Abstract
Approximately 30% of human cancers harbor oncogenic gain-of-function mutations in KRAS. Despite interest in KRAS as a therapeutic target, direct blockade of KRAS function with small molecules has yet to be demonstrated. Based on experiments that lower mRNA levels of protein kinases, KRAS-dependent cancer cells were proposed to have a unique requirement for the serine/threonine kinase STK33. Thus, it was suggested that small-molecule inhibitors of STK33 might have therapeutic benefit in these cancers. Here, we describe the development of selective, low nanomolar inhibitors of STK33’s kinase activity. The most potent and selective of these, BRD8899, failed to kill KRAS-dependent cells. While several explanations for this result exist, our data are most consistent with the view that inhibition of STK33’s kinase activity does not represent a promising anti-KRAS therapeutic strategy.
Keywords: KRAS dependence, synthetic lethality, targeted-therapy
Oncogenic mutations in the RAS family member KRAS are among the most common mutations in human cancer (1). For example, KRAS-mutation frequencies in lung, colon, and pancreas adenocarcinoma are 30%, 50%, and 90% respectively (2). These observations, coupled with functional studies, suggest that KRAS is a highly attractive therapeutic target for many cancers. Unfortunately, small-molecule targeting of KRAS has not yet been achieved, and no effective KRAS inhibitors have been described. Blockade of prenylation of the KRAS C-terminal membrane anchoring domain with farnesyltransferase inhibitors has met with limited success, due at least in part to increased expression of geranylgeranyl transferase (3). Similarly, targeting other steps in the processing of the KRAS C-terminal region through the inhibition of Ras-converting enzyme or isoprenylcysteine-carboxymethyltransferase has yet to be clinically validated (2). Efforts to target the downstream effector pathways of KRAS with MEK inhibitors alone or in combination with PI3K inhibitors have shown promising preclinical results and are currently being evaluated in the clinic (4). Nevertheless, the therapeutic targeting of KRAS remains one of the grand challenges in cancer research.
Recently, an alternative approach to targeting KRAS has been proposed—namely, the RNA interference (RNAi)-based screening for synthetic lethal gene/RNA interactions that might then suggest protein targets more “druggable” than the targeted mRNA or the KRAS protein itself (5–9). A recently reported RNAi screen suggested the serine-threonine kinase STK33 as such a target (9). Knock down of STK33 was reported to induce apoptosis in KRAS-dependent AML cancer cell lines but spare KRAS wild-type cells. While the normal function of the STK33 protein is unknown, the results led the authors to propose that a small-molecule inhibitor of STK33’s protein kinase activity would selectively kill KRAS-mutant cancer cells. As no such small-molecule inhibitors exist, we set out to discover them, and to characterize their activity as anti-KRAS agents.
Results
High Throughput Screening for STK33 Kinase Inhibitors.
We first established an assay suitable for screening for STK33 kinase inhibitors. Using baculovirus-expressed full length human recombinant STK33 and the general kinase substrate myelin basic protein (MBP), a biochemical assay was optimized that quantified the STK33 kinase-dependent generation of ADP (see Materials and Methods). Radiometric analysis and mass spectroscopy (MS) (10, 11) were used to corroborate that the ADP generated was quantitatively coupled to phosphorylation of MBP (SI Appendix, Fig. S1). The reaction rate was linear with respect to time and enzyme concentration and Michaelis-Menten constants were determined (SI Appendix, Fig. S2). The relatively nonspecific kinase inhibitor dimethyl fasudil (BRD7446) was identified in a pilot screen as a low micromolar STK33 inhibitor; it therefore served as a positive control for subsequent screening (SI Appendix, Fig. S2E). A high throughput screen of 27,500 compounds from diverse chemical collections (SI Appendix, Table S1) was performed in duplicate at 100 μM ATP (threefold above the KM,ATP). Coefficients of variation (CVs) of 4–6% for the DMSO vehicle control and corresponding Z’ factor values for the positive control between 0.64 and 0.76 were obtained (12). The hit rate was 0.4% at an inhibition threshold of 20% inhibition where Z’ factor equaled zero (SI Appendix, Fig. S3). The IC50 values of the 102 primary hits were measured at two ATP concentrations (100 and 25 μM) using both the ADP-Glo and HTRF assays.
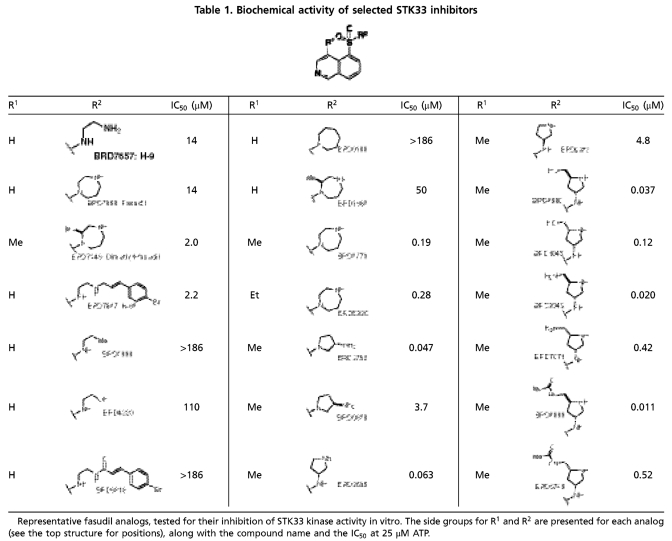
The STK33-inhibitory activity of 95/102 compounds (93%) was verified in replicate experiments, and these compounds clustered into 37 discrete chemotypes (SI Appendix, Fig. S4). Four groups appeared to be ATP-noncompetitive inhibitors but were either very weak or contained chemically reactive moieties, and were therefore not prioritized. All other hits were ATP-competitive inhibitors with IC50 values in the micromolar range, except the chemotype of staurosporine (a notoriously nonselective kinase inhibitor) that inhibited STK33 with low nanomolar potency. Fasudil (BRD7868) (Table 1), a known inhibitor of Rho-associated protein kinase (ROCK) (13) as well as seven of its analogs yielded a structure-activity relationship for inhibiting STK33. This series was therefore prioritized as the lead structure for chemical optimization due to its low micromolar potency, known selectivity for other kinases, physical properties, and synthetic feasibility.
Table 1.
Biochemical activity of selected STK33 inhibitors
Representative fasudil analogs, tested for their inhibition of STK33 kinase activity in vitro. The side groups for R1 and R2 are presented for each analog (see the top structure for positions), along with the compound name and the IC50 at 25 μM ATP.
STK33 Can Autophosphorylate In Vitro and Is Inhibited by a Fasudil Analog.
During the optimization of STK33 inhibitors, we sought to confirm that the candidate inhibitors were truly functioning as STK33 inhibitors, and were not simply an artifact of a screening assay based on an artificial kinase substrate (MBP). To address this concern, we assessed whether STK33, like many kinases, is autophosphorylated, and if so, whether our candidate STK33-inhibitory compounds blocked such autophosphorylation (SI Appendix, Fig. S1A) (9, 14). Mass spectrometry revealed several sites of phosphorylation on STK33 (Fig. 1). Two sites (Thr440 and Ser441) were found to be phosphorylated in the preparation of recombinant STK33 itself and the remaining sites were due to bona fide in vitro kinase activity of the enzyme. Among the sites observed were the Thr491/Ser493/Thr496 cluster that had been previously reported to be regulated during the cell cycle in HeLa cells (15). We then verified that the lead scaffold that emerged from the biochemical screen (represented in BRD3773, BRD3752, and BRD3695, see structures in Table 1), indeed inhibited the autophosphorylation of STK33 at these sites in a concentration-dependent manner (Fig. 1). These results suggested that the scaffold was a suitable starting point for further optimization with respect to STK33 inhibition.
Fig. 1.
Proteomic assessment of STK33 autophosphorylation. Sites on STK33 determined to be autophosphorylated are shown on the left. Increasing concentrations of the three fasudil analogs shown were added to an in vitro phosphorylation reaction in which the only substrate present was STK33 itself. The heatmap is colored according to how the level of phosphorylation at a given site changed in response to addition of compound at the specified concentration vs. DMSO control. Some compound concentrations were found to increase phosphorylations at given sites, leading to values > 100%. 1ambiguously localized phosphorylation within the span of bracketed amino acids. 2ambiguously localized double phosphorylation within the span of bracketed amino acids.
Chemical Optimization of STK33 Inhibitors.
For chemical optimization, 250 fasudil analogs were synthesized and tested in the biochemical assay (SI Appendix, Schemes S1–S5, Table S13, and SI Materials and Methods). Structurally, fasudil comprises two fragments: an isoquinoline ring and a homopiperazine ring, connected by the sulfonyl functionality on the 5-position of the isoquinoline. The sulfonamide functionality and nitrogen on the isoquinoline ring are critical for the potency against STK33 (SI Appendix, Table S2). Replacing the basic amine nitrogen with carbon or oxygen, or converting it to an amide, resulted in significantly reduced potency (BRD7657 vs. BRD2998 and BRD4220, BRD7868 vs. BRD0188, BRD7647 vs. BRD6818, Table 1, SI Appendix, Table S3). We observed that the two additional methyl groups in dimethyl fasudil lead to a sevenfold increase in potency compared to fasudil (BRD7446 vs. BRD7868). While one monomethyl fasudil isomer, namely BRD1869, was threefold less potent than fasudil (BRD7868), the other monomethyl fasudil isomer (BRD3773) was over 70-fold more potent against STK33 compared to fasudil (BRD7868). Therefore we explored the effect of monomethylation at several positions of the isoquinoline ring; only 4-methyl substitution significantly increased the potency (SI Appendix, Table S4). An SAR study performed on isoquinoline 4-substitution indicated that only a small alkyl group, such as ethyl (BRD5930), is tolerated, while polar or bulky groups resulted in decreased activity (SI Appendix, Table S5). Further optimization on the amine fragment led to two more nanomolar STK33 inhibitors (BRD3752 and BRD3695); their enantiomers were 60–100-fold less active (BRD0828 and BRD9573) (Table 1, SI Appendix, Table S6).
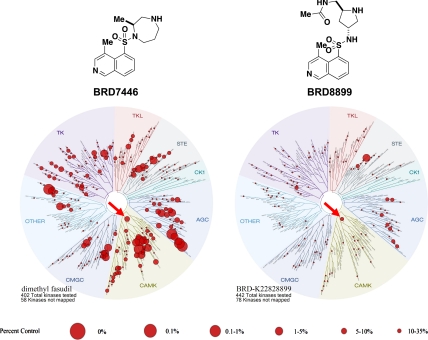
Further chemical optimization was guided by measures of STK33 selectivity. As measured by kinase activity profiling against 241 kinases (SI Appendix, Table S7), BRD3773 exhibited relative STK33 selectivity, inhibiting 36 kinases with estimated Ki values within 10-fold of that of STK33. Compound BRD3695 showed further selectivity with respect to those 36 kinases (SI Appendix, Fig. S5), with inhibition confined to the AGC subfamily of kinases, and therefore became the lead for further chemical optimization.
Modification of the pyrrolidine ring of BRD3695 (SI Appendix, Table S8) yielded more potent, low nanomolar analogs (BRD4980, BRD9949, and BRD8899). These compounds were more potent than their corresponding (2R, 4S) diastereomers (BRD1045, BRD7071, and BRD5749). The low nanomolar potency of BRD8899 against STK33 as well as other selected inhibitors was confirmed by competition binding assay (SI Appendix, Table S9) (16). Furthermore, the specificity of BRD8899 was retained, exhibiting significant off-target inhibition of a few other kinases, including RIOK1 (97% inhibition), MST4 (96%), RSK4 (89%), ATK1 (85%), KITD816V (85%), ROCK1 (84%), and FLT3 (81%), while STK33 was inhibited by 89% (SI Appendix, Table S10). BRD8899 thus represents a 200-fold improvement in potency compared to the initial screening hit (BRD7446), with increased selectivity for STK33 (Fig. 2).
Fig. 2.
Small molecule/kinase interaction maps for BRD7446 and BRD8899. The kinase profiling (16) was done at 5 μM for BRD7446 and 1 μM for BRD8899, resulting in equal inhibition of STK33 in a biochemical assay. Kinases found to bind are marked with red circles; larger circles indicate higher affinity binding. STK33 is indicated by the red arrow.
Characterization of BRD8899 in Cell-Based Assays.
Having demonstrated that BRD8899 is a potent and selective inhibitor of STK33 kinase activity in biochemical assays, we next turned to its characterization in cells. It has been previously reported that STK33 knock down via RNAi results in the selective killing of mutant KRAS-dependent cancer cells (9), a result that we confirmed using the same shRNAs in KRAS-mutant AML cell lines (NOMO-1 and SKM-1 AML cells) compared to KRAS-wild-type lines (THP-1 and U937) (SI Appendix, Fig. S7). We next asked whether BRD8899 could recapitulate KRAS-associated pattern of cell killing. We therefore tested BRD8899 at a range of doses across 35 cancer cell lines, but observed no effect on cell viability in any of the lines at concentrations as high as 20 μM (Fig. 3A, SI Appendix, Fig. S6 and Table S12). (9) Because BRD8899 (our most potent STK33 inhibitor) failed to kill KRAS-mutant cell lines as predicted by the RNAi experiments, we tested additional analogs for their ability to kill cancer cells. While some of these compounds reduced cell viability, such cytotoxicity was uncorrelated with KRAS-mutation status across the panel of cell lines (Fig. 3B, SI Appendix, Fig. S6). We therefore conclude that the observed cell death is most likely explained by off-target effects of these less potent compounds, rather than by STK33-inhibitory activity.
Fig. 3.
Cell viability assay for fasudil analogs BRD8899, BRD3773, BRD9573, and BRD3695. (A and B). Dose-response curves of KRAS and STK33 dependent NOMO-1 and SKM-1 cells and KRAS and STK33 independent THP-1 and U937 cells (as previously confirmed by RNAi), using CellTiter Glo as a measurement for viability. Cells were plated in 384-well plates and treated with compounds for 72 h. The luminescence values of each compound treatment divided by the median of all DMSO values for each individual cell line was taken as a measurement of relative viability. Error bars represent the standard deviation of three replicates. KRAS mutant and KRAS wild-type cell lines are depicted in red or blue, respectively. For 31 additional cell lines screened, see SI Appendix, Fig. S6.
Establishing Bioactivity of BRD8899 in Cells.
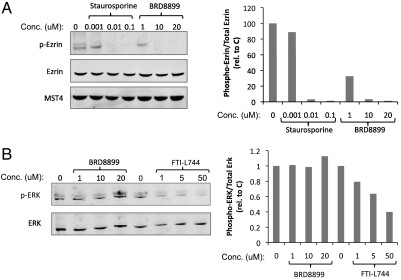
The experiments described above suggest that treatment of KRAS-mutant cancer cells with low nanomolar biochemical potency against STK33 does not result in cell death, arguing against the notion that the enzymatic function of STK33 represents an attractive therapeutic target in KRAS-mutant cancers. However, it is possible that while BRD8899 is highly potent in biochemical assays, it fails to inhibit STK33 in cells—even at 1,000-fold higher concentrations. To address this possibility, we sought a biomarker of BRD8899 activity in cells. We first looked for evidence of STK33 autophosphorylation in cells, given the observation that such autophosphorylation occurs with recombinant STK33, and is inhibited by BRD8899. However, looking across multiple cell lines, we found endogenous STK33 to be expressed at low levels, and phosphorylation was not detectable by mass spectrometry (for details, see Materials and Methods). We therefore turned to the kinase profiling data for BRD8899, where we observed similarly potent inhibition of the kinase MST4 in addition to STK33. Treatment of NOMO-1 cells with BRD8899 resulted in decreased phosphorylation of the MST4 substrate ezrin (Fig. 4A), but had no effect on ERK phosphorylation, as predicted from the kinase profiling data (Fig. 4B). This finding suggests that BRD8899 indeed gets into cells, inhibits the activity of a target (MST4) predicted by kinase profiling experiments, and yet does not kill KRAS-mutant cancer cells. These results are most consistent with BRD8899 likely inhibiting STK33 in cells, but STK33 kinase activity not serving as a compelling target for KRAS-mutant tumors. Of course in the absence of a direct biomarker of STK33 activity in cells, we cannot exclude that whereas BRD8899 inhibits MST4 (as an off-target effect), it does not effectively inhibit STK33 in cells.
Fig. 4.
MST4 and ERK kinase activity was used as surrogate biomarker and negative control for BRD8899, respectively. (A and B.) NOMO-1 cells were treated with the indicated concentrations of staurosporine, the farnesyl transferase inhibitor L744 (as positive controls) or the fasudil analog BRD8899 for 24 h. Samples were lysed and subjected to immunoblotting for phosphorylated and total levels of Ezrin (T576) and total MST4 (A) or phosphorylated and total levels of ERK (B). Quantifications for each lane were generated using ImageJ software. The normalized values to total protein levels are presented next to the blots.
Discussion
Cancer genomes have served as powerful guides to the development of cancer therapeutics that target oncogene dependencies. Several examples point to the utility of targeting mutant, oncogenic kinases with small-molecule inhibitors, with dramatic clinical success being seen in chronic myeloid leukemia (17), gastrointestinal stromal tumors (18), lung cancer (19), and melanoma (20). However, the therapeutic path for nonkinase mutations in cancer is less established. In particular, recurrent mutations in KRAS have been known to exist for over a quarter century, and yet KRAS has been considered “undruggable.”
A general solution to approaching cancer targets, however, has recently been suggested. Borrowing from the concepts of synthetic lethality first explored in yeast, “nononcogene addiction” has been proposed (8, 21). One avenue to identify nononcogene codependencies (proteins on which oncogenes are dependent yet not themselves mutated) relies on genetic screens to infer potentially more readily druggable proteins whose inhibition is selectively lethal in the context of particular mutations. Such approaches are highly attractive because they do not require prior knowledge of the biological basis of the synthetic lethal interaction—rather, the relationship is revealed through an unbiased screen such as an RNAi screen. On the other hand, while this approach might establish a synthetic relationship between a genetic feature of a cancer and the lowering of a target mRNA, they do not establish such a relationship involving a function of the encoded protein. Assuming RNAi-induced changes in mRNA levels and small-molecule-modulated protein function are equivalent, the report of STK33 essentiality in mutant KRAS-dependent cancer cells (9) was encouraging because STK33, as a serine/threonine kinase, was in principle an accessible target of small molecules, and such compounds could form the basis of anti-KRAS therapeutics. We therefore developed a biochemical STK33 kinase assay to identify specific STK33 inhibitors that could be evaluated in cellular proof-of-principle studies. A high throughput screen identified a series of STK33 kinase-inhibitory compounds, which were subsequently optimized for potency (representing 200 - 1,000-fold increase in potency) while minimizing off-target effects (inhibiting only three other kinases more potently than STK33). Specifically, our lead compound, BRD8899, exhibited an IC50 of 11 nM in an STK33 biochemical assay. However, contrary to essentiality of STK33 predicted by initial genetic studies of STK33, BRD8899 failed to kill KRAS-mutant cells at concentrations as high as 20 μM.
There are a number of explanations of the failure of a potent STK33 kinase inhibitor to kill KRAS-mutant cells, and several are offered here. It is conceivable that while BRD8899 (and its analogs) inhibit STK33 kinase activity in biochemical assays, it does not do so in living cells. For example, it is possible that BRD8899 does not penetrate cells. However, several pieces of evidence argue against this possibility. Ideally, one would have a direct measure of STK33 kinase activity in cells, but to date, no such biomarker has been developed for STK33. We attempted to identify such a direct biomarker, looking for evidence of the STK33 autophosphorylation we observed using recombinant STK33 protein. Unfortunately, owing to the low level of STK33 expression in the two cancer cell lines analyzed (NOMO-1 and SKM-1), we were unable to document such phosphorylation. We therefore turned to indirect measures of BRD8899 bioactivity, using the predicted off-target effects of the compound suggested by kinase specificity profiling. Indeed, we found that BRD8899 inhibits the kinase MST4 in cells (resulting in subsequent decreased phosphorylation of an MST4 substrate, ezrin). These biomarker studies, while indirect, suggest that BRD8899 does in fact enter cells, and can inhibit the activity of its kinase targets. While it is conceivable that BRD8899 inhibits MST4 but not STK33, we consider that scenario less likely. Although other explanations exist, such as a cellular context dependency recently reported with small-molecule inhibitors of PKC and PDK1 (23, 24), overall we surmise that inhibiting STK33 kinase activity is not likely to be an effective therapeutic strategy for KRAS-mutant cancer.
While inhibition of STK33 kinase activity may not result in KRAS-related cell killing, it is possible that nonkinase activities of STK33 may be responsible for its observed essentiality in RNAi-based studies. For example, STK33 could have a distinct function other than its kinase activity. It is conceivable that STK33 has a more structural, scaffolding function that is required for the proper function of a multiprotein complex. Whereas RNAi-mediated knock down of STK33 might disrupt such putative scaffolding activity, a small-molecule kinase inhibition may not. Such nonkinase activities of STK33 cannot be excluded by our present studies. A similar conclusion was reached in a recent independent study (22).
An alternative explanation for the failure of STK33 inhibitors to recapitulate STK33 the knock-down phenotype is that the shRNA reagents have off-target (non-STK33) effects. Arguing against this alternative explanation is the observation that a dominant-negative STK33 construct was shown to result in selective lethality to KRAS-dependent cell lines (9). Nevertheless, until clear rescue experiments using non-RNAi-inhibitable STK33 expression constructs demonstrate rescue of shRNA-mediated killing, an off-target mechanism must remain a formal possibility. We note that a recent study using STK33 siRNA oligonucleotides transiently transfected into AML cell lines failed to show a KRAS-specific killing effect (22). However, the transfection efficiency was not reported and only partial STK33 knock down was achieved in those experiments, making it difficult to interpret the results.
It has been recently suggested that while gain of function mutations within oncogenes lead to increases in rate-determining steps in signaling, their synthetic lethal partners may operate through relatively rapid signaling steps (8). Although these fast steps might be modulated by “irreversible” maneuvers such as RNAi knock down and dominant negative expression, achieving this same level of target modulation with reversible small molecule competitive inhibitors may be more difficult. The extent to which this phenomenon explains our STK33 results remains to be determined.
The experiments described here used cell viability as the readout of STK33 modulation. As such, these studies do not preclude the possibility of an important role of STK33 in other cancer phenotypes (e.g., migration, adhesion). However, our study indicates that the pharmaceutical targeting of STK33 kinase activity may not result in an effective strategy for patients with KRAS-mutant cancers. Nevertheless, the potential of synthetic-lethal RNAi screens should not be underestimated; they represent a powerful strategy for identifying promising new anticancer targets.
Materials and Methods
STK33 and Kinase Assays.
Human full length STK33 containing an amino terminal histidine tag expressed by baculovirus in Sf21 cells was purchased from Millipore Corporation (catalog # 14-671-K, purity > 93% by SDS-PAGE and Coomassie blue staining). Myelin basic protein (bovine) was purchased from Millipore Corporation (catalog # 13–104). ATP, ADP, 4-Morpholinepropanesulfonic acid (MOPS), MgCl2, Brij-35, glycerol, 2-mercaptoethanol, and BSA were purchased from Sigma-Aldrich. The 384-well general plates were purchased from VWR (Corning 3570); 384-well low volume plates were purchased from Greiner Bio-One (384W SV, HiBase, PS, LUMITRAC 200, Medium Binding, 30 μL/well, catalog # 784075). CyBi®-Well vario was purchased from CyBio AG. CyBi tips were purchased from CyBio AG (CyBi-Tip Trays 384 standard; catalog # OL 3800-25-513-N). HTRF Transcreener ADP assay was purchased from Cis-bio US (catalog # 62ADPPEC); ADP-Glo assays were purchased from Promega Corporation (catalog # V9103). The Envision 2012 multilabel reader was purchased from PerkinElmer, Inc. Multidrop Combi reagent dispenser and cassettes were purchased from Thermo Fisher Scientific, Inc.
Kinase reactions were performed under 10 mM MOPS-NaOH (pH 7.0), 10 mM MgCl2, 0.3 mM EDTA, 0.001% Brij-35, 0.5% glycerol, 0.01% 2-mercaptoethanol, and 0.1 mg/mL BSA. The enzyme concentration (STK33) and substrate concentration (MBP and ATP) were varied depending on the experiment. Reactions were initiated by the addition of ATP (final concentration 25 ∼ 500 μM depending on experiments) and incubated at 30 °C or room temperature for the indicated time. For additional details, see SI Appendix, SI Materials and Methods.
Mass Spectrometric Assessment of Phosphorylation State of MBP and STK33 for In Vitro Assays.
For MBP, assay conditions were as described in “STK33 and Kinase Assays” and SI Appendix, Fig. S1. For STK33, 1 μg of total STK33 was incubated with 100 μM ATP, in the absence or presence of the concentration of fasudil analog indicated in Fig. 1. The reaction was terminated by the addition of SDS-PAGE loading buffer containing 1% SDS containing 10 mM DTT and heating to 95 °C for 5 min. Samples were reduced at room temperature for 30 min and cysteines were subsequently alkylated with 30 mM iodoacetamide for 30 min. SDS-PAGE separation and gel band sample preparation was done as described (10). Liquid Chromatography—Mass spectrometry (LCMS) was performed on an Orbitrap mass spectrometer (ThermoFisher Scientific) coupled to a nano-flow chromatography system (Agilent 1100), as described (11). For each survey scan, the top 10 most abundant peptidic ions were selected for collision-induced dissociation (MS/MS sequencing), using intelligent sampling dynamic exclusion principles (21). The data were analyzed using SpectrumMill Proteomics Workbench (Agilent) as described (21). Extracted ion chromatograms corresponding to the phosphopeptides from either MBP or STK33 were generated using XCalibur (ThermoFisher Scientific) (± 7.5 ppm window width). Area under these curves was used as the basis for quantification of the effect of the fasudil analogs on site-specific phosphorylation. All peak areas were normalized to total protein amount for each condition, as determined by the peak area under a peptide that could not be phosphorylated in the assay.
Analysis of STK33 Expression in Cells.
NOMO-1 and SKM-1 cells were grown in SILAC (stable isotope labeling with amino acids in culture) heavy medium. Recombinant STK33 was added at known concentrations in SILAC light medium and proteins within lysates were separated on gel electrophoresis and then analyzed by MS. Based on the light standard, the levels of endogenous STK33 were estimated to approximately 10 ng/1.5 × 108 cells. Similar experiments were done using NOMO-1 cells stably expressing Flag-STK33 in a pLenti6.2 vector, and the levels of overexpressed protein were estimated to be approximately 10-fold higher. For the phosphorylation studies by MS, synthetic phospho-peptides for STK33 (based on known phosphorylation sites from the in vitro kinase assay) were added into the digest of Flag-STK33 expressed in NOMO-1 cells to estimate the detectable amounts of phospho-STK33 in cells.
Cell Lines and Culture Conditions.
NOMO-1, SKM-1, NB4, THP-1, U937, OCI-AML3, MM1S, and RPMI-8226, were kindly provided by Gary Gilliland (Brigham and Women’s Hospital, Boston). TF-1, P31/FUJ, MOLM-16, PL-21, EOL-1, GDM-1, Colo-205, HeyA8, OvCar-3, CaOV3, OvCar-8, and KYSE-405 cells were provided by the Broad-Novartis Cancer Cell Line Encyclopedia. The remaining cell lines were obtained from the American Type Culture Collection and the German Collection of Microorganism and Cell Cultures. All cell lines were maintained under the manufacturer’s recommended standard conditions.
STK33 and KRAS Knock Down.
Generation of virus, transduction, and selection of cells was done as previously described (9). In brief, cells were transduced with pLKO.1puromycin lentiviral shRNA vectors from the TRC shRNA library, and selected with 4 μg/mL puromycin for 48 h. Cell viability was measured at timepoints 0 and 72 h after selection. Hairpins targeting the following sequences in STK33 were used: GCAGTTCAAGTTTCACATCTA (2,078) GAACACATCATACATCTGGAA (2,079) and CTTGCCATTAACTTGCTGCTA (2,081). Hairpins targeting the following sequences in KRAS were used: GCAGACGTATATTGTATCATT (33,259) GAGGGCTTTCTTTGTGTATTT (33,260) and CCTATGGTCCTAGTAGGAAAT (33,262).
Cell Viability Assays.
Cells were plated in 384-well plates at optimized densities and incubated overnight prior to treatment. Compounds were added to cells using a CyBio Well Vario after which cells were cultured under standard condition for 72 h (unless stated otherwise). Cell viability was measured using CellTiter Glo luminescence (Promega) and readout using an LJL Biosystems Analyst microplate reader. Raw numbers were the normalized to the median of the DMSO treated wells for each plate.
Protein Assays and Western Blotting.
Whole-cell lysates were prepared using a lysis buffer from Cell Signaling Technology (# 9803) and subjected to Western blotting according to standard procedures. The membranes were developed using a LI-COR Odyssey Infrared Imaging system, and the images were analyzed by Adobe Photoshop and ImageJ softwares. The following antibodies were used: anti-MST4 (Cell Signaling, #3822), anti-ezrin (Cell Signaling, #3245), anti-phospho-ezrin (Cell Signaling, #3141), anti-phospho-ERK (Cell Signaling, #437) and anti-ERK (Santa Cruz, #135900).
Supplementary Material
Acknowledgments.
We thank members of the Broad Institute Chemical Biology Platform for high throughput screening assistance. We also thank D. Gary Gilliland at Merck Research Laboratories and Robert J. Gould at Epizyme for helpful advice. This study was supported by a Starr Cancer Consortium grant. In addition, T.L. was supported by a grant from the National Institute of General Medical Sciences (GM38627 awarded to S.L.S.), K.M. was supported by the Swedish Research Council (Vetenskapsrådet), and C.S. was supported by an Emmy Noether Fellowship from the German Research Foundation. Support for these studies was also provided by the National Institutes of Health (NIH) Genomics Based Drug Discovery-Driving Medical Projects Grant RL1-GM084437 and RL1-CA133834, administratively linked to NIH grants RL1-HG004671 and UL1-DE019585.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1120589109/-/DCSupplemental.
References
- 1.Friday BB, Adjei AA. K-ras as a target for cancer therapy. Biochim Biophys Acta. 2005;1756:127–144. doi: 10.1016/j.bbcan.2005.08.001. [DOI] [PubMed] [Google Scholar]
- 2.Karnoub AE, Weinberg RA. Ras oncogenes: split personalities. Nat Rev Mol Cell Biol. 2008;9:517–531. doi: 10.1038/nrm2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Downward J. Targeting RAS signalling pathways in cancer therapy. Nat Rev Cancer. 2003;3:11–22. doi: 10.1038/nrc969. [DOI] [PubMed] [Google Scholar]
- 4.Engelman JA, et al. Effective use of PI3K and MEK inhibitors to treat mutant Kras G12D and PIK3CA H1047R murine lung cancers. Nat Med. 2008;14:1351–1356. doi: 10.1038/nm.1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hartwell LH, Szankasi P, Roberts CJ, Murray AW, Friend SH. Integrating genetic approaches into the discovery of anticancer drugs. Science. 1997;278:1064–1068. doi: 10.1126/science.278.5340.1064. [DOI] [PubMed] [Google Scholar]
- 6.Barbie DA, et al. Systematic RNA interference reveals that oncogenic KRAS-driven cancers require TBK1. Nature. 2009;462:108–112. doi: 10.1038/nature08460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Luo J, et al. A genome-wide RNAi screen identifies multiple synthetic lethal interactions with the Ras oncogene. Cell. 2009;137:835–848. doi: 10.1016/j.cell.2009.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Luo J, Solimini NL, Elledge SJ. Principles of cancer therapy: oncogene and non-oncogene addiction. Cell. 2009;136:823–837. doi: 10.1016/j.cell.2009.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Scholl C, et al. Synthetic lethal interaction between oncogenic KRAS dependency and STK33 suppression in human cancer cells. Cell. 2009;137:821–834. doi: 10.1016/j.cell.2009.03.017. [DOI] [PubMed] [Google Scholar]
- 10.Kinter M, Sherman NE. Protein Sequencing and Identification Using Tandem Mass Spectrometry. New York: Wiley-Interscience; 2000. [Google Scholar]
- 11.Jaffe JD, et al. Accurate inclusion mass screening: a bridge from unbiased discovery to targeted assay development for biomarker verification. Mol Cell Proteomics. 2008;7:1952–1962. doi: 10.1074/mcp.M800218-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang JH, Chung TD, Oldenburg KR. A simple statistical parameter for use in evaluation and validation of high throughput screening assays. J Biomol Screen. 1999;4:67–73. doi: 10.1177/108705719900400206. [DOI] [PubMed] [Google Scholar]
- 13.Breitenlechner C, et al. Protein kinase A in complex with Rho-kinase inhibitors Y-27632, Fasudil, and H-1152P: structural basis of selectivity. Structure. 2003;11:1595–1607. doi: 10.1016/j.str.2003.11.002. [DOI] [PubMed] [Google Scholar]
- 14.Brauksiepe B, et al. The Serine / threonine kinase STK33 exhibits autophosphorylation and phosphorylates the intermediate filament protein Vimentin. BMC Biochem. 2008;23:9–25. doi: 10.1186/1471-2091-9-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Daub H, et al. Kinase-selective enrichment enables quantitative phosphoproteomics of the kinome across the cell cycle. Mol Cell. 2008;31:438–448. doi: 10.1016/j.molcel.2008.07.007. [DOI] [PubMed] [Google Scholar]
- 16.Karaman MW, et al. A quantitative analysis of kinase inhibitor selectivity. Nat Biotechnol. 2008;26:127–132. doi: 10.1038/nbt1358. [DOI] [PubMed] [Google Scholar]
- 17.Capdeville R, Buchdunger E, Zimmermann J, Matter A. Glivec (STI571, imatinib), a rationally developed, targeted anticancer drug. Nat Rev Drug Discov. 2002;1:493–502. doi: 10.1038/nrd839. [DOI] [PubMed] [Google Scholar]
- 18.Reichardt P, Reichardt A, Pink D. Molecular targeted therapy of gastrointestinal stromal tumors. Curr Cancer Drug Tar. 2011;11:688–697. doi: 10.2174/156800911796191042. [DOI] [PubMed] [Google Scholar]
- 19.Okamoto I, Mitsudomi T, Nakagawa K, Fukuoka M. The emerging role of epidermal growth factor receptor (EGFR) inhibitors in first-line treatment for patients with advanced non-small cell lung cancer positive for EGFR mutations. Therapeutic Advances in Medical Oncology. 2010;2:301–307. doi: 10.1177/1758834010370698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Flaherty KT, et al. Inhibition of mutated, activated BRAF in metastatic melanoma. N Engl J Med. 2010;363:809–819. doi: 10.1056/NEJMoa1002011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sharma SV, Settleman J. Oncogene addiction: setting the stage for molecularly targeted cancer therapy. Genes Dev. 2007;21:3214–3231. doi: 10.1101/gad.1609907. [DOI] [PubMed] [Google Scholar]
- 22.Babij C, et al. STK33 kinase activity is non-essential in KRAS-dependent cancer cells. Cancer Res. 2011;71:5818–5826. doi: 10.1158/0008-5472.CAN-11-0778. [DOI] [PubMed] [Google Scholar]
- 23.Prince JT, Ahn NG. The case of the disappearing drug target. Mol Cell. 2010;37:455–456. doi: 10.1016/j.molcel.2010.02.010. [DOI] [PubMed] [Google Scholar]
- 24.Hoshi N, et al. Interaction with AKAP79 modifies the cellular pharmacology of PKC. Mol Cell. 2010;37:541–550. doi: 10.1016/j.molcel.2010.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.