WT (black) and R515A (gray) solutions at (A) 25, (B) 30, (C) 35, and (D) 40 °C as measured by dynamic light scattering.
The number of hydrophobic domains determines the critical temperature (29–34) and rate (3) of coacervation, but this model does not explain the difference between WT and R515A. The unexpected increase in R515A coacervation temperature conceptually reflects a higher energy requirement for the disruption of bound water and therefore indicates an increase in protein hydration (35, 29), most likely due to a structural deviation from wild-type tropoelastin. The increase in R515A coacervation time also supports a model of a conformational change that affects its ability to associate through either steric hindrance and/or altered relative positions of associating hydrophobic domains.
Mutant Tropoelastin Species Form Abnormal Hydrogels.
The cross-linking of tropoelastin is dependent on proper alignment during coacervation and corresponds to its propensity to be incorporated into insoluble elastic fibers. Chemical cross-linking of tropoelastin using the amine-specific bifunctional cross-linker bis(sulfosuccinimidyl) suberate (BS3) approximates lysine-specific cross-linking in vivo to give allysine aldol and lysinonorleucine following oxidation by lysyl oxidase (1, 16). At physiological temperature, the addition of 10 mM BS3 enabled cross-linking of all WT and R515A and most M155n monomers in solution. Cross-linking of M155n was only partly temperature dependent, as evidenced by a comparable extent of monomer utilization at its higher coacervation temperature (Fig. S4). There were similar levels of free amine groups in all hydrogel constructs as determined by reaction with 2,4,6-trinitrobenzene sulfonic acid (Fig. S5). Although this result indicated a similar extent of lysine modification, the nature and functionality of the resulting cross-links are possibly different.
Microcomputed tomography (micro-CT) imaging of the cross-linked WT, R515A, and M155n hydrogels revealed visible morphological differences (Fig. S6). The WT hydrogel consisted of overlapping fibers interspersed with large channels, consistent with the filamentous nature of natural elastin (2, 30, 31). In contrast, the R515A and M155n hydrogels appeared nonfibrous and were composed of compact bead-like particles. Examination of various horizontal and vertical cross-sections within each hydrogel suggested structural homogeneity across each sample. Automated analyses of the micro-CT images calculated porosity of the WT, R515A, and M155n hydrogels at 80%, 64%, and 69%, respectively.
The hydrogels swelled substantially in water at 4, 25, and 37 °C (Fig. 4). WT, R515A, and M155n hydrogels were dominantly aqueous: They respectively absorbed 80.8 ± 8.8, 11.6 ± 3.3, and 15.8 ± 2.2 g H2O/g protein at 4 °C; 75.7 ± 11.3, 9.8 ± 2.7, and 14.3 ± 0.7 g H2O/g protein at 25 °C; and 57.9 ± 1.7, 11.3 ± 2.8, and 14.3 ± 0.9 g H2O/g protein at 37 °C. At each temperature, the mutant hydrogels swelled 75–86% less than the wild-type hydrogels, which was consistent with their reduced porosities. R515A showed comparable swelling to the M155n hydrogels. The extent of solvent influx is defined by the stretching of the hydrogel network junctions and the mixing of solvent and hydration water within the polymer (32, 33). The low porosity and decreased swelling of mutant hydrogels would be expected from atypical association and/or cross-linking (32, 34, 36).
Fig. 4.
Swelling of hydrogels in water at 4, 25, and 37 °C. The amount of water absorbed per gram of hydrogel was measured. Water absorption of the R515A and M155n hydrogels was compared against the WT hydrogels. Statistical significance is indicated by *** (p < 0.001) (n = 3).
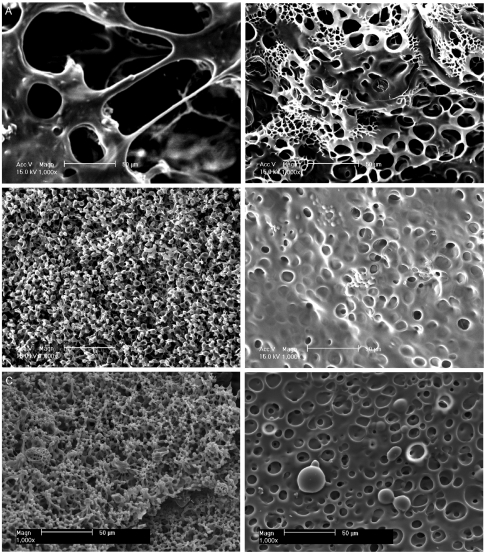
SEM of the hydrogels revealed distinct differences in the structure and composition of their surfaces (Fig. 5). The top and bottom surfaces of the WT hydrogel were flat, fibrous, and porous, consistent with the micro-CT images. Pore sizes were similar to those previously reported (37). A smoother but similarly porous layer was observed on the bottom surfaces of the R515A and M155n hydrogels. However, their top surfaces consisted entirely of approximately 5-µm globular clusters interlinked with fibers, which were similar in size to spherules observed during the early stages of cross-linking (28). These globules are believed to be partially cross-linked nascent elastin (1, 28, 38) and were also observed on the bottom surfaces, as would be expected if they arose from incomplete coalescence into the WT sheet-like structures. In contrast, these globules were only sparsely present on the WT hydrogel. Their abundance in the R515A and M155n hydrogels is consistent with a decreased ability of the mutants to form mature cross-linked structures (39).
Fig. 5.
Scanning electron micrographs of hydrogels constructed from (A) WT, (B) R515A, and (C) M155n taken at 1,000× magnification. The top (Left) and bottom (Right) surfaces of each hydrogel are shown.
The R515A Mutant Form of Tropoelastin Supports Decreased Fibroblast Attachment.
Binding of each tropoelastin species to tissue culture wells was detected by an ELISA using an elastin-specific antibody (BA4) in a concentration-dependent manner until saturation at 10 μg/mL tropoelastin. The BA4 antibody reacts with specific hexapeptides such as VGVAPG present within hydrophobic domains (40) and therefore allows a direct comparison of the amount of protein in each well. Equivalent levels of WT and R515A were detected at each coating concentration. As expected, the shorter M155n, which contains fewer epitopes per molecule, resulted in less detected bound protein (Fig. 6A).
Fig. 6.
(A) Attachment of tropoelastin to cell culture wells as detected by the BA4 anti-elastin antibody. The amount of bound R515A (□) and M155n (▴) was compared to WT (●) at each coating concentration. (B) Attachment of human dermal fibroblasts to wells coated with WT (●), R515A (□), or M155n (▴). Cell attachment to either mutant was compared to the wild-type species at each coating concentration. (C) Exposure of the tropoelastin C-terminal region on tropoelastin-coated wells as detected by an anti C-terminal peptide antibody. Values are expressed as a percentage of detected levels on 20 μg/mL WT-coated wells. C-terminal accessibility of R515A (□) and M155n (▴) were compared to that of WT (●) at the same concentration. In each figure, statistical significance is indicated by * (p < 0.05), ** (p < 0.01), or *** (p < 0.001) (n = 3).
Human dermal fibroblasts attached to wells coated with > 10 μg/mL WT or R515A tropoelastin (Fig. 6B). Fibroblast attachment saturated at 20 μg/mL protein, where 84 ± 5% of fibroblasts adhered to WT, whereas only 55 ± 8% attached to R515A. There was minimal cell attachment (10 ± 4%) to M155n, independent of concentration. Low attachment to M155n was expected because the region dictating fibroblast attachment is within the C-terminal region absent in M155n (8). Reduced cell attachment to R515A compared to WT is consistent with a decreased exposure of the C terminus.
The exposure of this C terminus on coated wells was determined using a polyclonal antibody specific for this region (41) (Fig. 6C). The negative control M155n-coated wells showed low antibody binding irrespective of coating concentration. At tropoelastin concentrations ≥10 μg/mL, 71 ± 4% of R515A C termini was detected, confirming reduced C terminus exposure compared to WT. This reduced exposure correlated in magnitude to the decreased cell attachment to R515A and points to a structural displacement of the C terminus in the R515A mutant.
R515A Tropoelastin Shows Impaired Fiber Assembly in a Cellular Environment.
Human retinal pigmented epithelium (ARPE-19) cells serve to model elastic fiber assembly, which depends on exogenously supplied tropoelastin, because they express major elastogenic components but do not produce tropoelastin (42). When WT and R515A tropoelastin were added to ARPE-19 cells with an established microfibrillar network, they progressively assembled into elastic fibers (Fig. 7). However, the resulting fibers of each tropoelastin species and the time course of fiber formation were visibly different. One day after tropoelastin addition, WT spherules aligned linearly to approximate a fibrillar morphology, whereas R515A particles were randomly dispersed. Four days after tropoelastin addition, WT and R515A had each formed distinct fibers, which persisted over the remainder of the experimental period. R515A fibers stained less intensely and displayed lower density than WT fibers at 4, 7, and 10 d after tropoelastin addition. Decreased immunofluorescence of R515A fibers would be expected to result from a reduced number or exposure of epitopes due to impaired or abnormal fiber assembly. However, the autofluorescence of R515A fibers (43) was also less intense than WT fibers and paralleled the decrease in immunofluorescence. These data suggest that fewer R515A molecules are assembled into stable elastic fibers, consistent with reduced cell binding and/or protein association.
Fig. 7.
(A–H) Confocal microscopy of ARPE-19 cells with 20 μg/mL WT or R515A tropoelastin added to the culture media 14 d after seeding. The cells were fixed at 1, 4, 7, and 10 d after tropoelastin addition. Elastin fibers were stained with mouse anti-elastin BA4 primary antibody and FITC-conjugated goat anti-mouse secondary antibody. Cell nuclei were stained with DAPI. (I) ARPE-19 cells with no tropoelastin in the culture media. (J) Fibrillin-1 fibers in ARPE-19 cells 14 d after seeding prior to tropoelastin addition, stained with mouse anti-fibrillin-1 primary antibody and FITC-conjugated goat anti-mouse antibody. Each pair of images represents the same field of view; the left image shows merged signals from the DAPI and FITC channels, and the right image shows fluorescence from the FITC channel only.
More striking differences in fiber formation were observed when WT and R515A were added to cultured human dermal fibroblasts. The effect was dramatic for GM3348 dermal fibroblasts where R515A spherules aligned linearly but did not assemble into distinct fibers (Fig. S7), whereas NHF8909 neonatal dermal fibroblasts only formed faint and sparse R515A fibers (Fig. S8). This reduced capacity for elastogenesis contrasts with the increasing network of WT fibers and confirms that R515A has a reduced ability to form elastin.
R515A Tropoelastin Displays a Structural Shift in the Bridge and C-Terminal Regions.
Small-angle X-ray scattering data of WT and R515A initially indicated differences in the distribution of interatomic spacings within the molecules (Fig. S9). Multiple particle shapes generated from the scattering images were superimposed to show congruence of the individual solutions (Fig. 8A). The overlaid WT models illustrated a structure consistent with previous findings (19), comprising an N-terminal coil region connected by a bridge region to the C-terminal foot. The overlaid R515A models, however, demonstrated greater conformational variability around the bridge and C-terminal regions.
Fig. 8.
(A) Ab initio models of (i) WT and (ii) R515A tropoelastin generated using GASBOR (WT chi = 0.98; R515A chi = 0.97). Eight representative solution structures of each tropoelastin construct were aligned and used to produce the average model. The elastic N-terminal coil region (NC), bridge region (BR), and cell-interactive C-terminal foot region (CF) are indicated. (B) Average solution structures of WT (green) and R515A (purple) tropoelastin derived from small-angle X-ray scattering analysis. Superimposition of the averaged models illustrates a conformational difference in the R515A bridge and C-terminal regions.
The averaged WT and R515A solution structures displayed similar structural features including an aligned N-terminal coil region (Fig. 8B). However, the conformational flexibility adopted by the R515A bridge and C-terminal regions resulted in an averaged model that exhibits substantial dislocation of the R515A C-terminal foot toward the central axis of the molecule. The mechanism behind this increased bridge/C-terminal flexibility remains unclear. R515 sits at the C terminus of an α-helical cross-linking region and may have stabilizing interactions with the helical dipole (44) that are abolished with alanine substitution. Although circular dichroism analyses of WT and R515A indicated essentially identical secondary structure content (Fig. S10 and Table S1), slight differences in helical content and/or conformation cannot be discounted.
We propose that the tropoelastin R515 residue is involved in maintaining the orientation of the bridge and, consequently, the C-terminal region in human tropoelastin. This unique native conformation is likely crucial to efficient participation of the tropoelastin molecule in elastic fiber assembly. The repositioning of the bridge and C-terminal regions of R515A tropoelastin appears to be associated with impaired function, including decreased self-assembly and cell-interactive properties.
Materials and Methods
Preparation of Tropoelastin Species.
The tropoelastin species WT (synthetic human tropoelastin without domain 26A, equivalent to the mature wild-type species corresponding to residues 27–724 of GenBank entry AAC98394), R515A, and M155n were produced as previously described (23, 45). R515A was confirmed by plasmid sequencing and comparative mass spectrometry of the purified protein against WT (Fig. S2). M155n was previously confirmed with electrospray mass spectrometry and N- and C-terminal sequencing (23). The integrity of all constructs was verified by SDS-PAGE to be full length.
Coacervation Assay.
A sample of 10 mg/mL WT, R515A, or M155n in PBS was placed in a Shimadzu UV-1601 spectrophotometer at set temperatures. Light scattering was monitored by measuring the absorbance at 300 nm over 600 s at 20–60 °C. Between each temperature shift, the sample was cooled at 4 °C for 10 min. The temperature above which there was no increase in sample turbidity was determined to be the coacervation temperature. The time taken to reach maximum sample turbidity at each temperature was set as the coacervation time. The tropoelastin species were assessed according both variables.
Particle Size Analysis.
Particle sizes in 10 mg/mL WT and R515A solutions were determined over a temperature range of 25–40 °C via dynamic light scattering using a Malvern Zetasizer Nano (Malvern Instruments). Tropoelastin solutions were equilibrated for 5 min at each temperature. Three runs of measurements, each with at least 12 data acquisitions, were taken and averaged to obtain the relative volume percentages of particle sizes in solution.
Hydrogel Construction.
WT, R515A, and M155n were dissolved to 100 mg/mL in PBS at 4 °C overnight and transferred to Chamber Slides (LabTek), then cross-linked with 10 mM bis(sulfosuccinimidyl) suberate at 37 °C for 16 h. The resulting hydrogels were taken off the slides and lyophilized.
Microcomputed Tomography.
Hydrogels were scanned with a SkyScan 1072 micro-CT system using a 60 kV X-ray beam at a resolution of 3.5 μm. The X-ray projection images were converted into a stack of cross-sections with the NRecon 1.4.4 cone-beam reconstruction program (SkyScan) and rendered into a three-dimensional structure with VGStudio Max 1.2.1 (Volume Graphics). Hydrogel porosity was estimated from the cross-section images using CTan software (SkyScan).
Swelling Assay.
Preweighed hydrogels were submerged in Milli-Q water (Millipore) for 24 h at 4, 25, and 37 °C. Excess water was drained and the hydrogels were weighed. The tared weight yielded the amount of water absorbed per gram of hydrogel.
Scanning Electron Microscopy.
Hydrogels were swelled in PBS then washed in phosphate buffer and lyophilized. Samples were mounted using Silver DAG, coated with a 20-nm gold layer, and imaged on a Philips XL-30 scanning electron microscope at 1,000× magnification.
Cell Attachment Assay.
Triplicate cell culture wells were coated with 1.25, 2.5, 5, 10, 20, or 30 μg/mL WT, R515A, and M155n at 4 °C overnight then washed with PBS to remove unbound tropoelastin. Wells were blocked for 1 h with 10 mg/mL denatured bovine serum albumin in PBS. GM3348 fibroblasts grown in DMEM with 10% (vol/vol) fetal calf serum were trypsinized at 37 °C for 2 min. The cells were centrifuged at 800 × g for 4 min and resuspended in serum-free DMEM. The tropoelastin-coated wells were seeded at a density of 1.56 × 105 cells/cm2 well surface. Standards with 10%, 20%, 50%, 80%, and 100% of the seeding density were added to uncoated and unblocked wells. Cells were allowed to attach at 37 °C for 1.5 h. After incubation, nonadherent cells in the tropoelastin-coated wells were removed with PBS. Cells were fixed with 3% (wt/vol) formaldehyde in PBS for 20 min and stained with 0.1% (wt/vol) crystal violet in 0.2 M MES, pH 5.0 for 1 h. Excess stain was washed away with water, and the crystal violet was solubilized with 10% (wt/vol) acetic acid. Absorbance at 570 nm from the standard wells were fitted to a linear regression and used to convert sample absorbances into percentage cell attachment.
Enzyme-Linked Immunosorbent Assay.
Wells were coated with 1.25, 2.5, 5, 10, 20, or 30 μg/mL WT, R515A, or M155n at 4 °C overnight and washed with PBS to remove unbound tropoelastin. Wells were blocked with 3% (wt/vol) BSA for 1 h. Bound tropoelastin was detected with 1∶2,000 BA4 mouse anti-elastin antibody (Sigma Aldrich) for 1 h and 1∶5,000 goat anti-mouse IgG conjugated with horseradish peroxidase for 1 h, then visualized with 2,2′-azino-bis(3-ethylbenzthiazoline-6-sulphonic) acid (ABTS) solution [1.04 mg/mL ABTS, 0.05% (vol/vol) H2O2, 10 mM CH3COONa, 5 mM Na2HPO4] at 37 °C for 1 h. Absorbance was measured at 405 nm.
To determine the exposure of the tropoelastin C-terminal region on coated wells, an ELISA was performed as above using 1∶500 rabbit anti-C-terminal peptide antibody (a gift of R. Mecham, Washington University, St. Louis, MO) and 1∶5,000 horseradish peroxidase-conjugated anti-rabbit IgG as the primary and secondary antibody, respectively. Sample absorbances were expressed as a percentage of the maximum absorbance of WT-coated wells. Net values that were just below the average background absorbance calculated from BSA-blocked wells were adjusted to zero.
Immunofluorescent Staining of Elastin Fibers.
Human retinal pigmented epithelium cells (ARPE-19; gift of M. Madigan, Save Sight Institute, New South Wales, Australia), human dermal fibroblasts (GM3348; obtained from the Coriell Research Institute), and human neonatal fibroblasts (NHF8909; gift of X. Q. Wang, University of Queensland, Queensland, Australia) were seeded on glass coverslips at a density of 18,400 cells/cm2 in DMEM:nutrient mixture F12 supplemented with 10% (vol/vol) fetal bovine serum, 2 mM L-glutamine, and 1% (vol/vol) penicillin/streptomycin. At 10 and 14 d after seeding, 20 μg/mL WT or R515A tropoelastin in PBS was added to triplicate ARPE-19 and fibroblast cultures, respectively. Culture media was changed every 2 d. At 1, 4, 7, and 10 d after tropoelastin addition, cells were fixed with 4% (wt/vol) paraformaldehyde for 20 min and quenched with 0.2 M glycine. The cells were incubated with 0.2% (vol/vol) Triton X-100 for 6 min, blocked with 5% bovine serum albumin at 4 °C overnight, and stained with 1∶500 BA4 mouse anti-elastin antibody for 1.5 h and 1∶100 anti-mouse IgG-FITC antibody (Sigma Aldrich) for 1 h. The coverslips were then mounted onto glass slides with ProLong Gold antifade reagent with DAPI (Invitrogen).
Confocal Microscopy.
Samples were visualized with an Olympus FluoView FV1000 confocal microscope under the same laser settings. Z stacks were taken from at least three areas distributed across each sample and converted to maximum projection images.
Small-Angle X-Ray Scattering.
WT and R515A tropoelastin were dissolved in PBS to 1.2 and 1.1 mg/mL, respectively. Small-angle X-ray scattering data were collected on European Molecular Biology Laboratory, beamline X33 at the light source facilities DORIS III at Hamburger Synchrotronstrahlungslabor/Deutsches Elektronen-Synchrotron (46). Data were collected using 4 × 30-s exposures and a 2.4-m sample-to-detector distance to cover a momentum transfer interval 0.008 < q < 0.54 Å-1. The modulus of the momentum transfer is defined as q = 4Π sin θ/λ, where 2θ is the scattering angle, and λ is the wavelength. The q range was calibrated using silver behenate powder based on diffraction spacings of 58.38 Å. The scattering images obtained were spherically averaged using inhouse software and buffer scattering intensities subtracted using PRIMUS software (47). Particle shapes were generated ab initio using GASBOR software (48). Multiple GASBOR runs were performed to generate 20 similar shapes that were combined and filtered to produce an averaged model using the DAMAVER software package (49).
Statistical Analyses.
Replicate values were reported as mean ± standard error. Statistical significance was calculated using analysis of variance. Significance was set at p < 0.05 or higher.
Supplementary Material
Acknowledgments.
A.S.W. acknowledges grant support from the Australian Research Council, the National Health and Medical Research Council, and the Defence Health Foundation. G.C.Y. is supported by an International Postgraduate Research/International Postgraduate Award PhD scholarship.
Footnotes
Conflict of interest statement: A.S.W. is founder and consultant scientist at Elastagen Pty., Ltd.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1111615108/-/DCSupplemental.
References
- 1.Mithieux SM, Weiss AS. Advances in Protein Chemistry. Vol 70. San Diego: Academic; 2005. Elastin; pp. 437–461. [DOI] [PubMed] [Google Scholar]
- 2.Ronchetti IP, Baccarani-Contri M, Fornieri C, Mori G, Quaglino D. Structure and composition of the elastin fibre in normal and pathological conditions. Micron. 1993;24:75–89. [Google Scholar]
- 3.Jensen SA, Vrhovski B, Weiss AS. Domain 26 of tropoelastin plays a dominant role in association by coacervation. J Biol Chem. 2000;275(37):28449–28454. doi: 10.1074/jbc.M004265200. [DOI] [PubMed] [Google Scholar]
- 4.Sato F, et al. Distinct steps of cross-linking, self-association, and maturation of tropoelastin are necessary for elastic fiber formation. J Mol Biol. 2007;369:841–851. doi: 10.1016/j.jmb.2007.03.060. [DOI] [PubMed] [Google Scholar]
- 5.Wachi H, et al. Domains 16 and 17 of tropoelastin in elastic fibre formation. Biochem J. 2007;402:63–70. doi: 10.1042/BJ20061145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Urry DW. Entropic elastic processes in protein mechanisms. I. Elastic structure due to an inverse temperature transition and elasticity due to internal chain dynamics. J Protein Chem. 1988;7:1–34. doi: 10.1007/BF01025411. [DOI] [PubMed] [Google Scholar]
- 7.Jamieson AM, Simic-Glavaski B, Tansey K, Walton AG. Studies of elastin coacervation by quasielastic light scattering. Faraday Discuss Chem Soc. 1976;61:194–204. doi: 10.1039/dc9766100194. [DOI] [PubMed] [Google Scholar]
- 8.Bax DV, Rodgers UR, Bilek MM, Weiss AS. Cell adhesion to tropoelastin is mediated via the C-terminal GRKRK motif and integrin alphaVbeta3. J Biol Chem. 2009;284:28616–28623. doi: 10.1074/jbc.M109.017525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Broekelmann TJ, et al. Tropoelastin interacts with cell-surface glycosaminoglycans via its COOH-terminal domain. J Biol Chem. 2005;280:40939–40947. doi: 10.1074/jbc.M507309200. [DOI] [PubMed] [Google Scholar]
- 10.Kozel BA, et al. Elastic fiber formation: A dynamic view of extracellular matrix assembly using timer reporters. J Cell Physiol. 2006;207:87–96. doi: 10.1002/jcp.20546. [DOI] [PubMed] [Google Scholar]
- 11.Sato F, et al. The characteristics of elastic fiber assembled with recombinant tropoelastin isoform. Clin Biochem. 2006;39:746–753. doi: 10.1016/j.clinbiochem.2006.02.017. [DOI] [PubMed] [Google Scholar]
- 12.Mecham RP. Elastin synthesis and fiber assembly. Ann N Y Acad Sci. 1991;624:137–146. doi: 10.1111/j.1749-6632.1991.tb17013.x. [DOI] [PubMed] [Google Scholar]
- 13.Pepe A, et al. Dissection of human tropoelastin: Supramolecular organization of polypeptide sequences coded by particular exons. Matrix Biol. 2005;24:96–109. doi: 10.1016/j.matbio.2005.01.004. [DOI] [PubMed] [Google Scholar]
- 14.Hirai M, et al. Fibulin-5/DANCE has an elastogenic organizer activity that is abrogated by proteolytic cleavage in vivo. J Cell Biol. 2007;176:1061–1071. doi: 10.1083/jcb.200611026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu X, et al. Elastin fiber homeostasis requires lysyl oxidase-like 1 protein. Nat Genet. 2004;36(2):178–182. doi: 10.1038/ng1297. [DOI] [PubMed] [Google Scholar]
- 16.Wise SG, Mithieux SM, Raftery MJ, Weiss AS. Specificity in the coacervation of tropoelastin: solvent exposed lysines. J Struct Biol. 2005;149:273–281. doi: 10.1016/j.jsb.2004.11.006. [DOI] [PubMed] [Google Scholar]
- 17.Debelle L, Tamburro AM. Elastin: Molecular description and function. Int J Biochem Cell Biol. 1999;31:261–272. doi: 10.1016/s1357-2725(98)00098-3. [DOI] [PubMed] [Google Scholar]
- 18.Bedell-Hogan D, Trackman P, Abrams W, Rosenbloom J, Kagan HM. Oxidation, cross-linking, and insolubilization of recombinant tropoelastin by purified lysyl oxidase. J Biol Chem. 1993;268:10345–10350. [PubMed] [Google Scholar]
- 19.Baldock C, et al. Shape of tropoelastin, the highly extensible protein that controls human tissue elasticity. Proc Natl Acad Sci USA. 2011;108:4322–4327. doi: 10.1073/pnas.1014280108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Holst J, et al. Substrate elasticity provides mechanical signals for the expansion of hemopoietic stem and progenitor cells. Nat Biotechnol. 2010;28:1123–1128. doi: 10.1038/nbt.1687. [DOI] [PubMed] [Google Scholar]
- 21.Dyksterhuis LB, Baldock C, Lammie D, Wess TJ, Weiss AS. Domains 17–27 of tropoelastin contain key regions of contact for coacervation and contain an unusual turn-containing crosslinking domain. Matrix Biol. 2007;26:125–135. doi: 10.1016/j.matbio.2006.10.002. [DOI] [PubMed] [Google Scholar]
- 22.Kozel BA, Wachi H, Davis EC, Mecham RP. Domains in tropoelastin that mediate elastin deposition in vitro and in vivo. J Biol Chem. 2003;278:18491–18498. doi: 10.1074/jbc.M212715200. [DOI] [PubMed] [Google Scholar]
- 23.Wu WJ, Weiss AS. Deficient coacervation of two forms of human tropoelastin associated with supravalvular aortic stenosis. Eur J Biochem. 1999;266:308–314. doi: 10.1046/j.1432-1327.1999.00891.x. [DOI] [PubMed] [Google Scholar]
- 24.Yeo GC, Keeley FW, Weiss AS. Coacervation of tropoelastin. Adv Colloid Interface Sci. 2011;167:94–103. doi: 10.1016/j.cis.2010.10.003. [DOI] [PubMed] [Google Scholar]
- 25.Toonkool P, Jensen SA, Maxwell AL, Weiss AS. Hydrophobic domains of human tropoelastin interact in a context-dependent manner. J Biol Chem. 2001;276:44575–44580. doi: 10.1074/jbc.M107920200. [DOI] [PubMed] [Google Scholar]
- 26.Vrhovski B, Jensen SA, Weiss AS. Coacervation characteristics of recombinant human tropoelastin. Eur J Biochem. 1997;250:92–98. doi: 10.1111/j.1432-1033.1997.00092.x. [DOI] [PubMed] [Google Scholar]
- 27.Wu WJ, Vrhovski B, Weiss AS. Glycosaminoglycans mediate the coacervation of human tropoelastin through dominant charge interactions involving lysine side chains. J Biol Chem. 1999;274:21719–21724. doi: 10.1074/jbc.274.31.21719. [DOI] [PubMed] [Google Scholar]
- 28.Clarke AW, et al. Tropoelastin massively associates during coacervation to form quantized protein spheres. Biochemistry. 2006;45:9989–9996. doi: 10.1021/bi0610092. [DOI] [PubMed] [Google Scholar]
- 29.Urry DW, Peng SQ, Xu J, McPherson DT. Characterization of waters of hydrophobic hydration by microwave dielectric relaxation. J Am Chem Soc. 1997;119:1161–1162. [Google Scholar]
- 30.Gotte L, Giro MG, Volphin D, Horne RW. The ultrastructural organization of elastin. J Ultrastruct Res. 1974;46:23–33. doi: 10.1016/s0022-5320(74)80019-5. [DOI] [PubMed] [Google Scholar]
- 31.Ronchetti IP, Baccarani-Contri M. Elastic fiber during development and aging. Microsc Res Tech. 1997;38:428–435. doi: 10.1002/(SICI)1097-0029(19970815)38:4<428::AID-JEMT10>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 32.Flory PJ, Rehner JJ. Statistical mechanics of cross-linked polymer networks II. Swelling. J Chem Phys. 1943;11:521–526. [Google Scholar]
- 33.Nickerson MT, et al. Some physical and microstructural properties of genipin-crosslinked gelatin-maltodextrin hydrogels. Int J Biol Macromol. 2006;38:40–44. doi: 10.1016/j.ijbiomac.2005.12.017. [DOI] [PubMed] [Google Scholar]
- 34.Lee J, Macosko CW, Urry DW. Swelling behavior of gamma-irradiation cross-linked elastomeric polypentapeptide-based hydrogels. Macromolecules. 2001;34:4114–4123. [Google Scholar]
- 35.Urry DW, et al. Temperature of polypeptide inverse temperature transition depends on mean residue hydrophobicity. J Am Chem Soc. 1991;113:4346–4348. [Google Scholar]
- 36.Vieth S, Bellingham CM, Keeley FW, Hodge SM, Rousseau D. Microstructural and tensile properties of elastin-based polypeptides crosslinked with Genipin and pyrroloquinoline quinone. Biopolymers. 2007;85:199–206. doi: 10.1002/bip.20619. [DOI] [PubMed] [Google Scholar]
- 37.Mithieux SM, Rasko JEJ, Weiss ASAS. Synthetic elastin hydrogels derived from massive elastic assemblies of self-organized human protein monomers. Biomaterials. 2004;25:4921–4927. doi: 10.1016/j.biomaterials.2004.01.055. [DOI] [PubMed] [Google Scholar]
- 38.Kozel BA, Ciliberto CH, Mecham RP. Deposition of tropoelastin into the extracellular matrix requires a competent elastic fiber scaffold but not live cells. Matrix Biol. 2004;23:23–34. doi: 10.1016/j.matbio.2004.02.004. [DOI] [PubMed] [Google Scholar]
- 39.Mithieux SM, Tu YD, Korkmaz E, Braet F, Weiss AS. In situ polymerization of tropoelastin in the absence of chemical cross-linking. Biomaterials. 2009;30:431–435. doi: 10.1016/j.biomaterials.2008.10.018. [DOI] [PubMed] [Google Scholar]
- 40.Wrenn DS, Griffin GL, Senior RM, Mecham RP. Characterization of biologically-active domains on elastin—identification of a monoclonal-antibody to a cell recognition site. Biochemistry. 1986;25:5172–5176. doi: 10.1021/bi00366a028. [DOI] [PubMed] [Google Scholar]
- 41.BrownAugsburger P, Broekelmann T, Rosenbloom J, Mecham RP. Functional domains on elastin and microfibril-associated glycoprotein involved in elastic fibre assembly. Biochem J. 1996;318:149–155. doi: 10.1042/bj3180149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wachi H, et al. Development of a new in vitro model of elastic fiber assembly in human pigmented epithelial cells. Clin Biochem. 2005;38:643–653. doi: 10.1016/j.clinbiochem.2005.04.006. [DOI] [PubMed] [Google Scholar]
- 43.Swatland HJ. Fiber-optic reflectance and autofluorescence of bovine elastin and differences between intramuscular and extramuscular tendon. J Anim Sci. 1987;64:1038–1043. [Google Scholar]
- 44.Forood B, Feliciano EJ, Nambiar KP. Stabilization of alpha-helical structures in short peptides via end capping. Proc Natl Acad Sci USA. 1993;90:838–842. doi: 10.1073/pnas.90.3.838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Martin SL, Vrhovski B, Weiss AS. Total synthesis and expression in Escherichia coli of a gene encoding human tropoelastin. Gene. 1995;154:159–166. doi: 10.1016/0378-1119(94)00848-m. [DOI] [PubMed] [Google Scholar]
- 46.Roessle MW, et al. Upgrade of the small-angle X-ray scattering beamline X33 at the European Molecular Biology Laboratory, Hamburg. J Appl Crystallogr. 2007;40:S190–S194. [Google Scholar]
- 47.Konarev PV, Volkov VV, Sokolova AV, Koch MHJ, Svergun D. PRIMUS—a Windows-PC based system for small-angle scattering data analysis. J Appl Crystallogr. 2003;36:1277–1282. [Google Scholar]
- 48.Svergun DI, Petoukhov MV, Koch MHJ. Determination of domain structure of proteins from X-ray solution scattering. Biophys J. 2001;80:2946–2953. doi: 10.1016/S0006-3495(01)76260-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Volkov VV, Svergun DI. Uniqueness of ab-initio shape determination in small-angle scattering. J Appl Crystallogr. 2003;36:860–864. doi: 10.1107/S0021889809000338. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.