Abstract
Human cytomegalovirus (HCMV) modulates numerous cellular signaling pathways. Alterations in signaling are evident from the broad changes in cellular phosphorylation that occur during HCMV infection and from the altered activity of multiple kinases. Here we report a comprehensive RNAi screen, which predicts that 106 cellular kinases influence growth of the virus, most of which were not previously linked to HCMV replication. Multiple elements of the AMP-activated protein kinase (AMPK) pathway scored in the screen. As a regulator of carbon and nucleotide metabolism, AMPK is poised to activate many of the metabolic pathways induced by HCMV infection. An AMPK inhibitor, compound C, blocked a substantial portion of HCMV-induced metabolic changes, inhibited the accumulation of all HCMV proteins tested, and markedly reduced the production of infectious progeny. We propose that HCMV requires AMPK or related activity for viral replication and reprogramming of cellular metabolism.
Keywords: herpesvirus, siRNA
Viruses are dependent on host cell signaling pathways for replication and spread. Infection with the prevalent β-herpes virus human cytomegalovirus (HCMV) induces increased levels of protein phosphorylation and markedly alters host cell signal transduction pathways (1). A portion of the phosphorylation changes induced by HCMV infection are attributed to the Ser/Thr kinase encoded by the viral genome, pUL97 (2), and others may derive from cellular kinase(s) packaged into virions (3) or from cellular kinases known to be activated by HCMV (4).
Here we sought to more completely delineate the effects of HCMV infection on kinase signaling by performing an siRNA screen of the entire cellular kinome. The screen identified 106 kinases predicted to influence the production of virus. The hits included the 5′-AMP–activated protein kinase (AMPK), a sensor of cellular energy homeostasis. AMPK is composed of three subunits: a catalytic subunit, AMPKα, and two regulatory subunits, AMPKβ and AMPKγ. Activation of the kinase requires cooperative AMP binding to AMPKγ, which occurs stochastically with shifts in the AMP:ATP ratio, and phosphorylation of AMPKα at Thr172 (5, 6). At least three different kinases are reported to phosphorylate Thr172 of AMPKα: Ca2+/calmodulin-dependent kinase kinase (CaMKK), TGF-β-activated kinase 1 (TAK1), and liver kinase B1 (LKB1) (7). Activated AMPK phosphorylates a number of substrates to effect changes in central carbon metabolism, lipid metabolism, physiological homeostasis, cell growth, apoptosis, and gene expression (5).
HCMV induces glycolysis (8–10) and also causes increased levels of the glucose transporter GLUT4 at the plasma membrane increasing glucose uptake (11).
AMPK controls GLUT4 relocalization to the plasma membrane (5), and this regulation likely links the kinase to altered metabolism in HCMV-infected cells. However, previous work indicates that pharmacological activation of AMPK during the early phase of HCMV infection can be deleterious to viral replication (12), yet CaMKK activity is required for HCMV replication (7). Thus, the connections between AMPK activity and metabolic changes during HCMV infection have remained unclear.
We confirmed the requirement for AMPK during infection, and we show that an AMPK antagonist, compound C, blocks HCMV-induced changes to glycolysis and inhibits viral gene expression. These studies argue that AMPK or a related, compound C-sensitive kinase is an essential contributor to metabolic changes initiated by HCMV and provide unique insight into potential antiviral strategies.
Results
Human Kinome Screen Identifies Putative Effectors of HCMV Replication.
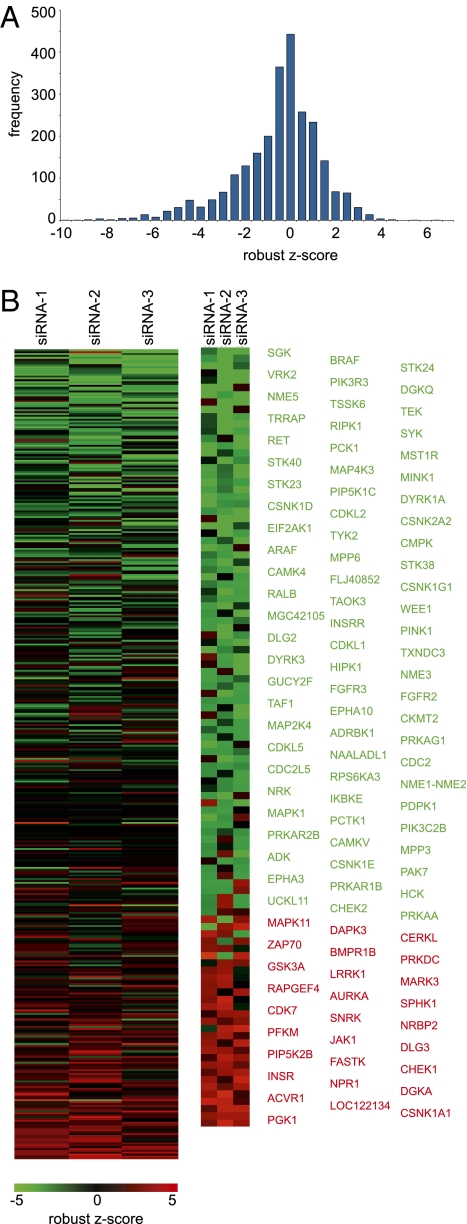
We conducted an siRNA screen of the human kinome to perform an unbiased search for effectors of HCMV replication (Fig. S1A). Three different siRNAs specific for each of 714 human kinases, kinase regulatory subunits, and hypothetical kinases, routinely achieved >98% transfection efficiency in fibroblasts. At 24 h posttransfection, cultures were infected with HCMV [0.1 infectious units (IU) per cell], and 96 h later, supernatants were harvested and assayed for virus. This schedule is designed so that the cellular kinase knockdown will be greatest during the peak period of viral replication and egress, and it permits detection of defects at any stage of the HCMV replication cycle. Each experiment included siRNA to GFP, with no effect on HCMV yield, and to the immediate-early viral gene product IE2, which reduced the yield by a factor of ≥100, as controls (Fig. S1B). The yield of virus in each sample was normalized to the plate median, log2 transformed, and a robust z score was calculated (13). The normalization strategy set the median robust z score at 0, with a median absolute deviation (MAD) of 1 (Fig. 1A).
Fig. 1.
Human kinome screen identifies candidate effectors of HCMV replication. (A) Yield of infectious HCMV following transfection of each siRNA was assayed, normalized by plate, and converted to a robust z score. These robust z scores were grouped into bins of 0.5 units and plotted as a histogram to show the range of robust z scores. (B) Robust z scores for identified hits were converted to a heat map for each of the three siRNAs tested for each kinase. Log2 scale ranges from green (decreased yield of HCMV) to red (increased yield). (Right) Enlarged view of candidate hits, which are identified at Far Right. See Fig. S2 for a blue-yellow version of this panel.
Kinases were considered hits if at least two of the three siRNAs altered the virus yield by ≥2 MAD; that is, a robust z score of ≥2 or ≤ −2. Using these criteria, we observed a false discovery rate of 2.6%, based on the spurious identification of the control siRNA as a hit. Our screen identified 77 kinases (10.7% of those screened) whose knockdown impaired HCMV replication and 29 (4.1% of those screened) whose knockdown increased the yield of infectivity (Fig. 1B and Fig. S2 and Dataset S1). The hits included eight kinases for which all three siRNAs tested gave significant effects on HCMV replication: knockdown of CSNK1A increased yield of HCMV, whereas targeting TAF1, PCK1, NME5, DYRK1A, CSNK1D, CDKL2, CDC2L5, or WEE1 decreased virus replication. Our list of hits included cyclin-dependent kinases (CDKs), multiple members of the extracellular signal-related kinase (ERK1/2) signaling pathway, and kinases regulating translation (including EIF2AK1 and RPS6KA3); each of these has previously been linked to HCMV replication (12, 14–21). Importantly, a role in the HCMV replication cycle has not been confirmed for the majority of the kinases identified in this screen. The hits, therefore, comprise kinases of potential importance for HCMV replication and spread.
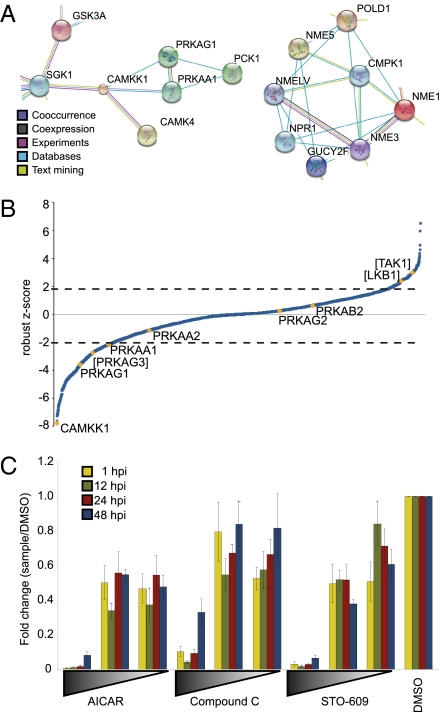
Hits predicted to influence HCMV replication were analyzed to identify signaling pathways and classified on the basis of known kinase families (22, 23), revealing an enrichment for kinases that regulate aspects of cellular metabolism and major cellular signaling pathways (Table S1). The identified kinases were also clustered into functional networks (24) (Fig. 2A and Fig. S3). A distinct cluster involved nucleotide diphosphate kinase family members NME1–NME2, NME3, and NME5, all of which were identified as hits whose knockdown decreases the yield of HCMV (Fig. 2A, Right). Nucleotide diphosphate kinases transfer a phosphate group from a nucleoside triphosphate to a nucleoside diphosphate, e.g., from GTP to ADP to yield GDP and ATP. Epstein-Barr virus modulates NME1 activity during infection (25). A second cluster focused on AMPK, the upstream kinase CAMKK, and downstream metabolic effectors (Fig. 2A, Left). Given that this cluster related to our recent exploration of glycolytic and tricarboxylic acid cycle (TCA) cycle changes during HCMV infection (8, 10, 26), we chose this avenue for further exploration.
Fig. 2.
Replication of HCMV is impaired by altered AMPK activity. (A) Clusters of hits related to AMPK (Left) and nucleotide metabolism (Right) were identified from a STRING analysis of the kinase hits. Connecting lines are color coded by the type of evidence used to build the cluster. For full analysis, see Fig. S3. (B) siRNA targeting AMPK-related subunits was assayed for effects on HCMV replication. The siRNA result with the greatest absolute difference from zero for each triplicate is plotted against the distribution of robust z scores for the entire kinome screen. Genes contained within brackets did not meet our criteria for inclusion as a hit; in these cases only one of the three tested siRNAs produced a robust z score >2 or <2. (C) Confluent, serum-starved fibroblasts were infected with HCMV at a multiplicity of 0.1 IU per cell and treated with AICAR (0.01–1 mM), compound C (0.2–20 μM), STO-609 (0.1–10 μg/mL), or DMSO alone (drug vehicle) at different times postinfection. Yield of infectious virus was assayed and normalized to DMSO control.
AMPK Pathway and Lipid Kinases Are Enriched Among Hits.
Multiple siRNAs targeting the α-1 isoform of the AMPKα catalytic subunit, PRKAA1, or the γ-1 isoform of the AMPKγ subunit, PRKAG1, decreased the yield of infectious HCMV (Figs. 1B and 2B). One siRNA targeting the γ-3 isoform of AMPKγ, PRKAG3, also caused a significant reduction in virus yield (Fig. 2B), but the other two siRNAs targeting this isoform did not score and consequently PRKAG3 did not meet our criteria for inclusion as a hit. Other AMPK subunits were not identified in the screen, perhaps due to tissue specificity of expression or redundancy (5, 27) or to less robust knockdown by siRNAs. Activation of AMPK function requires both AMP binding to the γ-subunit and phosphorylation of the regulatory Thr-172 residue on the α-subunit (5). Three kinases are known to phosphorylate Thr172 of AMPKα: CaMKK, LKB1, and TAK1, and it has been speculated that there may be other activators as well (5). Our screen identified CaMKKα (CAMKK1) as a kinase required for HCMV replication (Figs. 1B and 2B). This is consistent with a recent report that pharmacological inhibition of CaMKK impedes HCMV replication (7). In contrast, knockdown of LKB1 and TAK1 did not meet our criteria to be considered hits (Fig. 2B).
The screen also identified substrates of AMPK and downstream effectors. These downstream effectors included glycogen synthase kinase (GSK3A), PI3K family members, and one isoform of phosphofructokinase (PFKM) (Dataset S1).
Compound C Inhibits the Production of HCMV Progeny.
To further verify our siRNA results, we turned to pharmacological modulators of AMPK activity. Serum-starved, confluent fibroblasts were infected with HCMV (0.1 IU per cell) and treated with either the AMPK activator AICAR, the AMPK inhibitor compound C, or the CaMKK inhibitor STO-609. At various times after infection, supernatants were harvested and assayed for infectious virus (Fig. 2C). Treatment with compound C inhibited HCMV replication in a dose-dependent manner, in agreement with our PRKAA1 and PRKAG1 siRNA hits. STO-609 inhibited HCMV replication, consistent with previous results showing that CaMKK, an AMPK-activating kinase, is essential to HCMV (7). AICAR-mediated activation of AMPK also inhibited HCMV replication in a dose-dependent manner, as has been reported previously (12). The observation that either an AMPK activator or inhibitor impairs HCMV replication could mean that a precise level or specific subcellular localization of AMPK activation by CaMKK is essential to HCMV replication or that AMPK must be down-regulated at some stages of the viral replication cycle, and activated at others. Alternatively, it could result from off-target effects.
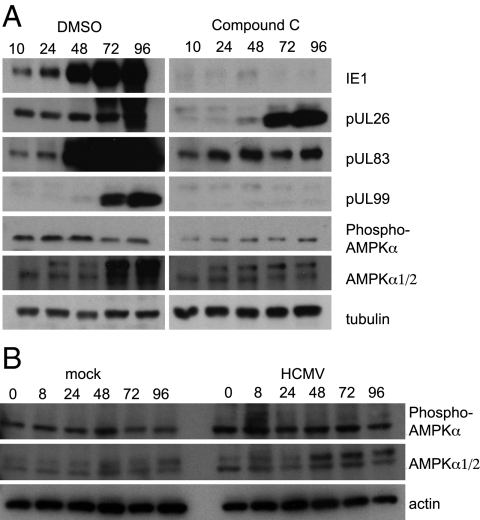
Maximal AMPK activity requires two steps of activation: AMP binding to AMPKγ and phosphorylation of AMPKα residue T172 (5). ATP inhibits phosphorylation of AMPK (28), and compound C is an ATP-competitive inhibitor of AMPK. To confirm that compound C altered AMPK phosphorylation, we monitored phospho-T172 AMPKα and total AMKPα levels in HCMV-infected cells treated with vehicle (DMSO) or compound C (Fig. 3A). In the absence of drug, total AMPKα levels increased, whereas phospho-T172 AMPKα levels decreased during the late phase of infection. This phenomenon could be caused by any combination of (i) decreased phosphorylation by LKB1, TAK1, CAMKK or other activating kinases; (ii) increased dephosphorylation by known effectors; and/or (iii) lack of accessible pools of AMP (as AMP binding promotes phosphorylation of AMPK) (5). At present we are unable to differentiate between these possibilities. In the presence of compound C, phospho-T172 AMPKα levels were reduced at all times assayed, and total AMPKα levels remained constant. To determine whether the increase in AMPK levels late after infection depends on the length of time that cells are maintained in stationary phase, uninfected cells were assayed for total and phospho-T172 AMPKα (Fig. 3B). Importantly, total AMPKα was unchanged in the uninfected cells, indicating that the increase is specific to infection.
Fig. 3.
HCMV infection modestly increases total AMPK but does not change the amount of phospho-T172 AMPK. (A) Western blot assays for expression of HCMV proteins IE1, pUL26, pUL83, and pUL99 and cellular total AMPKα and phospho-T172 AMPKα in fibroblasts infected with HCMV (3 IU per cell) and treated with compound C (20 μM) or DMSO (drug vehicle). (B) Western blot assays for total AMPKα and phospho-T172 AMPKα in mock-infected or HCMV-infected fibroblasts maintained in culture under conditions as in A.
To evaluate the effects of compound C on HCMV protein expression, we used the same set of cell lysates to assay the steady state levels of several HCMV proteins representing different classes of protein expression: immediate-early (IE1), early (pUL26), and late (pUL83 and pUL99) (Fig. 3A). Compound C instituted a block at the start of the viral replication cycle, blocking or delaying accumulation of each viral protein tested.
Compound C Blocks HCMV-Mediated Remodeling of the Metabolome.
Previous studies have demonstrated dramatic metabolic changes in HCMV-infected cells (8–10, 26), and AMPK is a master regulator of multiple steps in metabolism and glucose utilization (5, 29). We thus examined the effects of compound C on metabolites in fibroblasts that were either mock infected or HCMV infected by using liquid chromatography coupled to high-resolution mass spectrometry (Fig. 4 and Fig. S4).
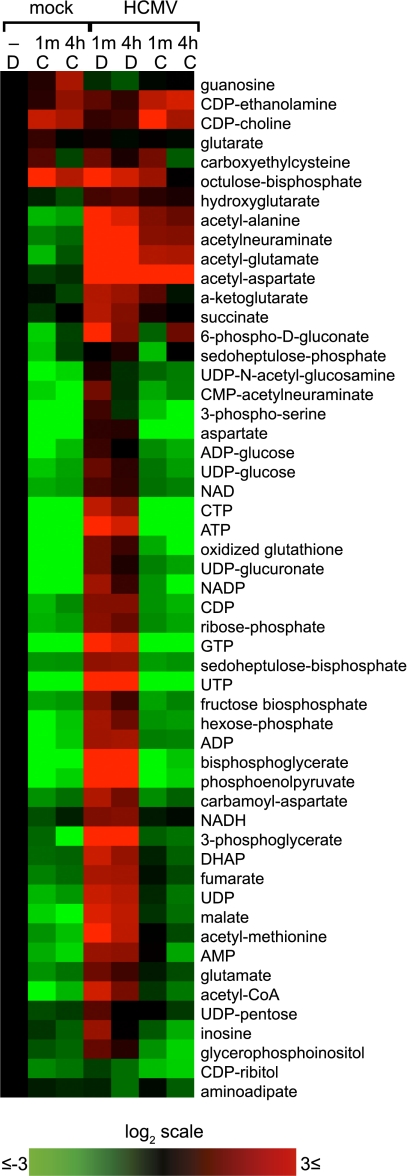
Fig. 4.
Compound C blocks establishment of the infected cell metabolome. (A) Cells were treated with compound C, (C, 20 μM) or DMSO alone (D, drug vehicle) beginning at 1 h after infection (3 IU per cell) or mock infection, and metabolites were assayed at 48 hpi. Heat map of drug-induced changes in metabolite pools is shown. Duplicate samples are shown, each normalized to mock-infected, DMSO-treated cells (Left column). Log2 scale of fold changes is shown. To view the same figure in blue-yellow color scale, see Fig. S4.
In mock-infected fibroblasts, compound C caused a massive depletion of most metabolites, including amino acids, nucleotides, glycolytic intermediates, and pentose phosphate pathway intermediates. This suggests a strong reliance of quiescent fibroblasts on constitutive AMPK (or related compound C-sensitive kinase) activity for metabolic homeostasis. Next, infected cultures were treated with vehicle control (DMSO) or compound C starting at 1 h postinfection (hpi), and metabolites were analyzed at 48 hpi. As previously observed (8, 10, 26), HCMV infection increased the levels of intermediates in glycolysis, pyrimidine biosynthesis, and the TCA cycle, as well as nucleotides and acetylated amino acids (Fig. 4). The effects of compound C were profound, with most of the metabolome changes due to HCMV reversed in the presence of compound C. One exception involved cytidine diphosphate (CDP)-ethanolamine and CDP-choline, which are used in the Kennedy pathway of phosphatidylcholine and phosphatidylethanolamine synthesis and remained elevated in the HCMV-infected, compound C-treated cells. Phosphatidylethanolamines are strongly enriched in HCMV virions (30). Another involved N-acetylated amino acids (e.g., acetylaspartate, acetylglutamate, and acetylalanine). The mechanism by which acetylated amino acid levels are induced and their possible role in infection are not yet clear.
HCMV induces altered metabolite fluxes in addition to increased metabolite levels (26). To examine potential flux changes induced by CpdC, we traced the incorporation of carbons from U-13C-glucose into cellular metabolites (Fig. S5A). These experiments involved a very short labeling (1 min) to probe initial rates of labeling of glycolytic intermediates; capturing the initial labeling rate before isotopic steady state, which occurs at ∼2 min for glycolytic intermediates (10), is essential to making inferences about metabolic flux. In addition, the experiments included a later time point (4 h) to characterize slower labeling events, particularly those in the TCA cycle. As reported previously, HCMV infection markedly accelerated the initial rate of labeling of fructose-1,6-bisphosphate (FBP) and dihydroxyacetone phosphate (DHAP), as measured by the labeled pool size 1 min after shifting into U-13C-glucose. This initial labeling rate is a reliable indicator of metabolic flux, and the two typically covary linearly (26). Compound C treatment of uninfected or HCMV-infected cells significantly decreased this rapid, presteady-state incorporation of labeled carbon into FBP and DHAP (Fig. S5 B and C), indicating that the decreased size of metabolite pools is correlated with decreased glycolytic activity.
To examine carbon flow from glycolysis to the TCA cycle, we monitored the accumulation of 13C-label from glucose to acetyl-CoA, which then feeds the TCA cycle by donating its two carbon atoms to oxaloacetate to form citrate, which subsequently cycles to malate (Fig. S5A). After a 4-h labeling period with U-13C-glucose, HCMV infection increased labeling of both malate (Fig. S5D) and acetyl-CoA (Fig. S5E), as reported previously (26), and compound C blocked these increases.
In uninfected fibroblasts, carbons from glucose predominantly enter the TCA cycle via pyruvate carboxylase-dependent conversion of pyruvate to oxaloacetate. This is evidenced by incorporation of three 13C labels from glucose into malate (Fig. S5A, pathway on Right). HCMV infection induces a switch toward increased carbon entry through conversion of acetyl-CoA to citrate (8, 26), which results in the incorporation of two 13C-labeled carbons from glucose into malate (Fig. S5A, pathway on Left). Compound C strongly blocked both routes of TCA cycle entry in both mock and HCMV-infected cells (Fig. S5D).
These results are consistent with the conclusion that AMPK activity is required for a substantial subset of the metabolic changes induced during HCMV infection, including enhanced carbon flux through upper glycolysis and the glycolytic–TCA cycle interface.
Discussion
We have performed an siRNA screen to identify cellular kinases that modulate the production of HCMV progeny in fibroblasts (Fig. S1) and focused on a cluster of hits related to AMPK signaling (Fig. 2A). siRNAs to either AMPKα (PRKAA1) or AMPKγ (PRKAG1) reduced the production of infectious HCMV progeny (Fig. 2B). The AMPK antagonist, compound C, inhibited the yield of HCMV in a dose-dependent manner (Fig. 2C), consistent with a role for AMPK in the HCMV replication cycle. The drug markedly inhibited the accumulation of the immediate-early IE1 protein (Fig. 3A), arguing that AMPK activity might be needed at the start of infection. Compound C also interfered with normal accumulation of early and late viral proteins, but this result is difficult to interpret because expression of the mRNAs encoding these proteins is dependent on IE1 protein (26). Compound C blocked many of the alterations to core metabolism that have been described for HCMV (Fig. 4 and Fig. S5). As for most kinase inhibitors, compound C inhibits multiple cellular kinases (31), and thus, results with the drug must be interpreted with caution. Nevertheless, the drug data, together with the fact that siRNAs to AMPK inhibit HCMV replication, are consistent with the view that HCMV remodels core metabolism through a mechanism that depends on AMPK or related kinase(s). This possibility is reinforced by the requirement for CaMKK, an AMPK activator, to generate optimal yields of HCMV (Fig. 2 B and C) and to elevate glycolysis after infection (7) and by the earlier observation that GLUT4, which is induced by AMPK (5), is substantially elevated after infection (11).
The mechanism of CaMKK activation during HCMV replication has not been identified, but may hinge upon the release of calcium stores into the cytosol that occurs in HCMV-infected cells in response to the viral immediate-early protein, pUL37x1 (32). Another AMPK activator may also be involved in maintaining an active AMPK pool during HCMV infection. In support of this possibility, treatment with STO-609 resulted in a less severe phenotype of viral protein expression than did treatment with compound C. Of note, neither LKB1 nor TAK1, the other established activators of AMPK via T172 phosphorylation, was identified as hits in the siRNA kinome screen. It remains possible that these kinases act redundantly in HCMV replication and thus were not identified by the screen. Alternatively, additional activators of AMPK may remain to be identified, or the cell type and growth conditions may alter the sensitivity of AMPK to AMP:ATP (33), or the increases in AMP that occur during HCMV infection may activate AMPK, despite ATP also increasing. Finally, it is possible that HCMV modulates the cycle of AMPK dephosphorylation to maintain specific levels of AMPK activity.
AMPK activity favors HCMV replication by phosphorylating multiple substrates that switch on catabolic pathways producing ATP, as is evident in Fig. 4, where compound C reduced ATP levels. In this regard, AMPK induces glucose uptake, providing fuel for glycolysis and the TCA cycle. However, activated AMPK also phosphorylates substrates that block ATP-consuming anabolic pathways. For example, active AMPK inhibits acetyl-CoA carboxylase (ACC), blocking fatty acid biosynthesis, and it activates the tuberous sclerosis protein complex (TSC1/2) to decrease mTOR signaling, disfavoring cell growth. Each of these alterations is exactly opposite to what is needed for HCMV replication (12, 34–38). How is active AMPK decoupled from some of its downstream substrates? The HCMV pUL38 protein binds to TSC2 and prevents TSC1/2 from responding to AMPK phosphorylation (39). Although ACC is induced and required for HCMV replication (34), the mechanism by which it is protected from inactivation by AMPK after infection is unknown. It has been suggested that distinct thresholds of AMPK activity are required for activity toward each of its different substrates (40). Specifically, phosphorylation of TSC1/2 by AMPK may require a higher level of AMPK activity than does AMPK activation of glucose transport and increased ATP production (40). Such a possibility is consistent with AICAR inhibitory effects on HCMV replication: overactive AMPK might lead to inhibition of ACC or mTOR. Additionally, it has been speculated that distinct subcellular pools of AMPK or perhaps changes to the composition of the heterotrimer subunits could play a role (6). Indeed, other viruses use a number of strategies to modulate AMPK activity and substrate selection (41). It is noteworthy that AMPK activity and substrates are modulated in other viruses (41). We therefore speculate that HCMV replication may use multiple mechanisms for directing the activity of AMPK.
In conclusion, a comprehensive RNAi kinome screen has identified many new potential effectors of the HCMV replication cycle. Two AMPK subunits were identified in the screen, providing a framework for interpretation of the virus’ ability to institute profound changes in cellular metabolism.
Methods
Cells, Virus, and Drugs.
Human MRC5 fibroblasts (ATCC) were maintained in DMEM with 4.5 g/L glucose (Sigma-Aldrich) supplemented with 10% FBS. BADwt (42), derived from HCMV strain AD169, was used in this study. Virus titers were determined by TCID50 assay. N1-(β-d-Ribofuranosyl)-5-aminoimidazole-4-carboxamide, AICAR (Tocris Bioscience), 6-[4-(2-piperidin-1-yl-ethoxy)-phenyl]-3-pyridin-4-yl-pyrazolo[1,5-a] pyrimidine, compound C (Calbiochem), and 7-Oxo-7H-benzimidazo[2,1-a]benz[de]isoquinoline-3-carboxylic acid acetate, STO-609 (Tocris Bioscience) were dissolved in DMSO and used at indicated concentrations.
siRNA Screen.
The screen is detailed in Fig. S1A (design of diagram modeled after ref. 43). Fibroblasts were cultured in 96-well dishes to 70–75% confluence and transfected with siRNA (0.1 nM) from the Mission siRNA Human Kinase Panel (Sigma-Aldrich) using oligofectamine (Invitrogen) following the manufacturer's protocol. siRNA against IE2, 5′-AAACGCAUCUCCGAGUUGGAC[dT][dT]-3′ (44), or GFP, 5′- GCAAGCUGACCCUGAAGUUCAU[dT][dT]-3′, were used as positive and negative controls, respectively. After incubation in medium containing 10% FBS for 24 h, cells were infected with HCMV (0.1 IU per cell), and 96 h later, supernatants were assayed for infectious virus (45) by using the Operetta High-Content Imaging system (Perkin-Elmer) or a Nikon Eclipse Ti microscopy system with NIS-Elements AR software, and ImageJ analysis (46) to image 1,500 cells per siRNA. We detected higher yields of HCMV in the edge rows of each plate, as reported in other multiwell screens (13). To minimize this problem, we relocated edge siRNAs to the center of another plate for analysis. Cytotoxicity of siRNAs targeting AMPKα subunits was assessed by trypan blue staining and counting cells at 72 h posttransfection. Transfection of negative control GFP siRNA gave 6.2 ± 7.1% dead cells, whereas the PRKAA1 siRNAs gave 10.1 ± 4.0 dead cells. These results were not statistically different (P = 0.367).
All P values were calculated by two-tailed, nonpaired t test. Robust z scores were calculated as described (13) with normalization to plate median values: robust z = [log2 (sample/plate median) − median log2 ratio]/median absolute deviation.
Western Blot Analysis.
Protein samples were harvested in buffer with 1× protease inhibitor (Roche, EDTA-free tablets) and phosphatase inhibitors (1 mM sodium fluoride; 1 mM β-glycerophosphate; 1 mM sodium orthovanadate; 10 mM sodium pyrophosphate), and analyzed by Western blot (3). Primary antibodies were mouse monoclonal anti–α-tubulin (Sigma-Aldrich), HRP-conjugated mouse monoclonal antiactin (AbCam), mouse monoclonal anti-IE1 (1B12) (45), mouse monoclonal anti-pUL99 (10B4-29) (47), mouse monoclonal anti-pUL83 (8F5) (48), mouse monoclonal anti-pUL26 (3), rabbit anti-AMPKα (Cell Signaling), and rabbit monoclonal antiphospho-T172 AMPKα (Cell Signaling). Secondary antibodies were HRP-conjugated goat antimouse and goat antirabbit (Jackson ImmunoResearch).
Analysis of Metabolites.
For glucose labeling and measurement of metabolite pools, fibroblasts were cultured to confluence in DMEM with 10% FBS and then maintained in serum-free DMEM. Cells were infected (3 IU per cell) and treated with DMSO or compound C (20 μM) at 1 hpi. At 46 hpi, 2 h before labeling, the medium and drugs on each plate were replaced with fresh serum-free DMEM. At the start of the experiment, cells received fresh medium containing U-13C-glucose. After labeling, metabolites were extracted, dried, and resuspended as previously described (26, 49). Metabolites were separated by liquid chromatography and analyzed using liquid chromatography coupled to an Exactive Orbitrap mass spectrometer (Thermo Fisher Scientific) according to established parameters (50). Compounds were identified on the basis of retention time and exact mass, measured to <2 ppm mass accuracy. Data were analyzed using the metabolomic analysis and visualization engine (51). Total metabolite pool sizes were calculated from the sum of isotopic forms observed.
Supplementary Material
Acknowledgments
We thank E. Koyuncu for sharing details of his design of the siRNA screening protocol, and we gratefully acknowledge critical comments and technical advice from S. Grady, J. Hwang, and J. Purdy. L.J.T. is supported by American Cancer Society Postdoctoral Fellowship PF-11-087-01-MPC. This work was supported by National Institutes of Health Grants CA82396 and AI78063.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1200494109/-/DCSupplemental.
References
- 1.Mocarski ES, Shenk T, Pass RF. Cytomegaloviruses. In: Knipe DM, Howley PM, editors. Fields Virology. Philadelphia: Lippincott Williams and Wilkins; 2007. pp. 2701–2772. [Google Scholar]
- 2.Prichard MN. Function of human cytomegalovirus UL97 kinase in viral infection and its inhibition by maribavir. Rev Med Virol. 2009;19:215–229. doi: 10.1002/rmv.615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Munger J, Yu D, Shenk T. UL26-deficient human cytomegalovirus produces virions with hypophosphorylated pp28 tegument protein that is unstable within newly infected cells. J Virol. 2006;80:3541–3548. doi: 10.1128/JVI.80.7.3541-3548.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yurochko AD. Human cytomegalovirus modulation of signal transduction. Curr Top Microbiol Immunol. 2008;325:205–220. doi: 10.1007/978-3-540-77349-8_12. [DOI] [PubMed] [Google Scholar]
- 5.Hardie DG. AMP-activated/SNF1 protein kinases: Conserved guardians of cellular energy. Nat Rev Mol Cell Biol. 2007;8:774–785. doi: 10.1038/nrm2249. [DOI] [PubMed] [Google Scholar]
- 6.Cantó C, Auwerx J. AMP-activated protein kinase and its downstream transcriptional pathways. Cell Mol Life Sci. 2010;67:3407–3423. doi: 10.1007/s00018-010-0454-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McArdle J, Schafer XL, Munger J. Inhibition of calmodulin-dependent kinase kinase blocks human cytomegalovirus-induced glycolytic activation and severely attenuates production of viral progeny. J Virol. 2011;85:705–714. doi: 10.1128/JVI.01557-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Munger J, et al. Systems-level metabolic flux profiling identifies fatty acid synthesis as a target for antiviral therapy. Nat Biotechnol. 2008;26:1179–1186. doi: 10.1038/nbt.1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chambers JW, Maguire TG, Alwine JC. Glutamine metabolism is essential for human cytomegalovirus infection. J Virol. 2010;84:1867–1873. doi: 10.1128/JVI.02123-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Munger J, Bajad SU, Coller HA, Shenk T, Rabinowitz JD. Dynamics of the cellular metabolome during human cytomegalovirus infection. PLoS Pathog. 2006;2:e132. doi: 10.1371/journal.ppat.0020132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yu Y, Maguire TG, Alwine JC. Human cytomegalovirus activates glucose transporter 4 expression to increase glucose uptake during infection. J Virol. 2011;85:1573–1580. doi: 10.1128/JVI.01967-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kudchodkar SB, Del Prete GQ, Maguire TG, Alwine JC. AMPK-mediated inhibition of mTOR kinase is circumvented during immediate-early times of human cytomegalovirus infection. J Virol. 2007;81:3649–3651. doi: 10.1128/JVI.02079-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Birmingham A, et al. Statistical methods for analysis of high-throughput RNA interference screens. Nat Methods. 2009;6:569–575. doi: 10.1038/nmeth.1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Johnson RA, Ma XL, Yurochko AD, Huang ES. The role of MKK1/2 kinase activity in human cytomegalovirus infection. J Gen Virol. 2001;82:493–497. doi: 10.1099/0022-1317-82-3-493. [DOI] [PubMed] [Google Scholar]
- 15.Johnson RA, Wang X, Ma XL, Huong SM, Huang ES. Human cytomegalovirus up-regulates the phosphatidylinositol 3-kinase (PI3-K) pathway: Inhibition of PI3-K activity inhibits viral replication and virus-induced signaling. J Virol. 2001;75:6022–6032. doi: 10.1128/JVI.75.13.6022-6032.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Johnson RA, Huong SM, Huang ES. Activation of the mitogen-activated protein kinase p38 by human cytomegalovirus infection through two distinct pathways: A novel mechanism for activation of p38. J Virol. 2000;74:1158–1167. doi: 10.1128/jvi.74.3.1158-1167.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen J, Stinski MF. Role of regulatory elements and the MAPK/ERK or p38 MAPK pathways for activation of human cytomegalovirus gene expression. J Virol. 2002;76:4873–4885. doi: 10.1128/JVI.76.10.4873-4885.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sun B, et al. Modulation of human cytomegalovirus immediate-early gene enhancer by mitogen-activated protein kinase kinase kinase-1. J Cell Biochem. 2001;83:563–573. doi: 10.1002/jcb.1251. [DOI] [PubMed] [Google Scholar]
- 19.Hertel L, Chou S, Mocarski ES. Viral and cell cycle-regulated kinases in cytomegalovirus-induced pseudomitosis and replication. PLoS Pathog. 2007;3:e6. doi: 10.1371/journal.ppat.0030006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Milbradt J, Auerochs S, Marschall M. Cytomegaloviral proteins pUL50 and pUL53 are associated with the nuclear lamina and interact with cellular protein kinase C. J Gen Virol. 2007;88:2642–2650. doi: 10.1099/vir.0.82924-0. [DOI] [PubMed] [Google Scholar]
- 21.Muranyi W, Haas J, Wagner M, Krohne G, Koszinowski UH. Cytomegalovirus recruitment of cellular kinases to dissolve the nuclear lamina. Science. 2002;297:854–857. doi: 10.1126/science.1071506. [DOI] [PubMed] [Google Scholar]
- 22.Manning G, Whyte DB, Martinez R, Hunter T, Sudarsanam S. The protein kinase complement of the human genome. Science. 2002;298:1912–1934. doi: 10.1126/science.1075762. [DOI] [PubMed] [Google Scholar]
- 23.Huang W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 24.Jensen LJ, et al. STRING 8—a global view on proteins and their functional interactions in 630 organisms. Nucleic Acids Res. 2009;37(Database issue):D412–D416. doi: 10.1093/nar/gkn760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Murakami M, Kaul R, Kumar P, Robertson ES. Nucleoside diphosphate kinase/Nm23 and Epstein-Barr virus. Mol Cell Biochem. 2009;329:131–139. doi: 10.1007/s11010-009-0123-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vastag L, Koyuncu E, Grady SL, Shenk TE, Rabinowitz JD. Divergent effects of human cytomegalovirus and herpes simplex virus-1 on cellular metabolism. PLoS Pathog. 2011;7:e1002124. doi: 10.1371/journal.ppat.1002124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Steinberg GR, Kemp BE. AMPK in Health and Disease. Physiol Rev. 2009;89:1025–1078. doi: 10.1152/physrev.00011.2008. [DOI] [PubMed] [Google Scholar]
- 28.Davies SP, Helps NR, Cohen PT, Hardie DG. 5′-AMP inhibits dephosphorylation, as well as promoting phosphorylation, of the AMP-activated protein kinase. Studies using bacterially expressed human protein phosphatase-2C alpha and native bovine protein phosphatase-2AC. FEBS Lett. 1995;377:421–425. doi: 10.1016/0014-5793(95)01368-7. [DOI] [PubMed] [Google Scholar]
- 29.Cantó C, Auwerx J. PGC-1alpha, SIRT1 and AMPK, an energy sensing network that controls energy expenditure. Curr Opin Lipidol. 2009;20:98–105. doi: 10.1097/MOL.0b013e328328d0a4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu ST, et al. Synaptic vesicle-like lipidome of human cytomegalovirus virions reveals a role for SNARE machinery in virion egress. Proc Natl Acad Sci USA. 2011;108:12869–12874. doi: 10.1073/pnas.1109796108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bain J, et al. The selectivity of protein kinase inhibitors: A further update. Biochem J. 2007;408:297–315. doi: 10.1042/BJ20070797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sharon-Friling R, Goodhouse J, Colberg-Poley AM, Shenk T. Human cytomegalovirus pUL37x1 induces the release of endoplasmic reticulum calcium stores. Proc Natl Acad Sci USA. 2006;103:19117–19122. doi: 10.1073/pnas.0609353103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mantovani J, Roy R. Re-evaluating the general(ized) roles of AMPK in cellular metabolism. FEBS Lett. 2011;585:967–972. doi: 10.1016/j.febslet.2010.12.015. [DOI] [PubMed] [Google Scholar]
- 34.Spencer CM, Schafer XL, Moorman NJ, Munger J. Human cytomegalovirus induces the activity and expression of acetyl-coenzyme A carboxylase, a fatty acid biosynthetic enzyme whose inhibition attenuates viral replication. J Virol. 2011;85:5814–5824. doi: 10.1128/JVI.02630-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Moorman NJ, Shenk T. Rapamycin-resistant mTORC1 kinase activity is required for herpesvirus replication. J Virol. 2010;84:5260–5269. doi: 10.1128/JVI.02733-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Clippinger AJ, Maguire TG, Alwine JC. Human cytomegalovirus infection maintains mTOR activity and its perinuclear localization during amino acid deprivation. J Virol. 2011;85:9369–9376. doi: 10.1128/JVI.05102-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Clippinger AJ, Maguire TG, Alwine JC. The changing role of mTOR kinase in the maintenance of protein synthesis during human cytomegalovirus infection. J Virol. 2011;85:3930–3939. doi: 10.1128/JVI.01913-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Buchkovich NJ, Yu Y, Zampieri CA, Alwine JC. The TORrid affairs of viruses: effects of mammalian DNA viruses on the PI3K-Akt-mTOR signalling pathway. Nat Rev Microbiol. 2008;6:266–275. doi: 10.1038/nrmicro1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Moorman NJ, et al. Human cytomegalovirus protein UL38 inhibits host cell stress responses by antagonizing the tuberous sclerosis protein complex. Cell Host Microbe. 2008;3:253–262. doi: 10.1016/j.chom.2008.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Alwine JC. Modulation of host cell stress responses by human cytomegalovirus. Curr Top Microbiol Immunol. 2008;325:263–279. doi: 10.1007/978-3-540-77349-8_15. [DOI] [PubMed] [Google Scholar]
- 41.Mankouri J, Harris M. Viruses and the fuel sensor: The emerging link between AMPK and virus replication. Rev Med Virol. 2011;21:205–212. doi: 10.1002/rmv.687. [DOI] [PubMed] [Google Scholar]
- 42.Yu D, Smith GA, Enquist LW, Shenk T. Construction of a self-excisable bacterial artificial chromosome containing the human cytomegalovirus genome and mutagenesis of the diploid TRL/IRL13 gene. J Virol. 2002;76:2316–2328. doi: 10.1128/jvi.76.5.2316-2328.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Brass AL, et al. Identification of host proteins required for HIV infection through a functional genomic screen. Science. 2008;319:921–926. doi: 10.1126/science.1152725. [DOI] [PubMed] [Google Scholar]
- 44.Wiebusch L, Truss M, Hagemeier C. Inhibition of human cytomegalovirus replication by small interfering RNAs. J Gen Virol. 2004;85:179–184. doi: 10.1099/vir.0.19453-0. [DOI] [PubMed] [Google Scholar]
- 45.Zhu H, Shen Y, Shenk T. Human cytomegalovirus IE1 and IE2 proteins block apoptosis. J Virol. 1995;69:7960–7970. doi: 10.1128/jvi.69.12.7960-7970.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Collins TJ. ImageJ for microscopy. Biotechniques. 2007;43(1, Suppl):25–30. doi: 10.2144/000112517. [DOI] [PubMed] [Google Scholar]
- 47.Silva MC, Yu Q-C, Enquist L, Shenk T. Human cytomegalovirus UL99-encoded pp28 is required for the cytoplasmic envelopment of tegument-associated capsids. J Virol. 2003;77:10594–10605. doi: 10.1128/JVI.77.19.10594-10605.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nowak B, et al. Characterization of monoclonal antibodies and polyclonal immune sera directed against human cytomegalovirus virion proteins. Virology. 1984;132:325–338. doi: 10.1016/0042-6822(84)90039-4. [DOI] [PubMed] [Google Scholar]
- 49.Yuan J, Bennett BD, Rabinowitz JD. Kinetic flux profiling for quantitation of cellular metabolic fluxes. Nat Protoc. 2008;3:1328–1340. doi: 10.1038/nprot.2008.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lu W, et al. Metabolomic analysis via reversed-phase ion-pairing liquid chromatography coupled to a stand alone orbitrap mass spectrometer. Anal Chem. 2010;82:3212–3221. doi: 10.1021/ac902837x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Melamud E, Vastag L, Rabinowitz JD. Metabolomic analysis and visualization engine for LC-MS data. Anal Chem. 2010;82:9818–9826. doi: 10.1021/ac1021166. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.