Abstract
Transcriptome profiling studies suggest that a large fraction of the genome is transcribed and many transcripts function independent of their protein coding potential. The relevance of noncoding RNAs (ncRNAs) in normal physiological processes and in tumorigenesis is increasingly recognized. Here, we describe consistent and significant differences in the distribution of sense and antisense transcripts between normal and neoplastic breast tissues. Many of the differentially expressed antisense transcripts likely represent long ncRNAs. A subset of genes that mainly generate antisense transcripts in normal but not cancer cells is involved in essential metabolic processes. These findings suggest fundamental differences in global RNA regulation between normal and cancer cells that might play a role in tumorigenesis.
Genome-wide unbiased assessment of RNA levels has been a useful tool to delineate physiological and pathological processes (1). Based on recent studies, a large fraction of the genome is transcribed, generating a vast array of RNA species of different lengths, protein coding potential, and regulatory function (2). Our understanding of the physiological function of noncoding RNAs (ncRNAs) is just beginning to emerge. Among all ncRNAs, micro-RNAs have been analyzed most extensively and their importance in tumorigenesis is well documented (3). Other classes of ncRNAs, including cis-natural antisense transcripts (cis-NATs), are poorly characterized. Initially, many of these ncRNAs were thought to be transcriptional noise, but the regulation of their expression in a cell type- and developmental stage-specific manner and their evolutionary conservation suggest physiological functions (4, 5). Indeed, several recent studies described a large set of ncRNAs associated with chromatin-modifying complexes, which modulate gene expression by regulating the localization of these complexes (6–8).
The role of ncRNAs in tumorigenesis has not been investigated comprehensively. Large-scale transcriptome analysis in head and neck cancer identified over 2,000 ncRNAs (9) that were down-regulated in clear-cell carcinoma (10). In prostate cancer, a subset of ncRNAs correlates with the degree of cellular differentiation (9). A recent report described altered expression of ncRNAs transcribed from ultraconserved genomic regions in multiple human cancer types (11). A family of mitochondrial ncRNAs is also differentially expressed between normal and cancer cells, but their biological function is unknown (12). The expression of HOTAIR, a large intervening ncRNA regulating the expression of the HOXD locus, is increased in primary breast tumors and correlates with the risk for distant metastasis (8).
Here, we describe significant and consistent differences in the distribution of sense and antisense transcripts between normal and neoplastic breast epithelial cells based on serial analysis of gene expression combined with high-throughput sequencing (SAGE-Seq) and identify cell type- and tissue-specific antisense transcripts. We also demonstrate by RT-PCR that several of these correspond to long ncRNAs (lncRNAs). Our results suggest that abnormalities in the regulation of antisense transcripts may play a role in breast tumorigenesis.
Results and Discussion
Analysis of SAGE-Seq Data.
We analyzed the transcriptomes of mammary epithelial cells purified from normal and neoplastic breast tissues (13, 14) using SAGE-Seq (15, 16). During the course of this analysis, we noticed substantial numbers of antisense tags that we investigated in further detail. We used cells purified utilizing different cell surface markers (e.g., CD24, CD44) (13, 14) (Table S1); however, because all normal cells and all cancer cells displayed the same phenotype with respect to antisense expression, the actual identify of the cells is irrelevant to the observations made in this study.
SAGE-Seq libraries were generated from 21 samples, and a total of 90,803,826 tags were sequenced. Tag counts were normalized to 5 million per sample sequencing depth based on maximum likelihood estimation of tag frequency. Tags were first mapped to sense strands, followed by mapping the no matches to antisense strands of exonic and intronic regions of Reference Sequence (RefSeq) genes (Dataset S1). Very low abundance genes with <5 tags in all samples combined were excluded to minimize potential noise. For global analysis, all unique sense and antisense tags were summed for each gene (Fig. S1A). The total filtered sense and antisense tag counts were not significantly different among samples (Fig. S1B and Dataset S1). The overall percentage of antisense tags was 14.3%, which is in good agreement with asymmetrical strand-specific analysis of gene expression (ASSAGE) data (17).
Differences in Antisense and Sense Transcriptomes Between Normal and Cancer Samples.
To assess the potential cell or tissue type specificity of strand-specific transcript distribution, we calculated the percentage of antisense tags for each gene. Many genes showed a high percentage of antisense tags (e.g., 1,931 genes showed >50% antisense tags in normal samples), indicating substantial antisense transcription in RefSeq genes (Fig. 1A). Interestingly, the number of genes showing a high percentage of antisense tags was much lower in tumors than in normal samples; this difference was consistently observed in each sample in both groups (Fig. 1B), and the percentage of antisense tags per gene showed a much broader distribution in the normal group than in the cancer group. The mean of the normal group was significantly larger than that of the cancer group (P = 6.804e-06, Wilcoxon rank-sum test). The distribution of the total number of sense tags per gene was significantly broader and larger in cancer (P = 6.804e-06, Wilcoxon rank-sum test), whereas the distribution of the number of antisense tags per gene was not different among samples (log mean value P = 0.4221, Wilcoxon rank-sum test) (Fig. S1C). Therefore, the overall difference in percentage of antisense tags might be attributable, in part, to increased sense transcript levels in tumors, because the sense-to-antisense transcript ratios were much higher in tumors compared with normal samples (Fig. 1C). A subset of genes clearly showed a high proportion of antisense transcripts only in normal samples, however, implicating potential functional relevance; thus, we analyzed these genes in further detail.
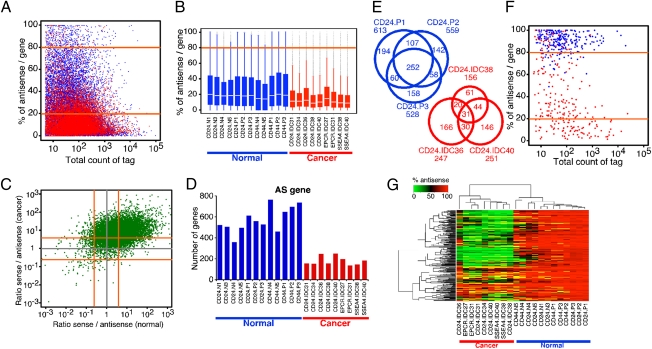
Fig. 1.
Comparison of antisense transcriptomes of normal and neoplastic breast epithelial cells. (A) Percentage of antisense tags relative to all tags in each gene is plotted using average tag counts in all normal (blue) and cancer (red) samples. Orange lines mark 80% and 20% values, which were used as criteria for gene classification into AS, S, and SAS groups. (B) Box plot depicting the percentage of antisense/gene ratio in each normal (blue) and cancer (red) sample. The box indicates the 25th and 75th percentiles; the white bar indicates the median; the whiskers extend to the most extreme data point, which is no more than 1.5 times the interquartile range from the box; and outliers are plotted as small dots. IDC, invasive ductal carcinoma. CD24, CD44, EPCR, and SSEA4 indicate the cell surface markers used for the isolation of epithelial cells (Table S1). (C) Ratio of sense to antisense tag counts in each gene is plotted using average tag counts in all normal (x-axis) and cancer (y-axis) samples. Orange lines mark 4.0 and 0.25 values corresponding to 20% and 80% values of percentage of antisense per gene, respectively. (D) Numbers of AS genes in each sample are shown. Breast cancer cells express a significantly (P = 6.804e-06; Wilcoxon rank-sum test) lower number of AS genes compared with normal samples. (E) Venn diagrams depicting the number of AS genes common among CD24+ samples. Significant overlap is observed among samples derived from the same tissue and cell type. Ratios of observed/expected overlaps are 139 and 321 for the CD24.P and CD24.IDC groups, respectively. (F) Scatterplot depicting percentage of antisense tags per gene relative to total tag counts in 252 AS genes common in CD24P cells in normal (blue) and cancer (red) samples. (G) Hierarchical clustering analysis of all samples is based on 252 AS genes common among CD24.P cells. The color scale indicates the percentage of antisense tag counts in each gene.
We categorized genes based on the percentage of antisense and sense transcripts derived from them into three groups: antisense (AS) genes (percentage of antisense >80%), sense (S) genes (percentage of antisense <20%), and sense-antisense (SAS) genes (between S and AS genes). Using this classification, the numbers of AS and SAS genes were significantly lower (P = 6.804e-06 and P = 0.00066, respectively, Wilcoxon rank-sum test) in cancer and the opposite was true for S genes (P = 6.804e-06, Wilcoxon rank-sum test) (Fig. 1D, Fig. S1D, and Dataset S1). The significant and consistent differences in the number of AS genes between normal and cancer samples and the presumed regulatory function of AS transcripts prompted us to analyze these AS genes in further detail.
AS genes demonstrated significant overlap among the same cell type both in normal and cancer samples (Fig. 1E). Interestingly, most of the 252 AS genes commonly detected in normal CD24.P samples (Dataset S2) were consistently converted to S genes in tumors (Fig. 1 F and G and Fig. S1 E and F), which is likely to reflect decreased antisense transcription in cancer because it cannot be explained simply by increased sense transcription. Similar observations were made for the 323 AS genes commonly detected in CD44.P samples (Fig. S2 A–D and Dataset S2) and for the 1,975 normal-specific AS genes (Fig. S2 E and F). These data suggested that AS genes observed here were not random and were unlikely to reflect transcriptional noise because their expression is regulated in a cell and tissue type-specific manner likely reflecting biological function. Correlating with this, normal-specific CD24.P and CD44.P common AS genes showed significant enrichment for genes involved in basic metabolic processes, such as nucleotide, RNA, and protein synthesis and processing using gene ontology (GO) term enrichment analysis (Figs. S1F and S2G and Dataset S3).
Potential Mechanisms Underlying Altered Antisense-to-Sense Levels in Tumors.
To investigate the relationship between sense and antisense transcript abundance within individual genes, we plotted antisense vs. sense tag counts for each gene. Based on this analysis, sense and antisense tag counts correlated very well in cancer samples; however, this correlation was less pronounced in normal cells (Fig. 2 A–C and Fig. S2H). One possible explanation for this observation is that higher numbers of antisense (AS) genes are detected in normal samples and many of these are converted to sense (S) genes in tumors.
Fig. 2.
Identity and functions of S and AS genes. Scatterplots depicting the correlation between average antisense and sense tag counts for each gene in normal (A) and cancer (B) samples. (C) Overlap of the two plots shows an overall shift toward increased sense-to-antisense tag ratios in cancer. (D) Pie charts depicting changes in antisense and sense transcript levels in genes differentially expressed between normal and cancer samples. (E) Functional annotation of concordant (green) and discordant (orange) genes from the left pie chart of D based on GO terms.
To analyze associations between antisense and sense transcript levels in genes differentially expressed between normal and cancer samples, we identified 7,308 genes that showed significant differences in sense transcripts between normal and cancer groups at a 1% false discovery rate (FDR) (Fig. 2D, Table S2, and Dataset S4). Overall, the direction of difference in sense and antisense transcript levels correlated well. There was a difference in the number of genes showing discordant antisense-sense changes between genes that showed increased or decreased sense transcript levels in tumors, however (Fig. 2D). The enrichment of concordant (high sense and antisense) and discordant (high sense and low antisense) genes in different GO terms suggests that they have distinct physiological functions (Fig. 2E). Furthermore, the altered antisense-to-sense transcript levels in these genes is unlikely to be a simple consequence of increased sense transcription of metabolic genes attributable to increased metabolism in cancer but might reflect fundamental differences in the regulation of ncRNAs between normal and cancer cells.
At this time, we can only speculate on why the levels of antisense transcripts may be decreased in cancer. One hypothesis is that they may be decreased because of the deregulation of nonsense-mediated decay (NMD) in tumors, because NMD activity correlates with ncRNA levels in Arabidopsis (18) and the Xist ncRNA in mouse embryonic stem cells is controlled by the NMD pathway (19). We attempted to investigate the role of NMD in antisense transcription in breast cancer cell lines using PTC-124, an inhibitor of NMD (20), and siRNAs targeting key NMD pathway components (i.e., UPF1, SMG1) but have not been able to obtain conclusive results, potentially because of technical limitations. Alternatively, aberrant epigenetic and transcriptional control mechanisms could also be involved. The transcription of sense and antisense transcripts might be regulated by the same factors, resulting in their coordinated expression, or there could be strand-specific transcriptional regulators. The former model is supported by the observation that the same transcription factors (e.g., c-myc, p53) have been detected at the transcriptional start sites of both sense and antisense transcripts (21).
Validation of Antisense Transcripts.
To validate the strand specificity of the transcripts we identified based on SAGE-Seq using independent platforms, we performed ASSAGE and custom oligonucleotide array experiments. In ASSAGE, the RNA is bisulfite-treated prior to reverse transcription, which converts all cytidine to uridine, allowing unique mapping only to one of the two possible DNA strands (17). Similarly, custom oligonucleotide arrays hybridized with direct-labeled RNA maintain strand specificity. Using both of these platforms, we confirmed the expression of antisense transcripts predicted based on SAGE-Seq (Fig. S3 A and B). For example, 5,442 predicted antisense transcripts (96.6% of those tested) showed significant expression on arrays in at least one tumor sample, whereas 74.0% was detected in all four tumors. We also observed that some genes showed consistent differences between normal and cancer samples both in sense and antisense transcript levels. The correlation between the three different platforms was rather small (Tables S3 and S4), however, likely reflecting platform-specific differences in quantitating transcript abundance.
The limitation of these three global platforms is that they cannot determine the length of individual transcripts with high accuracy, and the possibility of multiple overlapping transcripts of different lengths (which can complicate quantitation using short sequence tags) cannot be excluded. Thus, we also validated a few selected genes by strand-specific RT-PCR (17). We identified RICTOR as one of the genes displaying high antisense in normal and high sense transcript levels in cancer cells (Fig. S3C). First, we confirmed the presence of both sense and antisense RICTOR transcripts (Fig. 3 A and B) and verified that the antisense transcript is a long ncRNA because it was polyadenylated and >1.2 kb (∼11 kb based on Northern blot analysis; Fig. 3C). Moreover, it significantly overlapped with the sense RICTOR transcript; thus, it is likely to be a cis-NAT. Next, we analyzed the levels of sense and antisense transcripts in multiple normal and neoplastic breast epithelial cells. Because of the lack of appropriate controls for strand-specific RT-PCR and the semiquantitative nature of this procedure, the exact transcript ratios could not be calculated. Nevertheless, the results of these RT-PCR analyses showed good overall correlation with SAGE-Seq data, although some variability was observed (Fig. 3D).
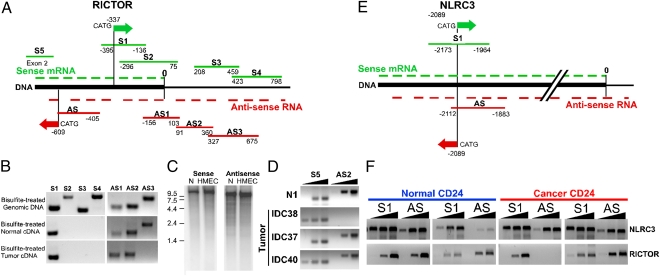
Fig. 3.
Validation of SAGE-Seq results. Schematic map of the 3′-ends of RICTOR (A) and NLRC3 (E) genes. The 3′-end of mRNA is marked with 0. Green and red arrows denote positions of sense and antisense SAGE-Seq tags, respectively. Green and red lines indicate sense and antisense PCR amplicons, respectively. (B) Mapping of sense and antisense RICTOR transcripts using strand-specific PCR. Bisulfite-treated genomic DNA (as control) or mRNA of normal and neoplastic CD24+ cells was used as a template. (C) Northern blot analysis of sense and antisense RICTOR transcripts. RNA prepared from normal breast organoids (N) and human mammary epithelial cells (HMEC) was hybridized with P32-labeled sense and antisense strand-specific probes. Numbers indicate the size of RNA markers (kilobase). (D) Relative expression levels of RICTOR 5′-end sense and 3′-end antisense transcripts in normal (N1) and neoplastic CD24+ cells based on semiquantitative PCR. S5 and AS2 indicate primer pairs for sense and antisense amplicons, respectively. Black triangles indicate increasing number of PCR cycles. (F) Validation of sense and antisense mRNAs for RICTOR and NLRC3 genes in normal and tumor cells by strand-specific PCR. Black triangles indicate increasing number of PCR cycles.
We also analyzed NLRC3, which showed similar antisense-to-sense transcript ratios between normal and cancer cells as control (Fig. S3D). We detected both sense and antisense polyadenylated transcripts transcribed from the NLRC3 locus, and this antisense transcript, again, was cis-NAT lncRNA because of its length (>200 bp) and overlap with the sense mRNA (Fig. 3E). In contrast to RICTOR, NLRC3 sense and antisense transcript levels did not differ between normal and neoplastic mammary epithelial cells (Fig. 3F).
To begin dissecting the function of the lncRNAs that we identified, we expressed siRNAs specifically targeting the RICTOR antisense transcript and evaluated Rictor and phospho-Akt protein levels in human mammary epithelial cells grown under different conditions but did not observe a consistent increase in Rictor protein levels (Fig. S3E). This could be attributable to technical difficulties (e.g., inefficient or not strand-specific siRNA targeting, change of cells attributable to culture conditions), however. Thus, further studies are required to understand the role these lncRNAs may play in the regulation of the Akt pathway and cellular metabolism.
In summary, we found altered ratios of antisense to sense transcripts between normal and neoplastic breast epithelial cells that may contribute to metabolic alterations associated with cellular transformation. The molecular mechanisms regulating the expression of these antisense transcripts and the role of these in tumorigenesis require further investigation.
Materials and Methods
Tissue Samples and Primary Culture.
Fresh normal and tumor specimens were collected at Harvard University-affiliated hospitals (Boston, MA) and the Johns Hopkins University (Baltimore, MD). All human tissue was collected using protocols approved by institutional review boards. Fresh tissue samples were immediately processed for immunomagnetic purification, followed by mRNA selection as previously described (13, 14).
SAGE-Seq Sample Preparation and Data Analysis.
SAGE-Seq libraries were generated essentially as previously described (13, 14), except that we used Illumina linkers for ligation and PCR amplification. SAGE-Seq libraries were sequenced using Illumina Genome Analyzer, and 17-bp tags were extracted from raw reads. We merged all libraries into one file and removed tags with only one tag count in any library, followed by normalization to 5 million total tag counts per sample. Tags were mapped to sense strand sequences of RefSeq genes at first. Tags that were not matched to any sense strand of RefSeq genes were then mapped to antisense strand sequences within RefSeq genes (examples of sense and antisense tag distribution within individual genes are depicted in Fig. S3 C and D). To calculate the ratio of sense and antisense tags for each gene, we summed all tags corresponding to each gene and calculated the percentage of antisense relative to total tag counts in each gene. Genes with very low (sum of all sense and antisense tag counts <5) expression levels in all samples combined were excluded from the analysis to minimize noise. GO term enrichment analyses were performed using DAVID bioinformatics resources (22); we used all RefSeq genes that we utilized for mapping as background for calculating enrichment. Differentially expressed sense and antisense transcripts were identified based on applying the significance analysis of microarray algorithm to the log2 ratio of normalized gene tag counts and using a 1% FDR as the cutoff for significance. Because antisense count gives another dimension to categorize the difference between normal and cancer, four kinds of differential gene expression patterns are identified as listed in Table S2 and Dataset S4.
ASSAGE Sample Preparation and Data Analysis.
ASSAGE libraries were generated essentially as described (17), except that we used polyA RNA. We generated a bisulfite-converted genome for both plus and minus strands (6.2 Gb) as a reference for ASSAGE reads. Using SeqMap (23), 16,762,174 reads were uniquely aligned to the bisulfite-converted genome and default conditions, allowing up to two mismatches in 32-bp reads. We used reads that uniquely mapped to either plus or minus strand-derived sequences. Samples were normalized to 5 million total tags. The number of reads aligned to specific regions (e.g., exons, introns, ±5 kb from 5′-end, ±5 kb from 3′-end) of each RefSeq gene was counted. To calculate the percentage of antisense counts per gene, we used total tag counts in all exonic and intronic regions of each gene.
Microarray Experiment and Data Analysis.
We used an Agilent Technologies Custom microarray (SurePrint G3 Custom CGH, 8 × 60 K), wherein each slide had eight arrays with ∼60,000 probes on each, in a two-color experimental design. We selected a total of 5,630 antisense tags, in which 5,030 showed a significant difference between normal and cancer, whereas 600 showed no difference but were abundant in both normal and cancer samples based on SAGE-Seq data. Using eArray software (Agilent Technologies), we designed 57,963 unique 60-mer oligonucleotide probes for sense and antisense transcripts. For each predicted antisense transcript, 5 probes were picked from the genomic region, ±1 kb from the antisense tag position. Similarly, five 60-mer probes (validated by Agilent Technologies) were selected for each corresponding sense transcript. For hybridization experiments, we used mRNA from four (N1 to N4) normal organoid and four breast tumor samples processed and purified as described previously (14). Two hundred nanograms of mRNA was directly labeled with Cy3 using a ULS micro-RNA labeling kit (Kreatech) and fragmented with an RNA fragmentation kit (Ambion). For the reference sample, equal amounts of mRNA from the four normal samples were mixed together, labeled with Cy5, and processed as above. For each individual array, 200 ng of Cy3-labeled normal or tumor sample was mixed with 200 ng of Cy5-labeled reference sample and two-color hybridization was performed according to the manufacturer's protocol (Agilent Technologies). Array processing and data extraction were performed by the Arthur and Rochelle Belfer Center for Cancer Genomics at the Dana–Farber Cancer Institute. Image analysis was conducted using Agilent Feature extraction software (version 10.5.1.1; Agilent Technologies), and processed signals were used for further analysis. To confirm the presence of antisense transcripts, a rigorous threshold was applied using the flag “IsWellAboveBG,” which first determines if the feature is significant (IsPosAndSignif fiag; determined by a two-sided t test) and then determines if the background-subtracted signal is >2.6-fold over the background SD for that feature. Significant expression was detected for 20,126 of 28,974 antisense probes [corresponding to 5,442 (96.6%) antisense transcripts of 5,603 tested] in at least one cancer sample, and 9,029 of these [corresponding to 4,165 (74.0%) antisense transcripts of 5,603 tested] were significantly expressed in all samples analyzed, whereas 18,900 of 28,989 sense probes [corresponding to 4,996 (88.7%) sense transcripts of 5,603 tested) had significant signal in all samples.
RT-PCR Analyses.
Bisulfite conversion of mRNA and cDNA synthesis and RT-PCR analyses using mRNA or bisulfite-treated mRNA were performed essentially as described (17).
Supplementary Material
Acknowledgments
We thank members of our laboratories for their critical reading of this manuscript and useful discussions. We greatly appreciate the help of Dr. Andrea Richardson (Brigham and Women's Hospital) in the acquisition of tissue samples and the Arthur and Rochelle Belfer Center for Cancer Genomics for array hybridization. This study was supported by the National Cancer Institute Specialized Program in Research Excellence in Breast Cancer at the Dana–Farber/Harvard University Cancer Center (Grant CA89393), the Department of Defense (Grant W81XWH-07-1-0294), and the Avon Foundation as well as by Breast Cancer Research Foundation grants (to K.P.), Susan G. Komen Foundation fellowships (to M.S. and R.M.), and a Terri Brodeur Breast Cancer Foundation Fellowship (to S.C.).
Footnotes
Conflict of interest statement: K.P. receives research support from and is a consultant to Novartis Pharmaceuticals, Inc. K.P. is also on the Scientific Advisory Board of Metamark Genetics, Inc. and Theracrine, Inc. Y.N. is a founder and employee of GeneGo, Inc.
Data deposition: SAGE-Seq, ASSAGE, and oligonucleotide array data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE 25292).
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1010559107/-/DCSupplemental.
References
- 1.Saha S, et al. Using the transcriptome to annotate the genome. Nat Biotechnol. 2002;20:508–512. doi: 10.1038/nbt0502-508. [DOI] [PubMed] [Google Scholar]
- 2.Carninci P, et al. FANTOM Consortium; RIKEN Genome Exploration Research Group and Genome Science Group (Genome Network Project Core Group) The transcriptional landscape of the mammalian genome. Science. 2005;309:1559–1563. doi: 10.1126/science.1112014. [DOI] [PubMed] [Google Scholar]
- 3.Malone CD, Hannon GJ. Small RNAs as guardians of the genome. Cell. 2009;136:656–668. doi: 10.1016/j.cell.2009.01.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: Insights into functions. Nat Rev Genet. 2009;10:155–159. doi: 10.1038/nrg2521. [DOI] [PubMed] [Google Scholar]
- 5.Guttman M, et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458:223–227. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huarte M, et al. A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell. 2010;142:409–419. doi: 10.1016/j.cell.2010.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Khalil AM, et al. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci USA. 2009;106:11667–11672. doi: 10.1073/pnas.0904715106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gupta RA, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071–1076. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Reis EM, Louro R, Nakaya HI, Verjovski-Almeida S. As antisense RNA gets intronic. OMICS. 2005;9:2–12. doi: 10.1089/omi.2005.9.2. [DOI] [PubMed] [Google Scholar]
- 10.Brito GC, et al. Identification of protein-coding and intronic noncoding RNAs down-regulated in clear cell renal carcinoma. Mol Carcinog. 2008;47:757–767. doi: 10.1002/mc.20433. [DOI] [PubMed] [Google Scholar]
- 11.Calin GA, et al. Ultraconserved regions encoding ncRNAs are altered in human leukemias and carcinomas. Cancer Cell. 2007;12:215–229. doi: 10.1016/j.ccr.2007.07.027. [DOI] [PubMed] [Google Scholar]
- 12.Burzio VA, et al. Expression of a family of noncoding mitochondrial RNAs distinguishes normal from cancer cells. Proc Natl Acad Sci USA. 2009;106:9430–9434. doi: 10.1073/pnas.0903086106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bloushtain-Qimron N, et al. Cell type-specific DNA methylation patterns in the human breast. Proc Natl Acad Sci USA. 2008;105:14076–14081. doi: 10.1073/pnas.0805206105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shipitsin M, et al. Molecular definition of breast tumor heterogeneity. Cancer Cell. 2007;11:259–273. doi: 10.1016/j.ccr.2007.01.013. [DOI] [PubMed] [Google Scholar]
- 15.Wood LD, et al. The genomic landscapes of human breast and colorectal cancers. Science. 2007;318:1108–1113. doi: 10.1126/science.1145720. [DOI] [PubMed] [Google Scholar]
- 16.JZ Wu, et al. Gene expression profiling of human breast tissue samples using SAGE-Seq. Genome Res. doi: 10.1101/gr.108217.110. 10.1101/gr.108217.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.He Y, Vogelstein B, Velculescu VE, Papadopoulos N, Kinzler KW. The antisense transcriptomes of human cells. Science. 2008;322:1855–1857. doi: 10.1126/science.1163853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kurihara Y, et al. Genome-wide suppression of aberrant mRNA-like noncoding RNAs by NMD in Arabidopsis. Proc Natl Acad Sci USA. 2009;106:2453–2458. doi: 10.1073/pnas.0808902106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ciaudo C, et al. Nuclear mRNA degradation pathway(s) are implicated in Xist regulation and X chromosome inactivation. PLoS Genet. 2006;2:e94. doi: 10.1371/journal.pgen.0020094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Linde L, Kerem B. Introducing sense into nonsense in treatments of human genetic diseases. Trends Genet. 2008;24:552–563. doi: 10.1016/j.tig.2008.08.010. [DOI] [PubMed] [Google Scholar]
- 21.Cawley S, et al. Unbiased mapping of transcription factor binding sites along human chromosomes 21 and 22 points to widespread regulation of noncoding RNAs. Cell. 2004;116:499–509. doi: 10.1016/s0092-8674(04)00127-8. [DOI] [PubMed] [Google Scholar]
- 22.Huang W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 23.Jiang H, Wong WH. SeqMap: Mapping massive amount of oligonucleotides to the genome. Bioinformatics. 2008;24:2395–2396. doi: 10.1093/bioinformatics/btn429. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.