The biological functions of RNA sequences are directly related to the folds they can adopt after transcription. Nowadays it is apparent that even coding RNA transcripts depend on complex architectures for their efficient translation (1). Furthermore, several noncoding RNA transcripts exert their biological functions through their folded states. RNA elements called riboswitches (2) are able to recognize a huge variety of ligands of various sizes and, thereby, exert their functions at the transcriptional or translational levels. In the absence of structural methods (like X-ray crystallography or NMR techniques) or chemical and biochemical solution probing methods, the prediction of RNA architecture is still a major challenge. However, attracted by the huge interest in noncoding RNAs, several groups of theoreticians have introduced original and innovative approaches to fold in silico RNA sequences. Such methods, highly complex, rely on force fields at various scales and computer algorithms with different levels of approximations for attempting to master the tremendous complexity of mixing locally atomic details and globally long-range contacts typical of large molecular assemblies. A study in PNAS by Sim et al. (3) introduces an original approach to deal accurately and efficiently with both long-range and short-range interactions during the modeling of large RNA architectures.
RNA transcripts, synthetized as single-stranded molecules, possess a strong tendency to fold back on themselves to form Watson–Crick pairs and double-stranded helical regions. The base pairings lead to hairpins of various lengths and complexities. The secondary structure enumerates or describes the Watson–Crick paired helical regions of an RNA molecule. The set of hairpins and internal helices defining the secondary structure can further assemble into intricate 3D architectures. The architectures are held together precisely through defined sets of RNA–RNA contacts with defined molecular interactions mediated by various types of RNA modules (for a recent overview see ref. 4). To an excellent approximation, RNA architecture can be viewed as a modular and hierarchical assembly of preformed building blocks (5). This approximation simplifies, at least in principle, the obstacles of dealing with multiple levels of complexity, but its automatic implementation with available computer tools still constitutes a daunting task.
Not surprisingly, a very efficient method has been to use human and manual intervention for the modeling of large RNA molecules. First, building blocks are preassembled automatically, and those are then manipulated and positioned on a computer graphics system until a satisfying architecture results (6, 7). The advantages are that large and global movements can be operated on the molecular objects without too much consideration of local atomic clashes that can afterward be relieved by standard refinement tools. Although such a method can be successful given an appropriate number of long-distance relationships, it has several drawbacks. A serious disadvantage is that a single model is obtained, and therefore the ensemble of compatible structures is only glimpsed by the human manipulator(s). In addition, most if not all of the rejected models are not saved. Even if they were not discarded, this set of models would still constitute a fraction of the full ensemble because of the amazing power of the human brain for screening 3D space. Another disadvantage is that the process is highly time-consuming and requires expert knowledge for using the assembly program. Most importantly, the approach relies on the available structural database and on dedicated tools for interrogating the databases in a relevant fashion, for example through topological relationships or relations in space. Otherwise, the method relies only on what has been acquired by the human manipulator(s) (in other words, the database is human-wired).
To alleviate those disadvantages, fragment assembly methods with knowledge-based libraries have been developed for automatic modeling (8–11). Such methods circumvent some of the disadvantages of human intervention. However, they present severe drawbacks, one of which is the difficulty of escaping from local conformational basins. Standard Monte Carlo or molecular dynamics methods are efficient locally only. Coarse-grained methods may sample globally more efficiently but in an unnatural and complex energy landscape with unsuspected barriers to cross. Further, although they produce many decoys, the resulting samplings are not adequate for deriving thermodynamic observables, as shown by Sim et al. (3).
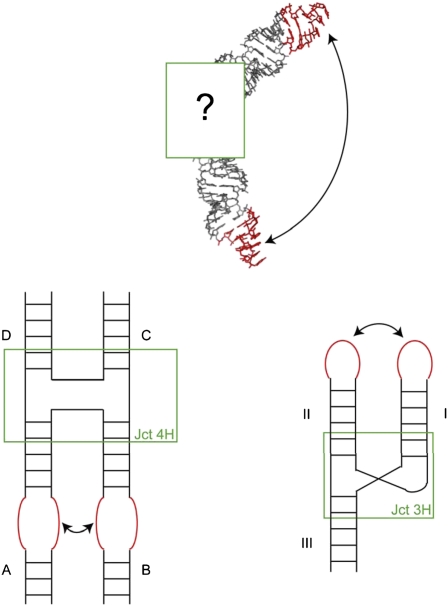
The power of the Sim et al. algorithm (3) stems from the facts that, first, it addresses these questions directly and, second, it offers an elegant and very natural solution anchored in the modular hierarchy of RNA architecture. Sim et al. develop an efficient sampling method that works at various hierarchical embedded levels. Thus, both global and local moves can be automatically performed. The inevitable loss in stereochemistry and covalent geometry is repaired afterward. The Hierarchical Natural Move Monte Carlo sampling can be used in conjunction with recent packages like Rosetta 3.0 (9) or MC-Sym (8). Canonical distributions of observables are produced whatever the starting structures. This latter advantage is particularly important for the design of nanostructures that rely on the assembly of RNA building blocks and modules (12, 13). Such large nanostructures are generally highly symmetric, at least globally but not at the local level (14), emphasizing the importance of being able to decouple global and local moves. Sim et al. (3) convincingly apply the method to four-way junctions. Junctions between helices constitute central RNA architectural elements in many RNAs (Fig. 1), and they are also key for building up RNA nanostructures.
Fig. 1.
Relative orientations of helices at multiple junctions are key for restricting the spatial ranges necessary to sample for correctly positioning the specific RNA–RNA interacting modules (Upper). For the four-way junction (Lower Left) there exist at least eight isomers [depending on the choice of the coaxially stacked helices (here is shown A with D and B with C), whether the strands cross or not at the junctions, and because the coaxial stacks are not parallel the resulting choice in the chirality of the two coaxial stacks (21)], and only one of them will allow the formation of a specific loop–loop contact. Three-way junctions (Lower Right) occur in three main families, with some conservation in the local junction folds [especially for one of the families, the most numerous one (22)], and generally a single conformation will allow for long-range contacts between two of the helical arms.
The recent increase in sophisticated automated algorithms for predicting secondary structure (15), detecting RNA 3D modules (16, 17), and modeling 3D structures of RNA (3, 8–11, 13, 18) is very
Sim et al. develop an efficient sampling method that works at various hierarchical embedded levels.
welcome by all those interested in the impact of RNA architecture on biological functions. RNA architecture, however, is the result of molecular Darwinian evolution and, consequently, contains various aspects of molecular contingencies coupled or not to the biological function of a particular RNA in a given cellular environment. One cannot expect that a single tool could manage such complexity. A recent blind experiment on undisclosed structures undertaken by eight groups (19) demonstrated the striking progress made in automatic modeling and, at the same time, the vast room for improvement that faces us. Further progress will be achieved through a concerted and hierarchical integration of the available tools so that the power of comparative sequence analysis can be coupled to knowledge-based databases and the highly efficient algorithms and sampling methods being developed. The boost obtained by mixing tools is illustrated by the work in PNAS (3) and, recently, by the work of others (20).
Footnotes
The author declares no conflict of interest.
See companion article on page 2890.
References
- 1.Martin F, et al. Cap-assisted internal initiation of translation of histone H4. Mol Cell. 2011;41:197–209. doi: 10.1016/j.molcel.2010.12.019. [DOI] [PubMed] [Google Scholar]
- 2.Breaker RR. Prospects for riboswitch discovery and analysis. Mol Cell. 2011;43:867–879. doi: 10.1016/j.molcel.2011.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sim AYL, Levitt M, Minary P. Modeling and design by hierarchical natural moves. Proc Natl Acad Sci USA. 2012;109:2890–2895. doi: 10.1073/pnas.1119918109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Butcher SE, Pyle AM. The molecular interactions that stabilize RNA tertiary structure: RNA motifs, patterns, and networks. Acc Chem Res. 2011;44:1302–1311. doi: 10.1021/ar200098t. [DOI] [PubMed] [Google Scholar]
- 5.Cruz JA, Westhof E. The dynamic landscapes of RNA architecture. Cell. 2009;136:604–609. doi: 10.1016/j.cell.2009.02.003. [DOI] [PubMed] [Google Scholar]
- 6.Westhof E, Masquida B, Jossinet F. Predicting and modeling RNA architecture. Cold Spring Harb Perspect Biol. 2011 doi: 10.1101/cshperspect.a003632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jossinet F, Ludwig TE, Westhof E. Assemble: An interactive graphical tool to analyze and build RNA architectures at the 2D and 3D levels. Bioinformatics. 2010;26:2057–2059. doi: 10.1093/bioinformatics/btq321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Parisien M, Major F. The MC-Fold and MC-Sym pipeline infers RNA structure from sequence data. Nature. 2008;452:51–55. doi: 10.1038/nature06684. [DOI] [PubMed] [Google Scholar]
- 9.Das R, Karanicolas J, Baker D. Atomic accuracy in predicting and designing noncanonical RNA structure. Nat Methods. 2010;7:291–294. doi: 10.1038/nmeth.1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sharma S, Ding F, Dokholyan NV. iFoldRNA: Three-dimensional RNA structure prediction and folding. Bioinformatics. 2008;24:1951–1952. doi: 10.1093/bioinformatics/btn328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cao S, Chen SJ. Physics-based de novo prediction of RNA 3D structures. J Phys Chem B. 2011;115:4216–4226. doi: 10.1021/jp112059y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chworos A, et al. Building programmable jigsaw puzzles with RNA. Science. 2004;306:2068–2072. doi: 10.1126/science.1104686. [DOI] [PubMed] [Google Scholar]
- 13.Kasprzak W, Bindewald E, Kim TJ, Jaeger L, Shapiro BA. Use of RNA structure flexibility data in nanostructure modeling. Methods. 2011;54:239–250. doi: 10.1016/j.ymeth.2010.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dibrov SM, McLean J, Parsons J, Hermann T. Self-assembling RNA square. Proc Natl Acad Sci USA. 2011;108:6405–6408. doi: 10.1073/pnas.1017999108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Deigan KE, Li TW, Mathews DH, Weeks KM. Accurate SHAPE-directed RNA structure determination. Proc Natl Acad Sci USA. 2009;106:97–102. doi: 10.1073/pnas.0806929106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cruz JA, Westhof E. Sequence-based identification of 3D structural modules in RNA with RMDetect. Nat Methods. 2011;8:513–521. doi: 10.1038/nmeth.1603. [DOI] [PubMed] [Google Scholar]
- 17.Sarver M, Zirbel CL, Stombaugh J, Mokdad A, Leontis NB. FR3D: Finding local and composite recurrent structural motifs in RNA 3D structures. J Math Biol. 2008;56:215–252. doi: 10.1007/s00285-007-0110-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rother M, Rother K, Puton T, Bujnicki JM. ModeRNA: A tool for comparative modeling of RNA 3D structure. Nucleic Acids Res. 2011;39:4007–4022. doi: 10.1093/nar/gkq1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cruz JA, et al. RNA-Puzzles: A CASP-like evaluation of RNA three-dimensional structure prediction. RNA. 2012 doi: 10.1261/rna.031054.111. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kladwang W, Chou FC, Das R. Automated RNA structure prediction uncovers a kink-turn linker in double glycine riboswitches. J Am Chem Soc. 2011 doi: 10.1021/ja2093508. [DOI] [PubMed] [Google Scholar]
- 21.Duckett DR, Murchie AI, Lilley DM. The global folding of four-way helical junctions in RNA, including that in U1 snRNA. Cell. 1995;83:1027–1036. doi: 10.1016/0092-8674(95)90218-x. [DOI] [PubMed] [Google Scholar]
- 22.Lescoute A, Westhof E. Topology of three-way junctions in folded RNAs. RNA. 2006;12:83–93. doi: 10.1261/rna.2208106. [DOI] [PMC free article] [PubMed] [Google Scholar]