Abstract
Human breast cancers are broadly classified based on their gene-expression profiles into luminal- and basal-type tumors. These two major tumor subtypes express markers corresponding to the major differentiation states of epithelial cells in the breast: luminal (EpCAM+) and basal/myoepithelial (CD10+). However, there are also rare types of breast cancers, such as metaplastic carcinomas, where tumor cells exhibit features of alternate cell types that no longer resemble breast epithelium. Until now, it has been difficult to identify the cell type(s) in the human breast that gives rise to these various forms of breast cancer. Here we report that transformation of EpCAM+ epithelial cells results in the formation of common forms of human breast cancer, including estrogen receptor-positive and estrogen receptor-negative tumors with luminal and basal-like characteristics, respectively, whereas transformation of CD10+ cells results in the development of rare metaplastic tumors reminiscent of the claudin-low subtype. We also demonstrate the existence of CD10+ breast cells with metaplastic traits that can give rise to skin and epidermal tissues. Furthermore, we show that the development of metaplastic breast cancer is attributable, in part, to the transformation of these metaplastic breast epithelial cells. These findings identify normal cellular precursors to human breast cancers and reveal the existence of a population of cells with epidermal progenitor activity within adult human breast tissues.
Keywords: cell of origin, epidermal progenitor cells, luminal progenitors
Invasive breast cancer is a multifaceted disease consisting of tumors that exhibit a wide spectrum of histological and molecular features. Much attention has been focused on identifying mutated or amplified genes in breast cancer to understand tumor heterogeneity and develop targeted therapies. This work has revealed that subclasses of breast tumors exhibit distinct constellations of genetic aberrations and was the driving force behind the development of targeted therapies for estrogen receptor (ER)+ and HER2+ breast cancers. Despite these successes, these findings fail to inform us about the details of how human breast cancers begin, including the identity of the cellular origins of breast cancer and how cell-intrinsic epigenetic programs interact with genetic alterations to affect tumor phenotype.
Ductal carcinomas are the most common type of breast cancer, accounting for ∼80% of all invasive tumors. They are broadly categorized into two types, ER+ and ER− tumors, but can be further subdivided molecularly and histologically into subclasses with different prognostic outcomes and therapeutic sensitivities (1). Molecular classification of tumors has shown that ER+ and ER− tumors generally retain expression of markers of the two major differentiation states of normal human breast tissue: luminal and basal/myoepithelial (ME). ER+ tumors express hormone receptors and genes characteristic of luminal epithelial cells [e.g., cytokeratin (CK)8/18, CK19, CD24, Mucin 1 (MUC1), GATA3, and epithelial cell adhesion molecule (EpCAM)]; in contrast, ER− tumors lack estrogen-responsive genes and express markers characteristic of basal/ME cells [e.g., α-smooth muscle actin (αSMA), CD49f, p63, CK14, EGF receptor, and CD44]. ER− tumors also encompass rare cancers, such as medullary, adenoid cystic, and metaplastic carcinomas, where the tumor cells not only lack ER-responsive and luminal genes but also exhibit features of alternate cell types not found in normal breast epithelium (2, 3). Although ER− tumors exhibit a broad range of characteristics, collectively they fall under the classification of HER2+ and basal-like, including the recently identified claudin-low subtype of breast cancers (2–4).
We have found that mutated tumor suppressors can act to disrupt the differentiation programs of progenitor populations and influence the type of tumor that will develop (5). However, studies of blood cancers provide strong evidence that distinct lineage-committed progenitors can give rise to different diseases (6, 7). Moreover, in the case of solid tumors, the intrinsic differentiation programs of cellular precursors were shown to contribute to the heterogeneity and behavior of tumor cells (8, 9). It has been hypothesized that ER+ luminal-type breast cancers may be derived from luminal progenitor cells, whereas ER− basal-like breast cancers may be derived from basal/ME progenitor cells (10). However, characterization of human breast luminal progenitor cells has revealed overlapping gene- and surface marker-expression profiles with basal-like tumors and cell lines (11, 12), suggesting that luminal-lineage cells could be the source of basal-like breast cancers. Thus, it remains unknown whether all human breast tumors are derived from the same cellular precursors or whether different cell types contribute to the heterogeneity of breast cancers. In this study, we sought to examine the relationship between luminal and basal/ME cells and breast cancer subtypes through use of the recently described human-in-mouse (HIM) model (13, 14). With this approach, we discovered unique properties of a cell population within human mammary basal/ME epithelial cells that provide important insights into the origins of rare breast cancers.
Results
Characterization of Breast Epithelial Cell Properties in Human Tissues.
Much recent effort has been focused on trying to define the hierarchal relationships among epithelial cells in the breast. This work has led to conflicting reports regarding the phenotype of stem/progenitor cells and their relationship to differentiated progeny (11, 15–18). EpCAM+MUC1− cells or EpCAM+CD49f+ cells were first reported to enrich for cells with bipotent potential in human tissues (15, 16). In contrast, studies have also suggested that EpCAM−CD49f+ cells enrich for stem/progenitor competency (11, 17, 18). Because EpCAM and CD49f are also used to identify populations of differentiated luminal and basal/ME cells no unique set of cell surface markers can currently discriminate progenitor cells from more mature cells. Given the ambiguities surrounding the precise identity of progenitor cells, we chose to evaluate mammary epithelial cells enriched within the two major breast epithelial lineages to determine whether they might give rise to distinct tumor subtypes.
Consistent with our previous reports (5, 12), flow cytometry analysis of reduction mammoplasty tissues for EpCAM and CD49f expression indicated that four populations of epithelial cells could be identified: EpCAMhiCD49f−, EpCAMhiCD49f+, EpCAMloCD49f+, and EpCAM−CD49f+ cells. We found that CD10, a well-established marker of basal/ME cells was enriched in the EpCAMloCD49f+ population, with an average of 74.4% CD10+ cells in this fraction (P < 0.00035; Fig. 1A and Fig. S1 A and B). In contrast, luminal populations with high expression of EpCAM (EpCAMhiCD49f− and EpCAMhiCD49f+) showed little CD10 expression (Fig. 1A and Fig. S1 A and B). Lineage commitment of EpCAM/CD49f-expressing populations was previously confirmed (5, 12).
Fig. 1.
Enrichment of basal/ME and luminal populations from primary human breast tissue. (A) Single-cell suspensions of human breast epithelial cells analyzed by flow cytometry for expression of EpCAM, CD49f, and CD10 (n = 5 patient samples). (Left) Representative dot plot of EpCAM and CD49f stained cells. Five fractions of cells were gated and analyzed for CD10 content (%) as shown in the histograms on the Right. (B) Schematic of immunomagnetic sorting strategy. (C) Unsorted cells and fractions recovered after immunomagnetic sorting were analyzed by flow cytometry for expression of EpCAM and CD49f (n = 3). Representative dot plots from unsorted (patient 641), CD10+, and depleted cells are shown. (Lower Right) Overlay of unsorted EpCAMhi, sorted CD10+, and depleted fractions. Sorted EpCAM+CD10− cells did not stain with the fluorescently conjugated EpCAM antibody because of occupation of antigen sites from bead sorting; however, the luminal EpCAMhi clouds from unsorted cells (red) are clearly missing from the depleted fraction (blue). Enrichment of the EpCAMloCD49f+ population within the sorted CD10+ fraction is shown in green.
Based on these findings, we used an immunomagnetic strategy to enrich for basal/ME-lineage cells (CD10+) and luminal-lineage cells (EpCAM+CD10−, hereafter referred to as EpCAM+) (Fig. 1B and Fig. S1C). Flow cytometry analysis of EpCAM and CD49f expression in sorted CD10+ cells and the fraction depleted of both EpCAM+ and CD10+ cells confirmed that bead sorting for CD10 efficiently enriches for basal/ME EpCAMloCD49f+ cells, and bead sorting for EpCAM efficiently recovers the EpCAMhi luminal populations containing both CD49+ and CD49f− cells (Fig. 1C and Fig. S1D). Lineage was confirmed by immunofluorescence on cytospun sorted CD10+ and EpCAM+ cells for markers of luminal (CK8/18) and basal/ME cells (CK14) (Fig. S1E).
To further confirm that our sorting strategy enriches for distinct populations of cells, we analyzed functional activities in vitro by growing cells under adherent, nonadherent, and 3D conditions. Unsorted primary human mammary epithelial cells (HMECs) plated in serum-free mammary epithelial growth medium (MEGM) formed colonies that grew in suspension as well as distinct luminal, ME, and mixed adherent colonies that could be distinguished morphologically and by expression of CK14 and CK8/18 (Fig. 2A and Fig. S2 A and B). Under adherent conditions, luminal EpCAM+ cells preferentially grew as suspension colonies (P = 0.015). Rare colonies that did arise from EpCAM+ cells were enriched for luminal-type CK8/18+ cells as well as two types of bipotent colonies: bipotent A, composed of a central core of tightly arranged CK8/18+ cells surrounded by CK14+ dispersed cells, and bipotent B, composed of dispersed cells that contained mixed single- and double-positive cells (Fig. S2 A and B). In contrast, basal/ME lineage CD10+ cells preferentially grew as adherent colonies, suggesting that CD10+ cells represent the greatest contribution of cells that expand in monolayer culture from unsorted HMECs under serum-free conditions (P = 0.004; Fig. 2A and Fig. S2 A and B). Notably, the majority of colonies derived from CD10+ cells were bipotent B colonies and pure basal/ME colonies that stained exclusively for CK14+ (Fig. S2 A and B).
Fig. 2.
EpCAM+CD10− luminal and CD10+ basal/ME sorted populations contain cells with distinct functional characteristics. (A and B) Unsorted or sorted fractions of cells were plated in adherent conditions (A) or nonadherent conditions (B) and allowed to grow for 7–10 d. Graphed data represent average fold ± SE from independently sorted patient samples (n = 4–5). P values were calculated by Student's t test. (A Left) Representative image of suspension colonies (arrows) and adherent colonies. (Bar: 200 μm.) (Right) Representative images of crystal violet-stained colonies. (B) Immunofluorescence staining for CK8/18 (red) and CK14 (green) in spheres formed in nonadherent conditions. (Bar: 100 μm.) (C) Outgrowth from unsorted cells or sorted fractions plated on collagen I gels. (Upper) Representative images of the three structures formed. (Bar: 100 μm.) Graphed data represent average number ± SE from independently sorted patient samples (n = 3). P values were calculated by Student's t test. (D) Immunohistochemistry for luminal (CK19 and ERα) and basal/ME (CK14 and αSMA) differentiation markers in bilayered structures formed in vivo in the HIM model by sorted fractions. (Bar: 25 μm.)
Under nonadherent conditions, EpCAM+ cells also preferentially grew in suspension compared with CD10+ cells, generating 6.8-fold more spheres that contained both CK8/18+ and 14+ cells (P = 0.0001; Fig. 2B and Fig. S2C). Spheres formed by CD10+ basal/ME-lineage cells stained more predominantly for CK14 and exhibited a compact morphology with a smooth outer surface (Fig. 2B and Fig. S2 C and D). Although we cannot exclude the possibility that aggregation in populations of EpCAM+ or CD10+ cells was responsible for colony formation under nonadherent conditions, these results combined with adherent colony-formation assays indicate that EpCAM+ and CD10+ cells exhibit distinguishable biological activities in vitro.
Mammary morphogenesis assays (3D collagen I gel overlays) were used to further assess EpCAM+ and CD10+ cells. In this assay, unsorted primary HMECs formed three types of morphologically distinct structures: flat monolayer colonies reminiscent of simple epithelium, round acinar colonies, and branching ductal structures; the latter two are reminiscent of glandular epithelium (Fig. 2C). Although there was considerable variation in the capacity to form acinar structures among patient samples, round acinar colonies were preferentially generated from cells enriched in the EpCAM+ fraction (Fig. 2C). In contrast, cells from the CD10+ fraction preferentially produced branching ductal structures and flat colonies compared with EpCAM+ cells (P = 0.0005 and P < 0.03, respectively; Fig. 2C).
Finally, sorted CD10+ and EpCAM+ cells were injected in vivo to evaluate their outgrowth competency in the HIM model (13, 14). Although EpCAM+ and CD10+ cells exhibited differential activities in vitro, within the humanized cleared fat-pad system, cells from both the CD10+ and EpCAM+ populations could generate bilayered mammary outgrowths comprised of an inner luminal epithelial layer that expressed CK8/18 and CK19 and an outer epithelial layer that expressed CK14 (Fig. 2D, Fig. S3, and Table S1). Both fractions were also able to generate differentiated mature luminal and ME cells, marked by expression of ERα and αSMA(Fig. 2D and Fig. S3). Altogether, these findings indicate that cells within the luminal and basal/ME lineages exhibit distinguishing phenotypic and progenitor-like functional activities and suggest that both lineages appear to contain cells with bipotent differentiation capacity.
Creation of Luminal-Like, Basal-Like, and Metaplastic Human Breast Cancers.
To evaluate the influence of breast epithelial precursor cells on tumor subtype, we modified the HIM model to create human breast cancer tissues in vivo by introducing oncogenes into freshly dissociated epithelial cells derived from reduction mammoplasty tissues before injection into humanized mammary fat pads. Importantly, the cells for these experiments were maintained in vitro for no more than 18–24 h after dissociation to avoid culture-adapted selection of cells.
Unsorted breast epithelial cells (n = 10 patient samples) were transduced with lentiviruses harboring two different combinations of transforming oncogenes (Fig. S4A): (i) mutant p53 (p53R175H), cyclin D1 (CCND1), myristoylated PI3K p110α (PI3KCA), and mutant K-ras (RasG12V), hereafter termed 4onc; or (ii) SV40 early region (encoding large and small T antigens) and mutant K-ras (RasG12V), hereafter termed SV40/Ras. Tumor formation was observed with either oncogenic combination across multiple patient samples (Table S2). Expression of the introduced genes was gauged by GFP expression, immunostaining, and quantitative RT-PCR (qRT-PCR) (Fig. S4 B–E). Genomic fluorescence in situ hybridization showed that the tumor cells were not of mouse origin, confirming derivation from human cells (Fig. S4F). Microscopic and immunohistochemical examination revealed that the tumors from unsorted cells were heterogeneous carcinomas with features of luminal, basal, and even rare squamous/metaplastic differentiation irrespective of the patient samples or oncogene model from which they were derived (Fig. 3A and Fig. S5 A–C). Immunostaining demonstrated that cancer cells in squamous regions expressed CK14, indicative of basal differentiation, and those within papillary/glandular regions expressed CK8/18, indicative of luminal differentiation (Fig. S5B).
Fig. 3.
Tumors formed from sorted fractions have distinct phenotypes. (A) Representative images from tumors derived from unsorted and sorted fractions transformed with the SV40/Ras oncogene combination stained for ERα, CK14 and CK8/18, and CK19. (Bars: 200 μm.) (B) Quantification of CK14 and CK19 expression (n = 8–9 tumors per group; immunofluorescence) and ERα expression (n = 4–7 tumors per group; immunohistochemistry) across tumors from 4onc and SV40/Ras models. Graphs represent average ± SE. P values were calculated by one-way ANOVA (CK19 and CK14) or Student's t test (ERα). (C) Heat map of hierarchical clustering of global gene-expression data collected from SV40/Ras tumors (n = 20) arising from unsorted (blue) or sorted CD10+ (green), EpCAM+ (red), EpCAM+/CD49f+ (yellow), or EpCAM+/CD49f− (magenta) cells. (D) Differentiation status of the various SV40/Ras tumors along a normal breast development axis using the Genomic Differentiation Predictor described in ref. 4. The P value was calculated by comparing gene-expression means across unsorted and sorted tumor groups. (E) Enrichment of the CD10 Signature derived from CD10+ tumors across a panel of breast tumor intrinsic subtypes from the UNC337 data set (Gene Expression Omnibus accession no. GSE18229). P value was calculated by comparing gene-expression means across all subtypes.
The finding that breast tumors from unsorted cells exhibited a mixed phenotype, containing both luminal and basal features, suggested the possibility that these tumors were derived from a mixture of transformed basal/ME- and luminal-lineage epithelial cells. To address whether transformation of luminal and basal/ME cells led to the formation of tumors with luminal and basal/ME-like features, respectively, epithelial cells from reduction mammoplasty tissues were sorted for EpCAM+ and CD10+ cells before oncogenic transduction as previously described (5). Examination of unsorted and sorted cells after infection in either 4onc or SV40/Ras models revealed similar gene-transduction efficiencies (Fig. S4 B–E).
Transformation of luminal EpCAM+ cells (n = 7 patient samples) with either combination of transforming oncogenes led primarily to the formation of ductal carcinomas with predominant luminal features, including expression of ERα, CK8/18, and CK19 (Fig. 3 A and B and Fig. S5C). Because EpCAM+ cells enrich for heterogeneous populations of luminal epithelial cells based on CD49f expression (11, 12, 16–18), we further sorted the EpCAM+ fraction into CD49f+ or CD49f− cells before lentiviral transduction (Fig. S6A). Interestingly, transformation of CD49f− luminal cells with SV40/Ras resulted in the development of tumors with significantly higher expression of ERα and reduced expression of basal CK14 compared with CD49f+ tumors (Fig. S6 B and C). In contrast to EpCAM+ tumors, tumors derived from CD10+ cells (n = 7 patient samples) exhibited pronounced squamous, metaplastic, and giant cell differentiation concomitant with a marked lack of ERα expression (P = 0.006; Fig. 3 A and B and Fig. S5C), significant decrease in luminal CK expression (CK19; P = 0.001), and robust expression of the basal marker CK14 (P = 0.0006) (Fig. 3 A and B and Fig. S5C).
To more comprehensively define the phenotype of the tumors generated, we performed global gene-expression analyses on RNA isolated from tumors arising from unsorted, EpCAM+, or CD10+ cells as well as from tumors derived from EpCAM+/CD49f+ and EpCAM+/CD49f− cells. Unsupervised hierarchical-clustering analysis indicated that tumors arising from EpCAM+ or CD10+ cells could be segregated from one another (Fig. 3C and Dataset S1). Interestingly, tumors derived from unsorted cells clustered more closely with tumors arising from CD10+ cells than with those derived from EpCAM+ cells. In addition, although tumors derived from EpCAM+/CD49f+ and EpCAM+/CD49f− cells could be distinguished from unsorted or CD10+ sorted cells, they could not be distinguished from tumors derived from bulk EpCAM+ cells (Fig. 3C and Dataset S1).
Gene set enrichment analysis (GSEA) showed significant enrichment of genes derived from pairwise comparisons of EpCAM+ and CD10+/unsorted tumors with genes associated with luminal, basal, and stem cell differentiation (Datasets S1, S2, and S3). Consistent with GSEA, when tumor differentiation was analyzed with the recently described Genomic Differentiation Predictor (4), tumors derived from EpCAM+ cells were more differentiated compared with CD10+ and unsorted-derived tumors (P = 0.0286; Fig. 3D).
We derived a “CD10 Signature” based on the genes that were differentially expressed between tumors derived from CD10+ sorted cells compared with tumors derived from all EpCAM+ cells (including EpCAM+/CD49f+ and EpCAM+/CD49f− cells; Dataset S4) and then examined this signature across the intrinsic breast cancer subtypes (UNC337, taken from ref. 4). Interestingly, the CD10 Signature was most enriched in claudin-low tumors (P =1.98 × 10−22; Fig. 3 E), which are associated with metaplastic, mesenchymal, and mammary stem cell-like characteristics (4, 19). Concordant with this finding, the CD10 Signature was also enriched in published gene-expression data from the mammary stem cell population (11) (P = 1.98 × 10−6; Fig. S7). Altogether, these results suggest that EpCAM+ epithelial cells serve as precursors for differentiated ER+ and ER− ductal carcinomas, whereas CD10+ cells serve as precursors for rare and undifferentiated metaplastic/claudin-low carcinomas.
Cells with Metaplastic Potential Reside Within Adult Human CD10+ Breast Epithelium.
Because transformation of CD10+ cells resulted in the formation of metaplastic breast cancers, we reasoned that breast epithelial cells within the CD10+ population might contain cells with metaplastic potential, i.e., reduced mammary specification, before neoplastic transformation. Ex vivo cultivation of HMECs selects for variant cells that exhibit significant differences in gene-expression profiles, lineage markers, and chromatin methylation states compared with primary HMECs (20, 21). We reasoned that these cells might be enriched within the CD10+ lineage and may exhibit features of alternate cell types. Indeed, we readily observed the emergence of variant HMEC (vHMEC) cells from cultured CD10+ cells (n = 5 patients); in contrast, EpCAM+ cells rarely gave rise to variant cells (Fig. 4A and Fig. S8 A and B).
Fig. 4.
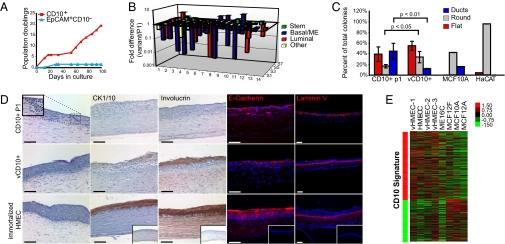
vCD10+ cells spontaneously lose mammary fate specification and gain ability to form skin tissues. (A) Representative graph of long-term culture of sorted CD10+ and EpCAM+ cells grown in MEGM showing the formation of vHMEC cells (vCD10+) preferentially from sorted CD10+ cells (n = 5 independently sorted patient samples). (B) Summary of mammary fate gene expression as analyzed by custom qRT-PCR array in P1 CD10+ compared with vCD10+ cells (n = 3 from independently sorted patient samples). (C) Outgrowth from P1 CD10+, vCD10+, MCF10A, and HaCAT cells plated on collagen I gels. (D) Representative images of sections from HSE assays stained for H&E, CK1/10, involucrin, E-cadherin, and laminin V (n = 3 from independently sorted or immortalized patient samples for each condition). (Bars: 100 μm.) (Inset) An example of ductal-like growths into the collagen gels from P1 CD10+ cells is shown. (E) Heat map of hierarchical clustering of global gene-expression data collected from a panel of HMECs: primary vHMECs (vHMEC-1, vHMEC-2, and vHMEC-3) isolated from three patients, immortalized HMECs (HME-CC and ME16C), and MCF10A, MCF12F, and MCF12A cells. Comparison with the CD10 Signature is shown on the right.
To determine whether these variant cells exhibited metaplastic features, short-term cultures of CD10+ cells harvested at the first passage (P1 CD10+ cells) as well as variant CD10+ cells harvested during exponential growth after escape from stasis/selection (vCD10+ cells) were examined for the expression of 84 genes associated with mammary differentiation by qRT-PCR (Table S3). Consistent with previous reports (20), P1 CD10+ cells expressed a range of ME, luminal, and progenitor lineage markers (αSMA, CK19, oxytocin receptor, progesterone receptor, CK8, and myosin light chain kinase), whereas vCD10+ lacked expression of many genes associated with mammary differentiation (Fig. 4B and Table S3).
Because the mammary gland is an epidermal appendage and metaplastic tumors are associated with features of epidermal differentiation, including squamous and apocrine differentiation, we reasoned that vCD10+ cells might exhibit features of epidermal cells. To investigate, we first assessed the morphogenesis competency of P1 CD10+, vCD10+, and vHMEC cells in collagen I gel overlay assays. HaCAT human epidermal-derived keratinocyte cells used as an epidermal control predominantly formed round structures in contrast to round and branching ductal structures formed by the MCF10A mammary cell line. In comparison to P1 CD10+ cells, vCD10+ cells produced significantly fewer branching ductal colonies and exhibited an increase in flat and round colony formation (Fig. 4C and Fig. S9A).
We next assessed whether vCD10+ or vHMEC cells had the ability to form epidermal tissues. P1 CD10+ cells, vCD10+ cells, and vHMECs were seeded onto collagen matrix containing adult human dermal fibroblasts and grown in a human skin equivalents (HSE) assay. In this assay, HaCAT cells formed stratified epidermal tissues displaying basal, spinous/granular, and cornified layers as well as expression of E-cadherin at cell junctions, localization of involucrin and CK1/10 to the spinous/granular layers, and laminin V deposition on the basement membrane (Fig. S9B). Remarkably, vCD10+ cells and immortalized vHMECs were able to form skin-like tissues in 3D HSE cultures that also displayed stratified layers and expression of skin markers (Fig. 4D and Fig. S9B). In contrast, P1 CD10+ cells formed ductal invaginations into the collagen matrix, reminiscent of rudimentary mammary structures, and did not stain for involucrin or CK1/10. MCF10A cells were not capable of forming stratified epidermal tissue, indicating that formation of skin tissues is not a property of all cultured HMECs (Fig. S9C).
In addition to epidermal differentiation capacity, gene-expression analysis of primary vHMECs (n = 3 patient samples), two immortalized vHMEC lines, and the MCF10A, MCF12F, and MCF12A cell lines revealed that vHMECs and immortalized vHMECs, but not MCF10A, MCF12F, and MCF12A cells, are enriched in the metaplastic CD10 Signature (Fig. 4E and Fig. S9D). Collectively, these results imply that cells within the CD10+ breast epithelial lineage have the capacity to exhibit metaplastic features before neoplastic transformation.
Creation of Metaplastic Tumors from HMECs.
To determine whether metaplastic breast epithelial cells were indeed precursors to metaplastic breast cancer, immortalized vHMECs were transformed with the SV40/Ras oncogenes as previously described (22), and tumors were examined by histopathological analysis. Consistent with previous reports (22), transformed vHMECs formed tumors that exhibited mixed epidermal and metaplastic features including squamous, spindle-cell, medullary, and giant cell differentiation (Fig. 5A and Fig. S10 A and B). Interestingly, transduced vHMECs that were unable to form expansive tumors created tissues that resembled human sebaceous glands, albeit somewhat disorganized, consistent with dedifferentiation into an early epidermal state (Fig. 5A and Fig. S10A). Concordant with these findings, we observed a significant enrichment of the CD10 Signature in a panel of human metaplastic, adenoid cystic, and medullary carcinomas from a microarray dataset of special histological breast cancer types (23) and remarkable similarity to histopathology of human metaplastic breast cancers (Fig. 5B and Fig. S10C). Given the histological and molecular similarities between tumors derived from CD10+ cells and vHMECs, these results strongly support the notion that the cellular precursors to rare metaplastic breast carcinomas may reside within the CD10+ cell population.
Fig. 5.
Transformed vHMEC cells give rise to metaplastic tumors. (A) Immortalized vHMECs from unsorted cells give rise to disorganized sebaceous-like growths (Upper Left) as well as tumors with medullary (Upper Right), spindle/giant-cell (Lower Left; arrows indicate giant cells), and squamous (Lower Right) histologies when transformed with SV40/Ras. (Bars: 100 μm.) (B) Enrichment of the CD10 Signature, derived from CD10+ tumors, across a panel of 11 special histological types of breast cancer from ref. 23.
Discussion
The enumeration of the cellular and functional activities of various cell types within human and mouse mammary tissues has been an area of intense investigation for understanding the cellular origins of breast cancers (10). Although breast stem/progenitor cells have been localized to cells within both main and terminal ducts (16), the precise identity of human mammary stem cells is an area of contentious debate and remains ill defined. Some studies claim that stem/progenitor cells in human breast tissues reside within the luminal EpCAM+/MUC1− or EpCAM+/CD49f+ population (15, 16), whereas other reports claim they reside within the basal/ME EpCAM−/CD49f+ population (11, 17, 18). Through in vivo and in vitro characterization of cells from EpCAM+ luminal and CD10+ basal/ME lineages, we found important functional distinctions between these two lineages and also that both populations retain some functional progenitor competency, which may explain why stem/progenitor activity has been difficult to exclusively measure from a unique and distinctive population of breast epithelial cells.
Through use of the HIM tumor model, we have also been able to address one of the major questions regarding human breast tumor heterogeneity, namely, how different pools of progenitor cells in normal human breast tissue contribute to tumor phenotype. Our results strongly imply that the great majority of human breast cancers are likely derived from EpCAM+ luminal epithelial cells because EpCAM+ cells were able to give rise to both ER+ and ER− tumors, indicating that basal-like tumors need not originate from basal/ME progenitor cells. This finding is in agreement with the previously speculated but untested hypothesis that human basal-like breast cancers may arise from luminal progenitor cells and the observation that most basal-like tumors lack CD10 expression (5, 11, 12, 24). Our data from the transformation of EpCAM+/CD49f− cells also supports the hypothesis that tumors can be derived from a pool of more mature luminal cells, as has been speculated based on similarities between gene-expression profiles of human luminal tumors and the differentiated luminal cell fraction of human breast tissue (11). Finally, our data indicate that basal/ME CD10+ cells are likely the source of rare types of breast tumors such as metaplastic tumors.
Although we have demonstrated that EpCAM+/CD49f− cells can serve as precursors to ER+ breast cancers, gene-expression analysis was unable to distinguish these tumors from those derived from EpCAM+/CD49f+ cells, suggesting that ER+ tumors could be derived from the same common luminal progenitor cell as ER− tumors and/or that the oncogenes used to transform EpCAM+/CD49f− cells affected their differentiation potential. Consistent with these notions are the recent findings that mutations in BRCA1 can affect the differentiation potential of luminal progenitor cells, leading to increased basal-like differentiation in tumors (5, 11). Because carcinoma cells from luminal ER+ and basal-like ER− breast cancers are reported to contain distinguishing and even mutually exclusive sets of mutated or misregulated genes (25), the genetic and epigenetic alterations sustained during early stages of transformation could alter differentiation commitment programs in a common cell of origin, leading to tumors with different phenotypes. In this model, undifferentiated tumors could result when the common cell of origin lost lineage commitment potential during the acquisition of malignancy. Additional studies will be needed with alternative combinations of transforming oncogenes to fully separate the contribution of the cell of origin from the role gene mutation has on differentiation programs and tumor phenotype. In addition, further studies that separate subpopulations of CD10+ cells and substitute other combinations of transforming oncogenes will be necessary to fully elucidate the contribution CD10+ cells on the development of other types of breast cancers.
Materials and Methods
Immunomagnetic bead sorting was used to isolate luminal and basal/ME populations from reduction mammoplasty tissue. Tumors were generated by lentiviral introduction of oncogenes and were characterized by protein and gene expression. All methods describing primary tissue isolation, flow cytometry, animal surgeries, in vitro assays, and gene-expression analysis are included in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Joslyn Mills for assistance with gene-expression experiments, Joe Gray and Sandy DeVries at the University of California, San Francisco, for genomic FISH analysis, and Annette Shepard-Barry at Tufts Medical Center in the Histology-Special Procedures Laboratory for assistance with histological and immunohistochemical staining. We thank Josh LaBaer at Harvard Medical School for providing us with cyclin D1, ras, p53, and PI3K cDNAs. Microarray processing was performed by Tufts Center for Neuroscience Research under National Institutes of Health Grant P30 NS047243 (to F. Rob Jackson). This work was supported by an American Cancer Society, New England Division, Broadway on Beachside Postdoctoral Fellowship (to P.J.K.); the Raymond and Beverly Sackler Foundation (P.J.K. and C.K.); the Breast Cancer Research Foundation (C.K. and C.M.P.); Department of Defense Breast Cancer Research Program Grant W81XWH-08-01-0653 (to P.J.K. and C.K.); National Institutes of Health, National Cancer Institute Grants CA125554 and CA125554 (to C.K. and L.M.A.) and CA58223 and CA148761 (to C.M.P.); and National Institute of Dental and Craniofacial Research Grant DE017413 (to J.A.G.). C.K. is a Raymond and Beverly Sackler Foundation scholar.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. K.P. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1017626108/-/DCSupplemental.
References
- 1.Peppercorn J, Perou CM, Carey LA. Molecular subtypes in breast cancer evaluation and management: Divide and conquer. Cancer Invest. 2008;26:1–10. doi: 10.1080/07357900701784238. [DOI] [PubMed] [Google Scholar]
- 2.Rakha EA, Reis-Filho JS, Ellis IO. Basal-like breast cancer: A critical review. J Clin Oncol. 2008;26:2568–2581. doi: 10.1200/JCO.2007.13.1748. [DOI] [PubMed] [Google Scholar]
- 3.Gluz O, et al. Triple-negative breast cancer—current status and future directions. Ann Oncol. 2009;20:1913–1927. doi: 10.1093/annonc/mdp492. [DOI] [PubMed] [Google Scholar]
- 4.Prat A, et al. Phenotypic and molecular characterization of the claudin-low intrinsic subtype of breast cancer. Breast Cancer Res. 2010;12:R68. doi: 10.1186/bcr2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Proia TA, et al. Genetic predisposition directs breast cancer phenotype by dictating progenitor cell fate. Cell Stem Cell. 2011;8:149–163. doi: 10.1016/j.stem.2010.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Thiel E. Cell surface markers in leukemia: Biological and clinical correlations. Crit Rev Oncol Hematol. 1985;2:209–260. doi: 10.1016/s1040-8428(85)80003-2. [DOI] [PubMed] [Google Scholar]
- 7.Foon KA, Todd RF., 3rd Immunologic classification of leukemia and lymphoma. Blood. 1986;68:1–31. [PubMed] [Google Scholar]
- 8.Gupta PB, et al. The melanocyte differentiation program predisposes to metastasis after neoplastic transformation. Nat Genet. 2005;37:1047–1054. doi: 10.1038/ng1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ince TA, et al. Transformation of different human breast epithelial cell types leads to distinct tumor phenotypes. Cancer Cell. 2007;12:160–170. doi: 10.1016/j.ccr.2007.06.013. [DOI] [PubMed] [Google Scholar]
- 10.Polyak K. Breast cancer: Origins and evolution. J Clin Invest. 2007;117:3155–3163. doi: 10.1172/JCI33295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lim E, et al. Aberrant luminal progenitors as the candidate target population for basal tumor development in BRCA1 mutation carriers. Nat Med. 2009;15:907–913. doi: 10.1038/nm.2000. [DOI] [PubMed] [Google Scholar]
- 12.Keller PJ, et al. Mapping the cellular and molecular heterogeneity of normal and malignant breast tissues and cultured cell lines. Breast Cancer Res. 2010;12:R87. doi: 10.1186/bcr2755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Proia DA, Kuperwasser C. Reconstruction of human mammary tissues in a mouse model. Nat Protoc. 2006;1:206–214. doi: 10.1038/nprot.2006.31. [DOI] [PubMed] [Google Scholar]
- 14.Kuperwasser C, et al. Reconstruction of functionally normal and malignant human breast tissues in mice. Proc Natl Acad Sci USA. 2004;101:4966–4971. doi: 10.1073/pnas.0401064101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gudjonsson T, et al. Isolation, immortalization, and characterization of a human breast epithelial cell line with stem cell properties. Genes Dev. 2002;16:693–706. doi: 10.1101/gad.952602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Villadsen R, et al. Evidence for a stem cell hierarchy in the adult human breast. J Cell Biol. 2007;177:87–101. doi: 10.1083/jcb.200611114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eirew P, et al. A method for quantifying normal human mammary epithelial stem cells with in vivo regenerative ability. Nat Med. 2008;14:1384–1389. doi: 10.1038/nm.1791. [DOI] [PubMed] [Google Scholar]
- 18.Stingl J, Eaves CJ, Zandieh I, Emerman JT. Characterization of bipotent mammary epithelial progenitor cells in normal adult human breast tissue. Breast Cancer Res Treat. 2001;67:93–109. doi: 10.1023/a:1010615124301. [DOI] [PubMed] [Google Scholar]
- 19.Hennessy BT, et al. Characterization of a naturally occurring breast cancer subset enriched in epithelial-to-mesenchymal transition and stem cell characteristics. Cancer Res. 2009;69:4116–4124. doi: 10.1158/0008-5472.CAN-08-3441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Garbe JC, et al. Molecular distinctions between stasis and telomere attrition senescence barriers shown by long-term culture of normal human mammary epithelial cells. Cancer Res. 2009;69:7557–7568. doi: 10.1158/0008-5472.CAN-09-0270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huschtscha LI, et al. Loss of p16INK4 expression by methylation is associated with lifespan extension of human mammary epithelial cells. Cancer Res. 1998;58:3508–3512. [PubMed] [Google Scholar]
- 22.Elenbaas B, et al. Human breast cancer cells generated by oncogenic transformation of primary mammary epithelial cells. Genes Dev. 2001;15:50–65. doi: 10.1101/gad.828901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Weigelt B, et al. Refinement of breast cancer classification by molecular characterization of histological special types. J Pathol. 2008;216:141e150. doi: 10.1002/path.2407. [DOI] [PubMed] [Google Scholar]
- 24.Livasy CA, et al. Phenotypic evaluation of the basal-like subtype of invasive breast carcinoma. Mod Pathol. 2006;19:264–271. doi: 10.1038/modpathol.3800528. [DOI] [PubMed] [Google Scholar]
- 25.Hollestelle A, et al. Distinct gene mutation profiles among luminal-type and basal-type breast cancer cell lines. Breast Cancer Res Treat. 2010;121:53–64. doi: 10.1007/s10549-009-0460-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.