Abstract
The antiviral potency of the cytokine IFN-α has been long appreciated but remains poorly understood. A number of studies have suggested that induction of the apolipoprotein B mRNA editing enzyme, catalytic polypeptide 3 (APOBEC3) and bone marrow stromal cell antigen 2 (BST-2/tetherin/CD317) retroviral restriction factors underlies the IFN-α–mediated suppression of HIV-1 replication in vitro. We sought to characterize the as-yet-undefined relationship between IFN-α treatment, retroviral restriction factors, and HIV-1 in vivo. APOBEC3G, APOBEC3F, and BST-2 expression levels were measured in HIV/hepatitis C virus (HCV)-coinfected, antiretroviral therapy-naïve individuals before, during, and after pegylated IFN-α/ribavirin (IFN-α/riba) combination therapy. IFN-α/riba therapy decreased HIV-1 viral load by −0.921 (±0.858) log10 copies/mL in HIV/HCV-coinfected patients. APOBEC3G/3F and BST-2 mRNA expression was significantly elevated during IFN-α/riba treatment in patient-derived CD4+ T cells (P < 0.04 and P < 0.008, paired Wilcoxon), and extent of BST-2 induction was correlated with reduction in HIV-1 viral load during treatment (P < 0.05, Pearson's r). APOBEC3 induction during treatment was correlated with degree of viral hypermutation (P < 0.03, Spearman's ρ), and evolution of the HIV-1 accessory protein viral protein U (Vpu) during IFN-α/riba treatment was suggestive of increased BST-2–mediated selection pressure. These data suggest that host restriction factors play a critical role in the antiretroviral capacity of IFN-α in vivo, and warrant investigation into therapeutic strategies that specifically enhance the expression of these intrinsic immune factors in HIV-1–infected individuals.
Despite nearly three decades of focused research since the discovery of HIV-1, to date, there is no cure or effective prophylactic vaccine for HIV-1 infection. Although the advent of highly active antiretroviral therapy has dramatically decreased the morbidity and mortality associated with HIV-1 infection, there is a pronounced demand for alternative clinical management strategies due to frequent evolution of antiretroviral resistance, toxicity, and access constraints in resource-limited settings (1). Recently, a number of innate immune factors have been identified in primates that suppress retroviral replication in vitro and therefore may constitute new avenues for therapeutic intervention (2–4). Three of these innate retroviral restriction factors—apolipoprotein B mRNA editing enzyme, catalytic polypeptide 3 (APOBEC3) (5), bone marrow stromal cell antigen 2 (BST-2/tetherin/CD317) (6, 7), and TRIM5α (8, 9)—have garnered substantial attention, since they specifically inhibit HIV-1 replication in vitro, and their patterns of diversification across primate lineages are suggestive of historical coevolutionary conflicts with retroviral pathogens (10–12). However, unlike variants found in nonhuman primates such as the rhesus macaque, the human allelic variant of Trim5α confers little, if any, inhibitory activity against HIV-1 and may, in fact, underlie our unique susceptibility to HIV-1 infection (13). The human APOBEC3 and BST-2 variants potently suppress HIV-1 replication in vitro and therefore represent promising candidates for innate immune-based therapeutic strategies (14).
Several members of the human APOBEC3 family of cytidine deaminases are capable of inhibiting HIV-1 replication to some degree (15), although evidence supporting an antiretroviral role of multiple members is often controversial and conflicting. Two family members, APOBEC3G (5) and APOBEC3F (16), are widely believed to exert strong inhibitory activity against HIV-1 (17). The human cytidine deaminases APOBEC3G and APOBEC3F serve as innate antiviral defense mechanisms by introducing C to U changes in the minus strand DNA of retroviruses and hepadnaviruses during replication (resulting in G to A mutations in the genomic sense strand sequence) (18). The HIV-1 genome, however, encodes the 23-kDa protein virion infectivity factor (Vif), which specifically counteracts this defense by promoting the proteolytic degradation of APOBEC3 in the host cell (19). In the absence of Vif expression, APOBEC3 is incorporated into virions, and the viral genome undergoes extensive G to A hypermutation in the coding strand, typically rendering it nonviable within a single replicative cycle (20). BST-2 is a type 2 integral membrane protein that inhibits retrovirus infection by restricting the release of fully formed progeny virions from infected cells (6, 7). Similar to the neutralization of APOBEC3 by HIV-1 Vif, BST-2 restriction is counteracted by an HIV-1 gene product, the 16-kDa viral protein U (Vpu). Vpu depletes BST-2 from the plasma membrane, allowing virions to detach from the cell and infect new targets (7). Consequently, the Vif-APOBEC3 and Vpu-BST-2 axes are emerging as attractive targets for therapeutic intervention (14).
The Vif-APOBEC3 and Vpu-BST-2 axes may be manipulated to increase cellular concentrations of these restriction factors and control HIV-1 infection in two basic fashions. The viral antagonist proteins Vif and Vpu could be targeted pharmacologically in host cells, abrogating their neutralization of APOBEC3 and BST-2, respectively (21, 22). Alternatively, the expression of these restriction factors may be induced to supraphysiologic levels, overriding the antagonistic activity of Vif and Vpu proteins in the producer cell (7, 23). In relation to the latter strategy, the cytokine IFN-α may hold important clues about the regulation and induction of these restriction factors in vivo.
Induction of IFN-α expression is a critical first step in the defense against a range of viral infections (24, 25). The antiretroviral activity of the IFN-α cytokine was demonstrated in vitro almost immediately after the discovery of HIV-1, and includes inhibition of HIV-1 reverse transcription, viral assembly, and virion release (26). IFN-α has been reported to suppress HIV-1 viremia in chronically infected individuals (27–29), and is known to induce APOBEC3 and BST-2 expression in a number of tissues and cell types in vitro (7, 30–32). However, to date, no data describe the effects of exogenous IFN-α treatment on the expression of these restriction factors in vivo, and the relevance of these factors to virologic control in chronically infected individuals is unknown. A legitimate evaluation of the therapeutic potential of APOBEC3 and BST-2 will require manipulation of these factors in vivo (14).
Our study makes use of a fortuitous synchronicity associated with the treatment of hepatitis C virus (HCV) disease in HIV/HCV-coinfected individuals. The current standard of treatment for HCV infection is combination therapy with ribavirin and pegylated IFN-α (IFN-α/riba) (28). In this study, we analyzed longitudinal clinical specimens from IFN-α/riba-treated, antiretroviral-naïve HIV/HCV-coinfected individuals to assess the extent to which IFN-α/riba treatment induces APOBEC3G, APOBEC3F, and BST-2 expression in vivo and characterize the influence of IFN-α/riba treatment on the replication and population genetics of HIV-1.
Results
IFN-α/Riba Treatment Potently Suppresses HIV-1 Viremia.
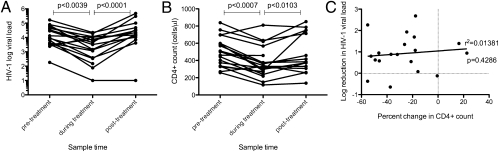
We initially examined the effects of IFN-α/riba treatment on HIV-1 plasma viral load in HIV/HCV-infected individuals enrolled in the Swiss HIV Cohort Study (SHCS) (33). Subject characteristics and IFN-α/riba treatment regimen are described in Table S1 (duration of HIV-1 infection was imputed by implementing an estimation method that was developed for SHCS data) (34). Median HIV-1 viral load and CD4+ count were 9,550 copies/mL and 501 cells/μL, respectively, at baseline. IFN-α/riba treatment resulted in a pronounced, transient reduction in HIV-1 viremia (Fig. 1A). This reduction is most probably driven entirely by IFN-α, since ribavirin exerts a negligible effect on HIV-1 viremia (28). Plasma viral load was reduced by −0.921 (±0.858) log10 copies/mL during treatment. Although CD4+ counts often declined during the treatment period (Fig. 1B), there was no correlation between change in viral load and CD4+ count, suggesting that target cell depletion (lymphopenia) is not the cause of plasma viral load reduction (Fig. 1C).
Fig. 1.
IFN-α/riba treatment strongly suppresses HIV-1 viremia. (A) Blood plasma HIV-1 viral load before, during, and after treatment. (B) CD4+ cell counts before, during, and after treatment. (C) Log10 reduction in HIV-1 viral load plotted against percent change in CD4+ lymphocyte count. Reported P values were obtained using a paired Wilcoxon test (A and B) or a Spearman's ρ (C).
IFN-α/Riba Treatment Induces APOBEC3G, APOBEC3F, and BST-2/Tetherin Expression in Vivo.
Since all previously published experiments describing the relationship between IFN-α and restriction factors involved laboratory-grade, nonpegylated IFN-α, we initially examined the effects of pegylated IFN-α on APOBEC3G expression in vitro to demonstrate that treatment with pegylated and nonpegylated versions of IFN-α yields similar results. Our data suggest that both versions of IFN-α exert nearly identical effects on expression in primary cells, and moreover, real-time PCR quantitation of APOBEC3G mRNA mirrors Western blot quantitation of cellular protein levels (Fig. S1).
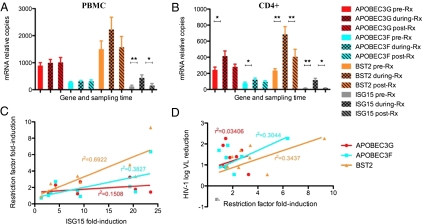
We measured the effects of IFN-α/riba treatment on APOBEC3G, APOBEC3F, and BST-2 expression in longitudinal peripheral blood mononuclear cell (PBMC) samples obtained from 19 HIV/HCV-coinfected individuals enrolled in the SHCS who were antiretroviral therapy (ART) -naïve during the entire observation period. Real-time PCR was used to measure restriction factor mRNA expression levels in unfractionated, unstimulated PBMCs before, during, and after IFN-α/riba treatment (Fig. 2A). The expression of IFN-stimulated gene 15 (ISG15) was measured as a positive control in these experiments to confirm IFN-α exposure and response during the treatment period. As expected, ISG15 showed a robust, significant elevation in expression during the treatment period with respect to pre- and posttreatment time points (P < 0.002, paired Wilcoxon test). However, in PBMCs, there were no statistically significant differences in levels of APOBEC3G, APOBEC3F, or BST-2 between the on-treatment samples and pre- or posttreatment time points.
Fig. 2.
APOBEC3G, APOBEC3F, and BST-2/tetherin are significantly induced in CD4+ T cells during IFN-α/riba treatment. (A) APOBEC3, tetherin/BST2, and IFN-stimulated gene 15 (ISG15) expressions in unfractionated PBMCs (mean expression is plotted, and error bars represent SEM). (B) APOBEC3, tetherin/BST2, and ISG15 expression in isolated CD4+ T cells. The expression of ISG15 was measured as a positive control in these experiments to confirm IFN-α exposure and response during the treatment period. *P < 0.05; **P < 0.005. (C) Relationship between restriction factor induction and ISG15 induction in CD4+ T cells during IFN-α/riba treatment. (D) Relationship between HIV-1 viral load reduction and restriction factor induction in CD4+ T cells during IFN-α/riba treatment.
We next examined the effects of IFN-α/riba treatment on expression levels in the CD4+ T-cell subset of PBMCs. CD4+ T cells are the primary HIV-1 target cells within peripheral tissues, and therefore, gene expression in this cellular subset is likely to be most relevant to viral production and propagation. CD4+ T cells were isolated from cryopreserved PBMCs with near 100% purity (Fig. S2), and restriction factor expression was assessed using real-time PCR (Fig. 2B). Similar to the observed pattern in PBMCs, ISG15 showed a robust, significant elevation in expression during the treatment period (7.5-fold mean induction) with respect to pre- and posttreatment time points. In contrast to PBMC-derived data, APOBEC3G and APOBEC3F exhibited moderate, statistically significant increases in expression levels during IFN-α/riba treatment in relation to the pretreatment time point (P < 0.05, paired Wilcoxon test), although comparison against posttreatment levels failed to achieve significance. BST-2 exhibited highly significant induction (2.9-fold mean induction) during IFN-α/riba treatment compared with both pre- and posttreatment time points in CD4+ T cells (P < 0.005, paired Wilcoxon test).
Relationships Between IFN-α Exposure, Restriction Factor Expression, and HIV-1 Viral Load.
Although the induction of APOBEC3 and BST-2 restriction factors in CD4+ T cells is provocative, the relevance of these host factors to control of viremia in chronically infected individuals during IFN-α/riba treatment requires additional substantiation. We investigated the correlations between extent of restriction factor induction in CD4+ T cells, extent of ISG15 induction (marker of IFN response), and HIV-1 viral load reduction during IFN-α/riba treatment (Fig. 2). Of the three restriction factors involved in our analysis, BST-2 induction showed the strongest significant correlation with ISG15 induction (r2 = 0.6922, P < 0.003, Pearson's r), suggesting that its expression is principally governed by IFN-α in vivo (Fig. 2C). APOBEC3F induction was also significantly correlated with ISG15 induction, whereas APOBEC3G exhibited only moderate IFN responsiveness (APOBEC3G: r2 = 0.1508, P < 0.151; APOBEC3F: r2 = 0.3827 and P < 0.038). Log10 HIV-1 viral load reduction showed the strongest significant correlation with BST-2 induction (r2 = 0.3437, P < 0.049, Pearson's r) and a similar but secondary correlation with APOBEC3F (r2 = 0.3044, P < 0.062); the correlation with APOBEC3G induction was minimal to nonexistent (r2 = 0.03406, P < 0.317) (Fig. 2D). These relationships between HIV-1 viral load reduction and restriction factor expression suggest that, of the three factors analyzed, BST-2 plays the most significant role in the IFN-α–mediated suppression of HIV-1 viremia.
HIV-1 Hypermutation Is Correlated with APOBEC3 Expression During IFN-α/Riba Treatment.
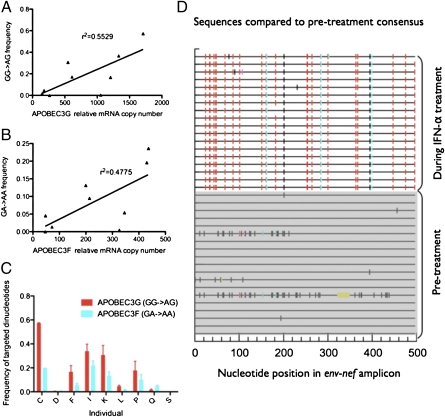
We generated and analyzed sequences of HIV-1 before and during IFN-α/riba treatment to examine the effects of treatment on viral population genetics, searching specifically for viral evolutionary patterns that would reflect enhanced selection pressure from the APOBEC3 and BST-2 restriction factors. In the case of APOBEC3G and APOBEC3F, enhancement of their antiviral activities should result in increased viral hypermutation during the IFN-α/riba treatment period (APOBEC3G and APOBEC3F principally target GG [GG → AG] and GA [GA → AA] dinucleotides, respectively) (18, 35). We generated clonal sequences of a 500-bp region of the HIV-1 DNA genome spanning the env and nef genes to examine levels of hypermutation. Replicate PCR products were proportionately pooled before cloning to minimize the probability of template resampling, and ∼20 clones per time point per individual were analyzed (36). This sequencing strategy was chosen for two principal reasons. PBMC-derived DNA was examined, because hypermutated genomes are difficult to sample in the RNA population; they are likely to be replication-incompetent and are expected to only appear transiently within the RNA compartment (17). The env-nef region of the viral genome was chosen since multiple studies have confirmed that APOBEC3 exhibits a pronounced 5′ to 3′ editing gradient, and therefore, the env and nef genes should be especially susceptible to APOBEC3 cytidine deaminase activity (18, 37). APOBEC3G-mediated hypermutation level during IFN-α/riba treatment was defined as the average proportion of GG dinucleotides in the pretreatment viral consensus sequence that was mutated to AG in treatment-associated clonal sequences. Similarly, APOBEC3F-mediated hypermutation level during treatment was defined as the average proportion of GA dinucleotides in the pretreatment viral consensus sequence that was mutated to AA in treatment-associated clonal sequences. Enforcing the GG/GA dinucleotide context ensures that the analyzed mutations are driven by APOBEC3G or APOBEC3F editing rather than simple viral polymerase error or the possible and controversial mutagenic effects of ribavirin therapy (38). Our results suggest that treatment-associated viral hypermutation levels were not correlated with fold induction of APOBEC3G and APOBEC3F during IFN-α/riba treatment compared with pretreatment expression. However, this lack of association may be due to the considerable interindividual variation observed in pretreatment APOBEC3 mRNA copy numbers. We therefore also compared viral hypermutation levels with peak copy number during treatment, a more direct measure of APOBEC3 potency and cytidine deaminase activity. Using this more biologically intuitive analytical approach, we determined that APOBEC3G- and APOBEC3F-associated viral hypermutation levels were significantly correlated with APOBEC3G and APOBEC3F mRNA copy numbers, respectively, during the IFN-α/riba treatment period (P < 0.034 and P < 0.019, Spearman's ρ) (Fig. 3 A and B). Additionally, in line with the higher relative expression of APOBEC3G in PBMCs and isolated CD4+ T cells (Fig. 2 A and B), APOBEC3G-associated G to A mutations were observed significantly more frequently than APOBEC3F-associated G to A mutations (P < 0.039, paired Wilcoxon test) (Fig. 3 C and D).
Fig. 3.
APOBEC3-induced HIV-1 hypermutation during IFN-α/riba treatment. (A) Relationship between GG to AG (APOBEC3G dinucleotide context) viral hypermutation level during treatment and APOBEC3G relative mRNA copy number. (B) Relationship between GA to AA (APOBEC3F dinucleotide context) viral hypermutation level during treatment and APOBEC3F relative mRNA copy number. (C) Relative frequency of APOBEC3G- and APOBEC3F-associated mutations (mean values are plotted and error bars represent SEM). (D) Example of treatment-associated viral hypermutation from a single individual. Red tick mark, GG → AG mutation (APOBEC3G pattern); cyan, GA → AA (APOBEC3F pattern); green, GC → AC; magenta, GT → AT; black, all other mutations; yellow, deletion (compared with pretreatment consensus sequence). Shaded sequences in the bottom half of the panel represent pretreatment PBMC-derived HIV-1 DNA clones, and the sequences in the top, unshaded one of the panel represent PBMC-derived HIV-1 DNA clones from the IFN-α/riba treatment period. Sequences span an ∼500-bp region of the env and nef genes, and ∼20 clones per time point per individual were analyzed.
Evolution of the HIV-1 Vpu Protein During IFN-α/Riba Therapy.
Unlike the APOBEC3 enzymes, BST-2 activity is not associated with a particular viral mutational signature. We looked instead for indirect effects of enhanced BST-2 pressure on the viral sequence by focusing on the evolution of the HIV-1 Vpu protein (viral antagonist of BST-2) in response to IFN-α/riba treatment. We hypothesized that enhanced BST-2 pressure would select for Vpu variants with greater BST-2 neutralization capacity. Vpu amino acid sequences acquired several substitutions during IFN-α/riba treatment (Fig. 4A). Two of these mutations, A11G and S61A, occurred in highly conserved, previously established Vpu sequence domains involved in BST-2 down-regulation (39, 40). We assessed the phenotypic consequence of these treatment-associated mutations by mutagenizing NL4-3 Vpu sequences at these positions and measuring the BST-2 down-regulation capacity of these mutant alleles in a subgenomic in vitro model (41). Flow cytometric analysis of the transfected HeLa cells, which express BST-2 constitutively, revealed that the two mutant alleles down-regulated BST-2 more efficiently than the WT HIV-1 NL4-3 allele, and the bimodal BST-2 expression pattern exhibited in the WT cultures suggests that the A11G and S61A treatment-associated mutations likely enhanced the expression, stability, and cellular concentration of the Vpu antagonist protein (Fig. 4B). Notably, whereas the A11G mutation fits a well-described GxxxG transmembrane interaction motif and could potentially enhance the interaction between Vpu and BST-2 (34), the S61A mutation has been reported to stabilize the Vpu protein (35). Taken together, our viral genotype and phenotype data are compatible with enhanced selection pressure from the APOBEC3 and BST-2/tetherin retroviral restriction factors during IFN-α/riba treatment.
Fig. 4.
Evolution of the HIV-1 Vpu protein during IFN-α/riba treatment. (A) Amino acid sequences of HIV-1 Vpu generated from nine individuals before and during IFN-α/riba treatment. Dashes indicate identity to the HXB2 reference sequence. Vpu positions 11 and 61 are shaded. (B) Effects of A11G and S61A treatment-associated Vpu mutations on Vpu-mediated down-regulation of BST-2 surface expression measured by flow cytometry. Shaded histograms represent mock DNA-transfected cells (negative control).
Discussion
The host gene expression and viral genetic data collected in this study suggest that the induction of APOBEC3 and BST-2/tetherin retroviral restriction factors plays a critical role in the suppressive effects of exogenous IFN-α treatment on HIV-1 replication in vivo. Moreover, the lack of correlation between HIV-1 viral load and restriction factor mRNA copy numbers before IFN-α/riba treatment (Fig. S3) suggests that variation in expression only has measurable virological consequences at supraphysiologic concentrations induced by pharmacological manipulation. Accordingly, the host gene expression profile associated with endogenous IFN-α expression is correlated with high viral load and poor disease outcomes, suggesting that the benefits of IFN-α may only be evident at concentrations achieved through exogenous administration (42). Antiretroviral strategies involving the enhancement of restriction factor expression in HIV-1–infected individuals may prove to be beneficial and should be explored in tandem with pharmacological strategies targeting viral antagonist proteins such as Vif and Vpu (21, 22). A thorough examination of IFN-α signal transduction pathways may allow us to identify specific mechanisms to enhance restriction factor expression, while avoiding the nonspecific, toxic cascade typically associated with IFN-α treatment.
Our viral sequence data exhibit a strong, intuitive positive correlation between extent of HIV-1 hypermutation and APOBEC3 mRNA expression levels in CD4+ T cells during IFN-α/riba treatment. Close scrutiny of these data, however, reveal a seemingly paradoxical relationship between the inferred antiviral potencies and observed hypermutation levels associated with APOBEC3G and APOBEC3F. HIV-1 viral load reduction during IFN-α/riba treatment was more strongly correlated with APOBEC3F induction, but curiously, HIV-1 DNA sequences from the treatment period exhibited higher levels of APOBEC3G-mediated editing. A possible explanation involves the noncytidine deaminase antiretroviral activity of these factors. Both factors are capable of suppressing HIV-1 replication to some extent in the absence of cytidine deaminase activity, and the accumulation of HIV-1 reverse transcription products in target cells is markedly inhibited, even when the proteins’ mutator domains are disrupted. Interestingly, mutator activity is more critical for APOBEC3G-mediated suppression of HIV-1, suggesting that nondeaminase capacity is a larger component of APOBEC3F antiretroviral effects (43). These observations are in alignment with our hypermutation data; this finding potentially explains how APOBEC3F had a greater inferred antiviral potency, while APOBEC3G-mediated editing was more pervasive in patient-derived sequences during IFN-α/riba treatment.
There are certain caveats associated with these data and their interpretation. Unlike studies involving in vitro or animal models, the relevance of restriction factors to IFN-α/riba-mediated control of HIV-1 cannot be explored through silencing or KO experiments. Therefore, it is difficult to unequivocally demonstrate a causal relationship between restriction factor expression and the suppression of HIV-1 replication using a patient-based study design. Consideration of our findings within the context of the robust literature describing a causal relationship between IFN-α, restriction factor induction, and retroviral suppression lends credibility to our in vivo observations. For instance, recent work by Liberatore and colleagues demonstrates that the suppression of a murine retrovirus by interferon-α is markedly diminished in BST-2/tetherin-deficient mice (44). Experiments by Peng et al. (32) and Chen et al. (31) show that siRNA-mediated knockdown of APOBEC3 in vitro largely abrogates the suppressive effects of IFN-α on HIV-1 replication in primary monocyte-derived macrophages and CD4+ T cells, respectively. Additional caveats stem from the basic biology of the restriction factors themselves. Although high levels of APOBEC3 expression may genetically erode the HIV-1 quasispecies in vivo to the point of population collapse, intermediate levels of induction may instead accelerate evolution of antiretroviral resistance, immune escape, and cellular tropism (coreceptor use phenotype), harming the host rather than the virus (45, 46). A true error or extinction threshold for HIV-1 is not determined, and additional experimentation is warranted to explore the effects of mutation rate modulation on HIV-1 fitness and persistence in vivo. However, recent data reveal that increased APOBEC3G and APOBEC3F expression levels in rhesus monkeys are associated with lower simian immunodeficiency virus viral loads and prolonged survival, reinforcing the concept that induction of these factors ultimately has beneficial effects on lentiviral disease progression (47). In regards to the antiretroviral effects of BST-2 induction, the impact of BST-2 surface expression on the cell to cell spread of HIV-1 may differ from its effect on the dissemination of free virus, and some reported data suggest that cell to cell transmission may, in fact, be enhanced by this restriction mechanism (48, 49). In addition, ISG15, our marker of IFN exposure in this study, has an emerging antiviral role in vitro. The possibility exists that ISG15 and other established (or unknown) IFN-induced cellular cofactors not measured in our study have contributed to the observed suppression of HIV-1 viremia during IFN-α/riba treatment (50). However, the weak, statistically insignificant correlation between HIV-1 viral load reduction and ISG15 induction during treatment (r2 = 0.2171, P = 0.1031, Pearson's r) suggests that APOBEC3 and BST-2 are likely to play a more important role in the IFN-α–mediated suppression of HIV-1 in vivo. Lastly, individuals with severe immunodeficiency are not represented in this study, because they would not be considered appropriate for deferral of ART (ART was an exclusion criterion). It will be necessary to determine the generalizability of these findings through the study of additional subjects reflecting a broader range of clinical stages.
The broad antiviral properties of IFN-α invite speculation that the relationships between APOBEC3 and BST-2 induction and the in vivo antiviral effects on HIV-1 observed in this study may translate to other viral infections. APOBEC3 expression and cyditine deaminase activity in hepatocytes are known to exert a strong antiviral effect against hepatic viral pathogens, namely hepadnaviruses such as hepatitis B virus (51). Influenza virus and HCV encode proteins thought to be important for late-stage infection events, including viral release, that have predicted porin-like features analogous to the HIV-1 Vpu protein. A number of other enveloped viruses, including HIV-2, simian immunodeficiency virus, Ebola, and KSHV, encode specific proteins as countermeasures to evade BST-2–mediated restriction (52). Therefore, up-regulation of APOBEC and BST-2 expression likely represents common, generic mechanisms mediating the antiviral properties of IFN-α, and development of interventions that enhance APOBEC3 and BST-2 expression may have applications that reach far beyond the treatment of HIV-1 disease.
Materials and Methods
Subjects and Specimens.
Nineteen HIV/HCV-coinfected, ART-naïve individuals from the SHCS who underwent IFN-α/riba treatment were studied retrospectively (Table S1). The SHCS is a nationwide, clinic-based prospective cohort study with continuous enrolment and at least semiannual study visits (www.shcs.ch) (33). Blood plasma and cryopreserved PBMC were collected from all subjects. CD4+ T cells were negatively selected from PBMC (Fig. S2). See SI Materials and Methods for further details.
Expression Profiling.
Total RNA was extracted from PBMC and CD4+ T cells using TRIzol. RNA was transcribed into cDNA using random primers and SuperScript III Reverse Transcriptase. TaqMan primers/probes were used to evaluate APOBEC3G, APOBEC3F, BST-2, and ISG15 expression. Raw cycle threshold numbers of amplified gene products were normalized to the housekeeping gene ribosomal protein L13a (RPL-13A) to control for cDNA input amounts. Fold induction was determined using the comparative cycle threshold (CT) method (53). See SI Materials and Methods for further details.
HIV-1 env-nef Clonal Sequencing.
Genomic DNA was extracted using the TRIzol method. The HIV-1 env-nef region (532 bp) was amplified by nested PCR using previously published primer sequences and PCR cycling conditions (36). Products were cloned and sequenced in both directions using universal M13 primers. See SI Materials and Methods for further details.
HIV-1 vpu Population Sequencing and Phenotypic Analysis.
Nine individuals were chosen for vpu analyses based on sample availability. Viral RNA was extracted and purified from 1 mL plasma using the QIAmp viral RNA kit (Qiagen) after an initial concentration step (ultracentrifugation at 53,000 × g). Reverse transcription was performed using SuperScript II Reverse Transcriptase (Invitrogen) using a gene-specific primer to generate cDNA. Nested PCR was performed using the Expand High-Fidelity PCR System (Roche). Generated PCR products were gel-purified using the QIAquick Gel Extraction Kit (Qiagen) and sequenced in both directions using the nested amplification primers. HeLa P4.R5 cells were transfected with pcDNA3.1-based plasmids expressing WT or mutagenized versions of HIV-1 vpu, and effects of Vpu proteins on BST-2/tetherin surface expression were assessed by flow cytometry as previously described (41). See SI Materials and Methods for further details.
Sequence and Statistical Analysis.
Phylogenetic reconstruction and BLAST were used to inspect sequences for interindividual contamination or contamination with HIV-1 laboratory strains. Sequences were aligned using Multalin (54), and analyses were performed using Hypermut (www.hiv.lanl.gov) (55) and the HyPhy software package (www.hyphy.org) (56). HIV-1 subtype was determined by applying the REGA HIV-1 subtyping tool (57) to pol sequences that were previously generated for drug resistance genotyping at the SHCS. A battery of nonparametric statistical tests (Spearman's rank correlation coefficient, Mann–Whitney u test, Paired Wilcoxon test) was applied to gene expression and genetic data using GraphPad Prism v5.0c. In cases where a (parametric) Pearson's r test was used, the Kolmogorov–Smirnov test was implemented beforehand to determine that the data were distributed normally.
Supplementary Material
Acknowledgments
We thank the patients who participate in the Swiss HIV Cohort Study and the physicians and study nurses for excellent patient care. This study was supported by National Institutes of Health Grant 1K01DA024654 (to S.K.P.), Grant AI081668, and an American Recovery and Reinvestment Act (ARRA) supplement (to J.G.). Additional support was provided by Swiss HIV Cohort Study Project 594; the Veterans Affairs Merit Review (K.F., S.Y., and J.K.W.); and Swiss National Science Foundation Grants 324730-130865 (to H.F.G.), R01NS051132, R21MH083573, and R56AI91573. The Swiss HIV Cohort Study is supported by Swiss National Science Foundation Grant 33CSC0-108787 and the Swiss HIV Cohort Study Research Foundation.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession nos. JQ479337–JQ480008).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1111573109/-/DCSupplemental.
References
- 1.Piot P, Bartos M, Ghys PD, Walker N, Schwartländer B. The global impact of HIV/AIDS. Nature. 2001;410:968–973. doi: 10.1038/35073639. [DOI] [PubMed] [Google Scholar]
- 2.Münk C, Brandt SM, Lucero G, Landau NR. A dominant block to HIV-1 replication at reverse transcription in simian cells. Proc Natl Acad Sci USA. 2002;99:13843–13848. doi: 10.1073/pnas.212400099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cowan S, et al. Cellular inhibitors with Fv1-like activity restrict human and simian immunodeficiency virus tropism. Proc Natl Acad Sci USA. 2002;99:11914–11919. doi: 10.1073/pnas.162299499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Simon JH, Gaddis NC, Fouchier RA, Malim MH. Evidence for a newly discovered cellular anti-HIV-1 phenotype. Nat Med. 1998;4:1397–1400. doi: 10.1038/3987. [DOI] [PubMed] [Google Scholar]
- 5.Sheehy AM, Gaddis NC, Choi JD, Malim MH. Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nature. 2002;418:646–650. doi: 10.1038/nature00939. [DOI] [PubMed] [Google Scholar]
- 6.Neil SJ, Zang T, Bieniasz PD. Tetherin inhibits retrovirus release and is antagonized by HIV-1 Vpu. Nature. 2008;451:425–430. doi: 10.1038/nature06553. [DOI] [PubMed] [Google Scholar]
- 7.Liberatore RA, Bieniasz PD. Tetherin is a key effector of the antiretroviral activity of type I interferon in vitro and in vivo. Proc Natl Acad Sci USA. 2011;108:18097–18101. doi: 10.1073/pnas.1113694108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stremlau M, et al. The cytoplasmic body component TRIM5alpha restricts HIV-1 infection in Old World monkeys. Nature. 2004;427:848–853. doi: 10.1038/nature02343. [DOI] [PubMed] [Google Scholar]
- 9.Towers G, et al. A conserved mechanism of retrovirus restriction in mammals. Proc Natl Acad Sci USA. 2000;97:12295–12299. doi: 10.1073/pnas.200286297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McNatt MW, et al. Species-specific activity of HIV-1 Vpu and positive selection of tetherin transmembrane domain variants. PLoS Pathog. 2009;5:e1000300. doi: 10.1371/journal.ppat.1000300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sawyer SL, Wu LI, Emerman M, Malik HS. Positive selection of primate TRIM5alpha identifies a critical species-specific retroviral restriction domain. Proc Natl Acad Sci USA. 2005;102:2832–2837. doi: 10.1073/pnas.0409853102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sawyer SL, Emerman M, Malik HS. Ancient adaptive evolution of the primate antiviral DNA-editing enzyme APOBEC3G. PLoS Biol. 2004;2:E275. doi: 10.1371/journal.pbio.0020275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kaiser SM, Malik HS, Emerman M. Restriction of an extinct retrovirus by the human TRIM5alpha antiviral protein. Science. 2007;316:1756–1758. doi: 10.1126/science.1140579. [DOI] [PubMed] [Google Scholar]
- 14.Neil S, Bieniasz P. Human immunodeficiency virus, restriction factors, and interferon. J Interferon Cytokine Res. 2009;29:569–580. doi: 10.1089/jir.2009.0077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bishop KN, et al. Cytidine deamination of retroviral DNA by diverse APOBEC proteins. Curr Biol. 2004;14:1392–1396. doi: 10.1016/j.cub.2004.06.057. [DOI] [PubMed] [Google Scholar]
- 16.Zheng YH, et al. Human APOBEC3F is another host factor that blocks human immunodeficiency virus type 1 replication. J Virol. 2004;78:6073–6076. doi: 10.1128/JVI.78.11.6073-6076.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Albin JS, Harris RS. Interactions of host APOBEC3 restriction factors with HIV-1 in vivo: Implications for therapeutics. Expert Rev Mol Med. 2010;12:e4. doi: 10.1017/S1462399409001343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yu Q, et al. Single-strand specificity of APOBEC3G accounts for minus-strand deamination of the HIV genome. Nat Struct Mol Biol. 2004;11:435–442. doi: 10.1038/nsmb758. [DOI] [PubMed] [Google Scholar]
- 19.Stopak K, de Noronha C, Yonemoto W, Greene WC. HIV-1 Vif blocks the antiviral activity of APOBEC3G by impairing both its translation and intracellular stability. Mol Cell. 2003;12:591–601. doi: 10.1016/s1097-2765(03)00353-8. [DOI] [PubMed] [Google Scholar]
- 20.Lecossier D, Bouchonnet F, Clavel F, Hance AJ. Hypermutation of HIV-1 DNA in the absence of the Vif protein. Science. 2003;300:1112. doi: 10.1126/science.1083338. [DOI] [PubMed] [Google Scholar]
- 21.Nathans R, et al. Small-molecule inhibition of HIV-1 Vif. Nat Biotechnol. 2008;26:1187–1192. doi: 10.1038/nbt.1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Khoury G, Ewart G, Luscombe C, Miller M, Wilkinson J. Antiviral efficacy of the novel compound BIT225 against HIV-1 release from human macrophages. Antimicrob Agents Chemother. 2010;54:835–845. doi: 10.1128/AAC.01308-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ejima T, Hirota M, Mizukami T, Otsuka M, Fujita M. An anti-HIV-1 compound that increases steady-state expression of apoplipoprotein B mRNA-editing enzyme-catalytic polypeptide-like 3G. Int J Mol Med. 2011;28:613–616. doi: 10.3892/ijmm.2011.737. [DOI] [PubMed] [Google Scholar]
- 24.Isaacs A, Lindenmann J. Virus interference. I. The interferon. Proc R Soc Lond B Biol Sci. 1957;147:258–267. [PubMed] [Google Scholar]
- 25.Stetson DB, Medzhitov R. Type I interferons in host defense. Immunity. 2006;25:373–381. doi: 10.1016/j.immuni.2006.08.007. [DOI] [PubMed] [Google Scholar]
- 26.Poli G, Orenstein JM, Kinter A, Folks TM, Fauci AS. Interferon-alpha but not AZT suppresses HIV expression in chronically infected cell lines. Science. 1989;244:575–577. doi: 10.1126/science.2470148. [DOI] [PubMed] [Google Scholar]
- 27.Aguilar Marucco D, et al. Antiretroviral activity of pegylated interferon alfa-2a in patients co-infected with HIV/hepatitis C virus. J Antimicrob Chemother. 2007;59:565–568. doi: 10.1093/jac/dkl497. [DOI] [PubMed] [Google Scholar]
- 28.Torriani FJ, et al. APRICOT Study Group Peginterferon Alfa-2a plus ribavirin for chronic hepatitis C virus infection in HIV-infected patients. N Engl J Med. 2004;351:438–450. doi: 10.1056/NEJMoa040842. [DOI] [PubMed] [Google Scholar]
- 29.Asmuth DM, et al. AIDS Clinical Trials Group A5192 Team Safety, tolerability, and mechanisms of antiretroviral activity of pegylated interferon Alfa-2a in HIV-1-monoinfected participants: A phase II clinical trial. J Infect Dis. 2010;201:1686–1696. doi: 10.1086/652420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tanaka Y, et al. Anti-viral protein APOBEC3G is induced by interferon-alpha stimulation in human hepatocytes. Biochem Biophys Res Commun. 2006;341:314–319. doi: 10.1016/j.bbrc.2005.12.192. [DOI] [PubMed] [Google Scholar]
- 31.Chen K, et al. Alpha interferon potently enhances the anti-human immunodeficiency virus type 1 activity of APOBEC3G in resting primary CD4 T cells. J Virol. 2006;80:7645–7657. doi: 10.1128/JVI.00206-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Peng G, Lei KJ, Jin W, Greenwell-Wild T, Wahl SM. Induction of APOBEC3 family proteins, a defensive maneuver underlying interferon-induced anti-HIV-1 activity. J Exp Med. 2006;203:41–46. doi: 10.1084/jem.20051512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schoeni-Affolter F, et al. Swiss HIV Cohort Study Cohort profile: The Swiss HIV Cohort study. Int J Epidemiol. 2010;39:1179–1189. doi: 10.1093/ije/dyp321. [DOI] [PubMed] [Google Scholar]
- 34.Taffé P, May M. Swiss HIV Cohort Study A joint back calculation model for the imputation of the date of HIV infection in a prevalent cohort. Stat Med. 2008;27:4835–4853. doi: 10.1002/sim.3294. [DOI] [PubMed] [Google Scholar]
- 35.Liddament MT, Brown WL, Schumacher AJ, Harris RS. APOBEC3F properties and hypermutation preferences indicate activity against HIV-1 in vivo. Curr Biol. 2004;14:1385–1391. doi: 10.1016/j.cub.2004.06.050. [DOI] [PubMed] [Google Scholar]
- 36.Gandhi SK, Siliciano JD, Bailey JR, Siliciano RF, Blankson JN. Role of APOBEC3G/F-mediated hypermutation in the control of human immunodeficiency virus type 1 in elite suppressors. J Virol. 2008;82:3125–3130. doi: 10.1128/JVI.01533-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Suspène R, Rusniok C, Vartanian JP, Wain-Hobson S. Twin gradients in APOBEC3 edited HIV-1 DNA reflect the dynamics of lentiviral replication. Nucleic Acids Res. 2006;34:4677–4684. doi: 10.1093/nar/gkl555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vignuzzi M, Stone JK, Andino R. Ribavirin and lethal mutagenesis of poliovirus: Molecular mechanisms, resistance and biological implications. Virus Res. 2005;107:173–181. doi: 10.1016/j.virusres.2004.11.007. [DOI] [PubMed] [Google Scholar]
- 39.Estrabaud E, et al. Regulated degradation of the HIV-1 Vpu protein through a betaTrCP-independent pathway limits the release of viral particles. PLoS Pathog. 2007;3:e104. doi: 10.1371/journal.ppat.0030104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vigan R, Neil SJ. Determinants of tetherin antagonism in the transmembrane domain of the human immunodeficiency virus type 1 Vpu protein. J Virol. 2010;84:12958–12970. doi: 10.1128/JVI.01699-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Skasko M, et al. BST-2 is rapidly down-regulated from the cell surface by the HIV-1 protein Vpu: Evidence for a post-ER mechanism of Vpu-action. Virology. 2011;411:65–77. doi: 10.1016/j.virol.2010.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rempel H, Sun B, Calosing C, Pillai SK, Pulliam L. Interferon-alpha drives monocyte gene expression in chronic unsuppressed HIV-1 infection. AIDS. 2010;24:1415–1423. doi: 10.1097/QAD.0b013e32833ac623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Holmes RK, Koning FA, Bishop KN, Malim MH. APOBEC3F can inhibit the accumulation of HIV-1 reverse transcription products in the absence of hypermutation. Comparisons with APOBEC3G. J Biol Chem. 2007;282:2587–2595. doi: 10.1074/jbc.M607298200. [DOI] [PubMed] [Google Scholar]
- 44.Liberatore RA, Bieniasz PD. Tetherin is a key effector of the antiretroviral activity of type I interferon in vitro and in vivo. Proc Natl Acad Sci USA. 2011;108:18097–18101. doi: 10.1073/pnas.1113694108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mulder LC, Harari A, Simon V. Cytidine deamination induced HIV-1 drug resistance. Proc Natl Acad Sci USA. 2008;105:5501–5506. doi: 10.1073/pnas.0710190105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pillai SK, Wong JK, Barbour JD. Turning up the volume on mutational pressure: Is more of a good thing always better? (A case study of HIV-1 Vif and APOBEC3) Retrovirology. 2008;5:26. doi: 10.1186/1742-4690-5-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mussil B, Sauermann U, Motzkus D, Stahl-Hennig C, Sopper S. Increased APOBEC3G and APOBEC3F expression is associated with low viral load and prolonged survival in simian immunodeficiency virus infected rhesus monkeys. Retrovirology. 2011;8:77. doi: 10.1186/1742-4690-8-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Casartelli N, et al. Tetherin restricts productive HIV-1 cell-to-cell transmission. PLoS Pathog. 2010;6:e1000955. doi: 10.1371/journal.ppat.1000955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jolly C, Booth NJ, Neil SJ. Cell-cell spread of human immunodeficiency virus type 1 overcomes tetherin/BST-2-mediated restriction in T cells. J Virol. 2010;84:12185–12199. doi: 10.1128/JVI.01447-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Skaug B, Chen ZJ. Emerging role of ISG15 in antiviral immunity. Cell. 2010;143:187–190. doi: 10.1016/j.cell.2010.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Suspène R, et al. Extensive editing of both hepatitis B virus DNA strands by APOBEC3 cytidine deaminases in vitro and in vivo. Proc Natl Acad Sci USA. 2005;102:8321–8326. doi: 10.1073/pnas.0408223102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Douglas JL, et al. The great escape: Viral strategies to counter BST-2/tetherin. PLoS Pathog. 2010;6:e1000913. doi: 10.1371/journal.ppat.1000913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vandesompele J, et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3:RESEARCH0034. doi: 10.1186/gb-2002-3-7-research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Corpet F. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 1988;16:10881–10890. doi: 10.1093/nar/16.22.10881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rose PP, Korber BT. Detecting hypermutations in viral sequences with an emphasis on G —> A hypermutation. Bioinformatics. 2000;16:400–401. doi: 10.1093/bioinformatics/16.4.400. [DOI] [PubMed] [Google Scholar]
- 56.Pond SL, Frost SD, Muse SV. HyPhy: Hypothesis testing using phylogenies. Bioinformatics. 2005;21:676–679. doi: 10.1093/bioinformatics/bti079. [DOI] [PubMed] [Google Scholar]
- 57.de Oliveira T, et al. An automated genotyping system for analysis of HIV-1 and other microbial sequences. Bioinformatics. 2005;21:3797–3800. doi: 10.1093/bioinformatics/bti607. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.