Abstract
High expression of the oncoprotein Myc has been linked to poor outcome in human tumors. Although MYC gene amplification and translocations have been observed, this can explain Myc overexpression in only a subset of human tumors. Myc expression is in part controlled by its protein stability, which can be regulated by phosphorylation at threonine 58 (T58) and serine 62 (S62). We now report that Myc protein stability is increased in a number of breast cancer cell lines and this correlates with increased phosphorylation at S62 and decreased phosphorylation at T58. Moreover, we find this same shift in phosphorylation in primary breast cancers. The signaling cascade that controls phosphorylation at T58 and S62 is coordinated by the scaffold protein Axin1. We therefore examined Axin1 in breast cancer and report decreased AXIN1 expression and a shift in the ratio of expression of two naturally occurring AXIN1 splice variants. We demonstrate that this contributes to increased Myc protein stability, altered phosphorylation at S62 and T58, and increased oncogenic activity of Myc in breast cancer. Thus, our results reveal an important mode of Myc activation in human breast cancer and a mechanism contributing to Myc deregulation involving unique insight into inactivation of the Axin1 tumor suppressor in breast cancer.
Keywords: Ras, ERK, GSK3β, PP2A, Fbw7
The c-Myc (Myc) oncoprotein is a pleiotropic transcription factor involved in controlling many cellular functions, including cell proliferation, cell growth, and cell differentiation, as well as pathways that regulate genome stability and cell death (1–5). High levels of Myc expression occur in a wide variety of human tumors, and animal models exhibit Myc-induced tumorigenesis in many tissues (6–8). These tumors are often dependent on continued high expression of Myc and withdrawal of Myc can induce tumor regression (8, 9), highlighting the importance of understanding how Myc expression is regulated. In breast cancer, Myc protein is reported to be overexpressed in approximately 50% to 100% of breast tumors depending on the study, whereas only approximately 16% show Myc gene amplification and 22% show increased mRNA expression (6, 10–13). Mechanisms for high Myc expression in human breast tumors lacking gene amplification or elevated mRNA expression have not been reported.
Cells have evolved an elegant signaling pathway to help regulate turnover of Myc so that Myc protein levels are kept low when not needed (1, 14). In this pathway, sequential and interdependent phosphorylation events on Myc at serine 62 (S62) and threonine 58 (T58) influence Myc stability. Initial phosphorylation of S62 by ERK or CDK kinases in response to mitogen signaling transiently increases Myc stability, whereas subsequent phosphorylation of T58 by GSK3β triggers dephosphorylation of S62 by protein phosphatase 2A-B56α (PP2A-B56α), ubiquitination by the SCF-Fbw7 E3 ligase, and proteasomal degradation (15, 16). Burkitt lymphoma-derived Myc mutations usually occur at or around T58, generally resulting in loss of T58 phosphorylation, elevated S62 phosphorylation, and increased Myc protein stability. These Myc mutants have increased oncogenic activity compared with WT Myc when expressed at similar levels, suggesting the importance of the two phosphorylation events in regulating Myc oncogenic activity (16–19). However, mutations in Myc have not been found in any epithelial cancer. We have been exploring whether WT Myc can be stabilized in cancer. Here we report that increased stability of WT Myc is a prominent mechanism for Myc overexpression in breast cancer and that this is associated with a change in the ratio of phospho-S62 (pS62) and phospho-T58 (pT58) to more closely match mutant Myc that has increased oncogenic activity. Exploration into the mechanism behind the altered ratio of S62 and T58 phosphorylation in breast cancer suggests a prominent role for deregulation of the Axin1 tumor suppressor.
Axin1 is a multidomain scaffold protein that coordinates several different protein complexes involved in regulating Wnt, TGFβ, SAPK/JNK, and p53 signaling (20–24). Recently, we found that Axin1 promotes Myc degradation and decreases levels of Myc S62 phosphorylation by coordinating the formation of a Myc destruction complex that includes GSK3β, PP2A, and other proteins involved in degrading Myc (24). The AXIN1 gene expresses two naturally occurring splice variants, variant 1 (v1) and variant 2 (v2). AXIN1V1 encodes an 862-aa protein, whereas the protein encoded by AXIN1V2 lacks the 36 aa from exon 9. Whether Axin1v2 functions differently from Axin1v1 has not been reported to our knowledge. Here we show that decreased expression of total AXIN1 and differential expression of AXIN1V1 and AXIN1V2 contribute to increased Myc protein stability, altered phosphorylation at S62 and T58, and increased Myc oncogenic activity in human breast cancer.
Results
Myc Protein Stability Is Increased in Breast Cancer Cell Lines and Is Associated with Altered Phosphorylation at S62 and T58.
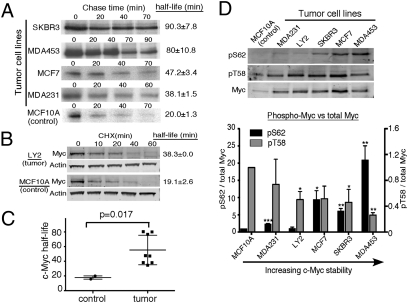
To study mechanisms that underlie elevated Myc expression in breast cancer, we initially focused on five breast cancer cell lines—MCF7, MDA231, SKBR3, LY2, and MDA453—and compared them with MCF10A cells, a nontransformed human mammary epithelial cell line. Relative to MCF10A cells, all five breast cancer cell lines showed increased Myc protein expression, whereas MYC mRNA was only modestly elevated in two of the cell lines, SKBR3 and LY2 (Fig. S1 A and B). We analyzed the turnover rate of Myc protein in these cell lines plus three additional breast cancer cell lines and an additional control cell line, human mammary epithelial cells immortalized with hTERT (hMEC-hTERT). Myc half-life was significantly longer in all eight breast cancer cell lines, ranging from 34 to 90 min, compared with Myc in the controls, which ranged from 16 to 20 min, consistent with a variety of other nontransformed proliferating cell types (25, 26) (Fig. 1 A–C and Fig. S1 C and D). These results indicate that increased Myc half-life may be an important mechanism contributing to increased Myc expression in breast cancer.
Fig. 1.
Increased Myc protein stability associated with increased pS62 and decreased pT58 in breast cancer cell lines. (A) [35S]Methionine pulse/chase analysis shows the decay of radiolabeled Myc. Myc half-life was determined as described in SI Methods. (B) Western analysis of Myc decay in cells treated with 10 μg/mL cycloheximide (CHX). (C) Summary of Myc half-life in control cell lines and in breast cancer cell lines. Bars represent SD. P value was calculated by using a one-tailed Student t test. (D) Western analysis of pS62 and pT58 levels in the indicated cells. Total Myc antibody and pS62-Myc antibody were probed on one Western blot and detected by two-channel imaging with a LI-COR scanner. pT58 was probed separately. Expression of pS62/total Myc (left y axis) and pT58/total Myc (right y axis) was quantified and normalized to MCF10A. Bars represent SD (*P < 0.05, **P < 0.01, and ***P < 0.001).
Although translocated MYC genes in Burkitt lymphoma can harbor coding sequence mutations involving T58 that lead to mutant Myc with increased stability, this has not been reported in any solid cancers to our knowledge. We found no coding mutations in MYC in the breast cancer cell lines under study. We then investigated whether dysfunction of the Myc degradation pathway involving T58 and S62 phosphorylation could account for the increased Myc stability. As part of this pathway, in normal cells, Myc is dephosphorylated at S62 soon after T58 is phosphorylated, leading to rapid Myc turnover and an overall relatively low level of pS62 and high level of pT58 (14, 25). In contrast, deregulation of this degradation pathway leads to an overall high level of pS62 and low level of pT58. We examined phosphorylation at T58 and S62 by using phospho-specific antibodies (24, 25, 27) (Fig. S2A). When comparing to MCF10A cells, we found that the pS62/total Myc ratios were significantly higher in many of the breast cancer cell lines, particularly those with the longer Myc half-lives (Fig. 1D). In contrast, pT58 levels trended lower in many of the breast cancer cell lines when calculated relative to total Myc, again correlating with increased Myc stability. Together, these data indicate a shift of Myc phosphorylation status from high pT58/low pS62 in nontransformed cells to high pS62/low pT58 in breast cancer cells with increased Myc stability. These results are consistent with our previous observations in leukemia cell lines (25).
Myc Has Altered pS62 and pT58 Levels in Primary Human Breast Tumors.
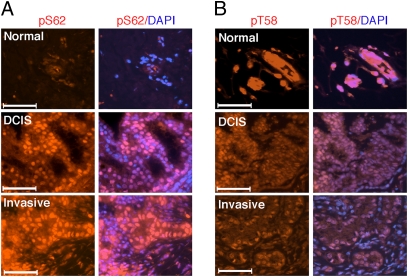
To study whether this shift of pS62 and pT58 levels occurs in primary human breast tumors, we first validated the use of our phospho-specific antibodies on formalin-fixed/paraffin-embedded tissues (Fig. S2B). We then stained slides of patient-matched normal and breast tumor with the pS62- and pT58-specific antibodies. Consistent with our observations in the breast cell lines (Fig. 1D), normal mammary epithelial cells showed very low pS62 staining whereas cells of ductal carcinoma in situ (DCIS) and invasive adenocarcinoma in the same sample showed high pS62 staining (Fig. 2A, red). In contrast, and again consistent with our data in the breast cell lines, serial sections from the same patient samples showed very high pT58 staining in normal mammary gland acini (Fig. 2B, red), whereas matched DCIS and invasive adenocarcinoma showed relatively lower pT58 staining. The same trend was observed in other matched normal and breast tumor samples (Fig. S3 A and B), and can be appreciated when adjacent normal and tumor tissue are under the same microscope field (Fig. S3C). Quantification of the pS62 and pT58 staining from multiple patient-matched normal and breast tumor samples showed increasing levels of pS62 staining from normal to DCIS and invasive adenocarcinoma (Fig. S3D), and a corresponding decrease in pT58 staining (Fig. S3E). These results indicate that normal mammary epithelial cells and breast tumor cells express different forms of Myc as a result of different posttranslational modifications, and suggest that increased Myc protein stability occurs in primary human breast tumors. Moreover, our results indicate that deregulation of the Myc T58/S62 degradation pathway is common in primary breast tumors. We later expanded our pS62 staining to a total of 22 cases, and we found that 16 of the 22 cases showed an increase in pS62 from normal to invasive carcinoma. Of interest, all the pS62-negative cases were also negative for estrogen receptor (ER) and progesterone receptor (PR; Table S1).
Fig. 2.
Increased pS62 and decreased pT58 levels of Myc in human breast cancer. (A) Matched sections of tumor and normal tissue from patient 1 were placed on the same slide and simultaneously stained with pS62 antibody (red) and DAPI (blue). (Scale bars: 50 μM.) (B) Serial sections from patient 1 were stained with pT58 antibody (red) and DAPI (blue). (Scale bars: 50 μM.).
Axin1 Expression Is Decreased in Breast Cancer.
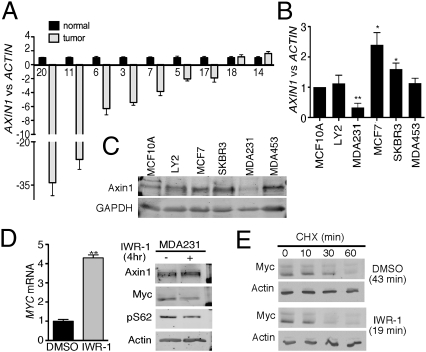
Phosphorylation of T58 by GSK3β and dephosphorylation of S62 by PP2A-B56α are important steps in the T58/S62 Myc degradation pathway (14). We examined expression levels of GSK3β and PP2A-B56α in the breast cancer cell lines relative to the MCF10A cells and did not see any obvious difference. Our recent work identified Axin1 as a scaffold protein that coordinates Myc's interaction with GSK3β and PP2A (24). Alterations of Axin1 including AXIN1 gene mutations and decreased Axin1 expression have been reported in several types of solid tumors (28–30), but so far we are aware of no evidence of these alterations of Axin1 reported in breast cancer. We analyzed AXIN1 mRNA expression in primary breast cancer and adjacent matched normal breast tissue (Fig. 3A). Of the nine sample pairs with sufficient cDNA, seven breast cancer samples showed decreased AXIN1 mRNA levels compared with their adjacent normal tissues. Analysis of AXIN1 mRNA and protein expression in the five breast cancer cell lines relative to MCF10A cells showed a reduction in Axin1 expression only in the MDA231 cells (Fig. 3 B and C).
Fig. 3.
Decreased Axin1 in human breast cancer. (A) Quantitative PCR (qPCR) analysis of AXIN1 versus ACTIN expression tumor samples relative to matched normal samples was graphed in the order of most down-regulated AXIN1. In this case only, bars represent SD from the triplicate qPCR reactions. Missing sample numbers are a result of a lack of sufficient cDNA. The matched normal/tumor ratios were log-transformed, and a P value for AXIN1 down-regulation significance was calculated by single-tailed t test (P = 0.005). (B) qPCR analysis of AXIN1 mRNA levels in human breast cancer cell lines relative to MCF10A cells. (C) Western analysis of Axin1 protein levels in breast cancer cell lines and control MCF10A cells. (D) MDA231 cells were treated with 5 μM IWR-1 for 4 h and harvested for qRT-PCR analysis of Myc and Western analysis of the indicated proteins. MYC mRNA from equal total RNA input was graphed based on changes in Ct and is shown ±SD. (E) MDA231 cells were starved in 0.1% FBS for 48 h, treated with DMSO or 10 μM IWR-1 for 1 h, and stimulated with 10% FBS for another 3 h in the presence of DMSO or IWR-1. Protein stability was analyzed as described in Fig. 1 (*P < 0.05, **P < 0.01, and ***P < 0.001).
Given the low expression of Axin1 in the MDA231 cells, we examined whether increasing Axin1 expression would affect Myc protein levels by using a newly published small chemical compound, IWR-1, that can increase Axin1 protein stability (31). IWR-1 treatment of MDA231 cells for 24 h increased Axin1 protein levels, and this corresponded with a decrease in Myc (Fig. S4). However, we also observed a decrease in MYC mRNA that likely reflects Axin1 regulation of β-catenin, which can transcriptionally activate the MYC gene. To avoid this complication, we treated MDA231 cells with IWR-1 for 4 h. At this time, IWR-1 caused a consistent increase in MYC mRNA (Fig. 3D, graph), but a reduction in Myc protein and pS62 that correlated with a small increase in Axin1 (Fig. 3D, Western blot). The increase in MYC mRNA here might reflect a relief of Myc's negative autoregulation on its own transcription when Myc protein levels are decreased (32). Nonetheless, these results demonstrate that MYC mRNA and protein expression are strongly uncoupled upon increasing Axin1 expression. Indeed, IWR-1 treatment decreased Myc protein half-life from 43 min to 19 min in MDA231 cells (Fig. 3E). Taken together, these data show that restoring Axin1 expression in tumor cells can promote removal of the stabilizing pS62 and decrease Myc stability and expression.
Axin1 Regulates Myc's Oncogenic Activity in Breast Cancer.
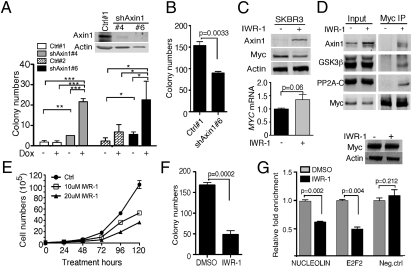
To test whether Axin1 can affect Myc's oncogenic activity, we made stable cell lines that express doxycycline-inducible Myc and either shRNA against Axin1 or the corresponding empty vector in MCF10A cells. Knocking down Axin1 expression in these cells increased Myc levels, consistent with Axin1's role in regulating Myc protein stability (Fig. S5A, lane 4 vs. lane 2). We then performed soft agar assays with two Axin1 shRNA clones (Fig. 4A, Western blot) and two control clones to test if Axin1 loss affects Myc's ability to transform MCF10A cells. As expected, control clones produced very few colonies in soft agar, and overexpression of Myc (plus doxycycline) increased this modestly in only one clone (Fig. 4A, graph). Knocking down Axin1 alone also modestly increased colony numbers. However, when we knocked down Axin1 in the presence of ectopic Myc, the numbers of colonies increased dramatically (Fig. 4A). In contrast, Axin1 knockdown did not increase the colony number in cells expressing the Axin1-insensitive Myc phospho-mutant, MycT58A (Fig. 4B) (24). These results indicate that Axin1 knockdown cooperates with overexpression of Myc in mammary epithelial cell transformation dependent on regulatable phosphorylation of Myc at S62 and/or T58, suggesting that a direct effect on Myc phosphorylation underlies the cooperation.
Fig. 4.
Axin1 regulates Myc oncogenic activity in breast cancer. (A) The MCF10A stable clones Ctrl#1,2 (empty vector) and shAxin1#4,6 were grown in soft agar for 4 wk with or without 1 μg/mL doxycycline (Dox), and colonies were counted. The degree of Axin1 knockdown in different clones is shown in the Western analysis. (B) Stable clones were infected with adenovirus Ad-MycT58A for 18 h. Soft agar assay was done as in A. (C) SKBR3 cells were treated with 10 μM IWR-1 for 24 h and analyzed for Myc and Axin1 expression by Western analysis. MYC mRNA expression was analyzed as in Fig. 3D. (D) SKBR3 cells were starved in 0.2% FBS for 24 h and treated with 5 μM IWR-1 or DMSO for another 30 h. Cells were treated with 10 μM MG132 for 4 h before harvesting for coimmunoprecipitation (co-IP) with Myc antibody C33. Western analysis of input and IP is shown. (E) Cell growth curve of SKBR3 cells with or without IWR-1 treatment. (F) Soft agar assay of SKBR3 cells treated with 10 μM IWR-1 or DMSO as described in Methods. (G) SKBR3 cells were treated with 10 μM IWR-1 for 4 h, and ChIP was done with N262 antibody followed by qPCR. GAPDH internal primers were used as negative control (Neg. ctrl). Western blot of ChIP inputs is shown at the top right corner (*P < 0.05, **P < 0.01, and ***P < 0.001).
We next examined the effects of increasing Axin1 expression in breast cancer cells besides MDA231. In our previous study characterizing the effects of Axin1 on Myc, we showed that the SKBR3 breast cancer cell line had decreased interaction between Myc and Axin1, PP2Ac, and GSK3β, compared with the control MCF10A cells (24). Thus, even though we did not observe decreased expression of Axin1 in the SKBR3 cells, this result suggested a dysfunction of the Axin1 scaffold protein in these cells. To test whether we could overcome this dysfunction by increasing Axin1 expression, we treated SKBR3 cells with IWR-1 for 24 h. Similar to the results in the MDA231 cells, we observed increased Axin1 and decreased Myc protein upon IWR-1 treatment and, in this case, no significant difference in MYC mRNA expression was seen (Fig. 4C). In addition, IWR-1 treatment increased interaction between Myc and its regulators Axin1, GSK3β, and PP2Ac, indicating an increased scaffold function of Axin1 (Fig. 4D). Consistently, we saw decreased Myc protein stability in SKBR3 cells upon IWR-1 treatment (Fig. S5B). Moreover, SKBR3 cells treated with IWR-1 grew slower in proliferation studies (Fig. 4E) and formed less colonies in soft agar (Fig. 4F). Interestingly, short-term treatment with IWR-1 that did not result in decreased Myc levels in these cells (Fig. 4G, Western blot) still resulted in an inhibition of Myc promoter binding at its target genes determined by ChIP (Fig. 4G). This inhibition could underlie, at least in part, the reduced transformation of these cells upon IWR-1 treatment. Taken together, these data demonstrate that increasing Axin1 expression in breast cancer cells can decrease Myc expression, stability, and transactivation activity associated with reduced cell transformation, providing a biological relevance of decreased Axin1 expression in breast cancer.
Breast Cancer Cells Have a Switch in AXIN1 Splice Variant Expression That Contributes to Myc Activation.
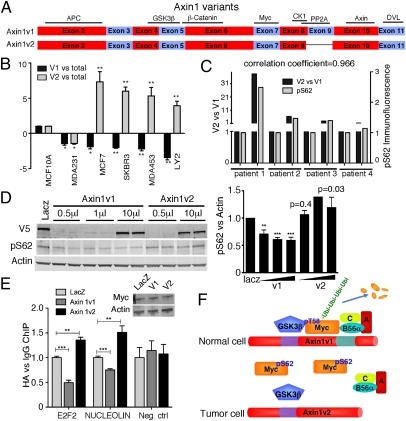
Although many of the primary human breast cancer samples showed decreased Axin1 expression relative to their adjacent normal tissue, only one of the five breast cancer cell lines showed reduced Axin1 expression. As cells like SKBR3 that have a normal level of Axin1 also have an Axin1 dysfunction (24), we examined whether AXIN1 could be mutated in these cells as well as the rest of the breast cancer cell lines under study. We sequenced AXIN1 cDNA and did not find any mutation that would affect the Axin1 coding sequence. Interestingly, the AXIN1 cDNA sequences that we obtained were all from a naturally occurring splice variant of AXIN1, termed AXIN1V2, suggesting an enrichment in this variant. AXIN1, also known as AXIN1V1, encodes an 862-aa protein, whereas the protein encoded by AXIN1V2 lacks 36 aa encoded by exon 9 (Fig. 5A). The function of this domain is unknown, but the PP2A binding domain has been mapped to this region (33). We previously demonstrated that Axin1v2 has a significantly reduced ability to interact with Myc and PP2Ac, suggesting that Axin1v2 has a decreased ability to regulate S62 phosphorylation (24). We are aware of no other published report on the functional significance of this splice variant.
Fig. 5.
Switch from AXIN1V1 to AXIN1V2 expression in breast cancer. (A) Schematic diagram of Axin1v1 and Axin1v2 coding exons modified from a previous publication (28). (B) qRT-PCR analysis of AXIN1V1 and AXIN1V2 in breast cancer cell lines. Ratios of AXIN1V1 versus total AXIN1 and AXIN1V2 versus total AXIN1 normalized to those of MCF10A were graphed ±SD. (C) Sets of normal (N) human breast tissue and matched tumor (T) tissue were prepared as snap-frozen samples for extracting RNA for qRT-PCR of AXIN1V1 and AXIN1V2 expression or paraffin-embedded samples for pS62 immunofluorescence. The ratio of V2 versus V1 and the pS62 staining intensity in tumor tissue relative to matched normal tissue were graphed. Correlation coefficient was calculated by using Excel. (D) Top: SNU475 cells were infected with lentivirus expressing V5-tagged LacZ as a control or V5-tagged Axin1v1 or v2 protein for 72 h and subjected to Western analysis. Graph shows pS62-Myc versus Actin levels from multiple experiments. (E) SNU475 cells were infected as in D for 48 h and then infected with adenovirus expressing HA-tagged Myc for 18 h. ChIP studies were done with HA antibody as described for Fig. 4G. (F) A model showing deregulation of Axin1 contributes to stabilization and accumulation of pS62-Myc in breast cancer. In normal cells, Axin1 coordinates a Myc destruction complex including GSK3β, PP2A, and other proteins, and promotes Myc degradation. In tumor cells, a switch in Axin1 splice variants and/or decreased Axin1 total level contributes to disruption of the Myc destruction complex and resulting accumulation of pS62-Myc. Fig. S8 shows a more detailed model (*P < 0.05, **P < 0.01, and ***P < 0.001).
We examined AXIN1V1 and AXIN1V2 expression in the breast cancer cell lines and found that the ratios of AXIN1V1 versus total AXIN1 were decreased in the breast cancer cell lines except MDA231 in which total AXIN1 was reduced, whereas the ratios of AXIN1V2 versus total AXIN1 levels were increased compared with their expression in MCF10A cells (Fig. 5B). This result is likely to at least partially explain the reduced association between Myc and Axin1 and the increased Myc stability that we reported in the SKBR3 cells (24). As IWR-1 treatment can affect Myc in SKBR3 cells (Fig. 4), we tested its ability to stabilize Axin1v1 and Axin1v2. Although IWR-1 is reported to interact with the C-terminal part of Axin1, including the region absent in Axin1v2 (31), we found that IWR-1 could increase both Axin1v1 and Axin1v2 ectopically expressed (for detection purposes) in SKBR3 cells (Fig. S6A). Thus, it appears that IWR-1 treatment can recover Axin1 function even in tumor cells with enhanced AXIN1V2 and reduced AXIN1V1 expression. In addition, similar alterations in splice variant expression were found in primary human breast tumors, in which the ratio of AXIN1V2 versus AXIN1V1 was significantly increased compared with their expression ratio in patient matched normal samples in seven of 11 cases (Fig. S6B). Moreover, we also observed an enhanced expression of axin1v2 versus axin1v1 in Myc/Neu-induced mouse mammary gland tumors relative to normal mammary gland (Fig. S6C). More importantly, we found that the switch in AXIN1V2 versus V1 expression correlated with increased pS62-Myc in sets of patient matched breast tumor relative to normal, as well as in the Myc/Neu-driven tumors (Fig. 5C and Table S2). Thus, not only can breast cancer exhibit a reduction in total AXIN1 levels (Fig. 3A), but also, in many cases, increased AXIN1V2 versus AXIN1V1 expression relative to normal cells. To our knowledge, this is the first report to demonstrate altered expression of these splice variants. Taken together, these data show that a shift toward enhanced Axin1v2 expression is common in human breast cancer, suggesting a functional significance of this switch for breast oncogenesis.
As Axin1 facilitates PP2A-mediated dephosphorylation of Myc at S62 and Axin1v2 shows decreased interaction with PP2A (24), we tested whether Axin1v2 differs in its ability to promote dephosphorylation of S62. For these experiments, we used SNU475 cells, a human hepatocellular carcinoma cell line with homozygous deletion of exons 1 and 2 of AXIN1 and no Axin1 expression. Our previous work showed that SNU475 cells have high pS62-Myc levels compared with the HCC cell line HepG2, which has WT Axin1 (24). Expression of Axin1v1 in SNU475 cells consistently decreased pS62-Myc levels as expected, whereas expression of Axin1v2 did not (Fig. 5D). No changes in MYC mRNA levels were observed with expression of either splice variant. We also infected these cells with adenovirus expressing CMV-driven Myc as another way to rule out indirect effects via β-catenin. Similar to the results with endogenous Myc, we observed a decrease in pS62-Myc in cells expressing Axin1v1 but not in cells expressing Axin1v2 (Fig. S7A). We did not observe a significant change in total Myc in either of these experiments, probably because of other unknown defects downstream of Axin1 in the Myc degradation pathway in this cancer cell line (Fig. S7A). Besides affecting Myc protein stability, Myc phosphorylation also affects its oncogenic activity in that the high-pS62 form of Myc is more oncogenic than the form of Myc lacking pS62 (17, 19), suggesting an effect of altering this phosphorylation on Myc target gene regulation. Indeed, recent studies in other laboratories and in our laboratory have demonstrated an important role for pS62 in Myc binding to target gene promoters (34, 35) (Fig. S7B). Thus, we analyzed the effects of the two Axin1 splice variants on Myc promoter binding and found that Axin1v1 consistently reduced Myc binding at the NUCLEOLIN and E2F2 promoters whereas Axin1v2 did not inhibit, but rather significantly increased, Myc promoter binding, suggesting that Axin1v2 might have a dominant-negative role in regulating Myc promoter binding (Fig. 5E). Consistent with the ChIP results, NUCLEOLIN and E2F2 mRNA levels were decreased with Axin1v1 expression and increased with Axin1v2 expression (Fig. S7C). From this, it appears that the negative effects of IWR-1 in SKBR3 cells (Fig. 4 C–G) likely result from increased expression of Axin1v1, rather than Axin1v2. Together, our results show that breast cancer cells commonly express a splice variant of Axin1 that has lost its ability to negatively regulate Myc, and that compounds that stabilize Axin1 can overcome this.
Discussion
Myc is a well known oncoprotein that regulates many cellular activities important for tumorigenesis. Several mechanisms have been shown to regulate Myc oncogenic activity. These include changes in Myc protein level, which is commonly elevated in human cancer (36, 37), and phosphorylation changes at T58 and S62 (17, 19, 37). Importantly, these two mechanisms can be linked in that phosphorylation at T58 and S62 also helps to regulate Myc protein stability and expression level. Although it has become clear that T58 and S62 phosphorylation play important roles in Myc biology, analysis of Myc protein stability and/or T58 and S62 phosphorylation levels has not been reported in most human cancer types. In this study, we have examined Myc stability and phosphorylation in human breast cancer cell lines as well as patient samples, and we have uncovered a mechanism to increase pS62-Myc and stability in breast cancer, involving down-regulation of the Axin1 tumor suppressor protein and newly identified changes in Axin1 splice variant expression (Fig. 5F and Fig. S8).
Increased Stability and Altered Ratios of T58 and S62 Phosphorylation in Human Breast Cancer.
Our analysis of Myc protein stability and pT58 and pS62 levels in human breast cancer cell lines and primary patient breast tumor samples demonstrates that stabilization of Myc associated with altered S62 and T58 phosphorylation ratios is common in breast cancer. Careful examination of the effects of different Myc expression levels in mice has revealed different activities at different expression levels (36). In addition, studies have shown that tumor-derived Myc mutants, mutations of which affect the phosphorylation status at S62 and T58, are more tumorigenic than WT Myc (17). Moreover, we have shown, by using a unique mouse model that, at near-physiological levels of expression in the mammary gland, the MycT58A mutant, which lacks T58 phosphorylation and has constitutively high S62 phosphorylation, similar to the phosphorylation pattern we report here in breast cancer, is tumorigenic whereas deregulated near-physiological levels of WT Myc is not (19). Recent reports have demonstrated that pS62 is important for Myc binding to a number of transactivated target genes important for cell proliferation, growth, and survival (34, 35). Thus, increased Myc protein stability and altered T58/S62 phosphorylation are likely to play important roles in breast tumorigenesis.
Aberrant Expression of Axin1 in Human Breast Cancer.
Axin1 has been characterized as a tumor suppressor, and multiple mutations have been identified throughout AXIN1 in a number of different cancers (28). Overexpression of Axin1 in a transgenic mouse model causes increased apoptosis in the mouse mammary gland (38), suggesting a tumor suppressor function of Axin1 there. However, thus far, no mutation that affects the Axin1 coding sequence or deregulation of Axin1 expression has been reported in human breast cancer. The present study demonstrates that decreased AXIN1 expression and altered ratios of two splice variants are common occurrences in breast cancer cell lines and primary tumor samples. Moreover, we show that this contributes to increased Myc protein stability and oncogenic activity in breast cancer. Specifically, knocking down Axin1 cooperates with Myc in promoting cell transformation in immortalized cells, and increasing Axin1 levels with a small molecule, IWR-1, in breast cancer cells decreases cell transformation associated with suppressed Myc binding to target gene promoters. Thus, our study reveals another mechanism of action for the Axin1 tumor suppressor, whereby, in addition to its other targets such as β-catenin, SAPK/JNK, TGFβ, and p53, Axin1 can suppress cell transformation through inhibition of Myc. Further research on how Axin1 coordinately regulates all of its targets will help our understanding of Axin1's role in suppressing breast tumorigenesis.
In breast tumor cells, besides decreased expression of total AXIN1, we found that a shift from AXIN1V1 to AXIN1V2 was common and is correlated with high pS62. The biological role of these two splice variants has not been reported. We previously found that Axin1v2's interaction with Myc and PP2Ac is lower than that of Axin1v1 (24). Here we showed that ectopic Axin1v1 decreased pS62-Myc levels in Axin1-null cells, whereas Axin1v2 did not. Moreover, whereas Axin1v1 can decrease Myc promoter binding, Axin1v2 does the opposite, suggesting a potential oncogenic side of this splice variant. Clearly, Axin1v2 and Axin1v1 differ significantly in their regulation of Myc phosphorylation and activity. It is possible that the two splice variants might also differentially regulate other Axin1 targets such as β-catenin and p53. Thus, increasing expression of Axin1v2 represents another way of deregulating Axin1 to promote tumorigenesis. In addition, Axin1 activity is regulated posttranslationally, including phosphorylation, protein stability, and subcellular localization, and it would be interesting to know in the future if deregulation through these mechanisms contributes to Myc stabilization in cancer. However, it is also clear that down-regulation of Axin1 or a switch in splice variant expression did not occur in all the tumors we examined, and other mechanisms are likely to contribute to altered Myc T58/S62 phosphorylation, such as deregulation of GSK3β and PP2A.
Therapeutic and Diagnostic Implications of Our Results.
Myc is overexpressed in many human tumors, and turning off Myc expression has been shown to be an effective and efficient cancer therapy in mouse models (8, 9, 39). In addition, animal modeling has demonstrated that inhibition of some Myc activity can be tolerated by many organs (39). Thus, understanding how Myc phosphorylation and stability are deregulated in different cancer types becomes critically important to help design rational therapies. Indeed, we have shown that treatment of breast cancer cells with a small molecule that increases Axin1 expression and suppresses Myc activity repressed their oncogenic potential substantially. Furthermore, with the pS62-specific antibody we have developed for Myc, it should be possible to screen human breast tumors for those expressing more stable and oncogenically active Myc protein to help direct treatment.
Methods
Cell Culture.
Cell lines used were purchased from ATCC except LY2, which was a gift from Lawrence Berkeley National Laboratory (Berkeley, CA). Cells were cultured and maintained as described in SI Methods. Patient samples were obtained from the Oregon Health and Science University Cancer Pathology Shared Resource (institutional review board approval nos. 4918 and 2086). cDNA samples used in Fig. 3A and Fig. S6B were provided by D.C.
Statistics.
SD results were taken from three independent experiments. P value was analyzed by Student t test, with a two-tailed method unless otherwise indicated. Western blot was done as described previously (24). Immunoblots were visualized via Odyssey IR imager (LI-COR) that can simultaneously detect Fluor 680 and IRDye 800 secondary antibodies. Quantification of Western blots was done by using Odyssey IR software, version 1.2 (LI-COR). [35S]Methionine pulse/chase experiments were done as described previously (25).
Immunofluorescence.
Matched normal and tumor sections were placed on the same slide and stained simultaneously or adjacent normal was present in the tumor block and thus on the same section. Immunofluorescence density was analyzed with OpenLab 5.5 software (see SI Methods for details).
Cell Proliferation Assay.
A total of 80,000 SKBR3 cells treated with 10 μM IWR-1 compound or DMSO were cultured for 5 d and counted with a hemacytometer.
Soft Agar Assays.
The bottom and top agar layers were 0.8% and 0.35% Nobel agar, respectively. Cells (2 × 104) were plated for each six-well plate. For SKBR3, 20 μM IWR-1 was used. ChIP methods were modified from previous report (24). SI Methods provides detailed information on soft agar assays.
Supplementary Material
Acknowledgments
We thank Dr. Xiaoyan Wang for providing mouse tumor samples, Dr. Chris Corless for providing breast cancer samples, and Carl Pelz for help with statistics analysis. This study was supported by Department of Defense Breast Cancer Research Program Award BC061306 (to R.S.), Susan. G. Komen Breast Cancer Foundation Awards BCTR0201697 and BCTR0706821 (to R.S.), and National Institutes of Health Award 1 R01 CA129040-01 (to R.S.), and Welch (I-1665) and CPRIT (RP100119) (to L.L.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. K.P. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1100764108/-/DCSupplemental.
References
- 1.Meyer N, Penn LZ. Reflecting on 25 years with MYC. Nat Rev Cancer. 2008;8:976–990. doi: 10.1038/nrc2231. [DOI] [PubMed] [Google Scholar]
- 2.Mai S, et al. Chromosomal and extrachromosomal instability of the cyclin D2 gene is induced by Myc overexpression. Neoplasia. 1999;1:241–252. doi: 10.1038/sj.neo.7900030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Felsher DW, Bishop JM. Transient excess of MYC activity can elicit genomic instability and tumorigenesis. Proc Natl Acad Sci USA. 1999;96:3940–3944. doi: 10.1073/pnas.96.7.3940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yin XY, Grove L, Datta NS, Long MW, Prochownik EV. C-myc overexpression and p53 loss cooperate to promote genomic instability. Oncogene. 1999;18:1177–1184. doi: 10.1038/sj.onc.1202410. [DOI] [PubMed] [Google Scholar]
- 5.Prochownik EV, Li Y. The ever expanding role for c-Myc in promoting genomic instability. Cell Cycle. 2007;6:1024–1029. doi: 10.4161/cc.6.9.4161. [DOI] [PubMed] [Google Scholar]
- 6.Nesbit CE, Tersak JM, Prochownik EV. MYC oncogenes and human neoplastic disease. Oncogene. 1999;18:3004–3016. doi: 10.1038/sj.onc.1202746. [DOI] [PubMed] [Google Scholar]
- 7.Pelengaris S, Littlewood T, Khan M, Elia G, Evan G. Reversible activation of c-Myc in skin: Induction of a complex neoplastic phenotype by a single oncogenic lesion. Mol Cell. 1999;3:565–577. doi: 10.1016/s1097-2765(00)80350-0. [DOI] [PubMed] [Google Scholar]
- 8.D'Cruz CM, et al. c-MYC induces mammary tumorigenesis by means of a preferred pathway involving spontaneous Kras2 mutations. Nat Med. 2001;7:235–239. doi: 10.1038/84691. [DOI] [PubMed] [Google Scholar]
- 9.Felsher DW, Bishop JM. Reversible tumorigenesis by MYC in hematopoietic lineages. Mol Cell. 1999;4:199–207. doi: 10.1016/s1097-2765(00)80367-6. [DOI] [PubMed] [Google Scholar]
- 10.Blancato J, Singh B, Liu A, Liao DJ, Dickson RB. Correlation of amplification and overexpression of the c-myc oncogene in high-grade breast cancer: FISH, in situ hybridisation and immunohistochemical analyses. Br J Cancer. 2004;90:1612–1619. doi: 10.1038/sj.bjc.6601703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Blancato JK, Williams MS, Dickson RB. Fluorescence in situ hybridization assessment of c-myc gene amplification in breast tumor tissues. Methods Mol Med. 2006;120:297–307. doi: 10.1385/1-59259-969-9:297. [DOI] [PubMed] [Google Scholar]
- 12.Agnantis NJ, Mahera H, Maounis N, Spandidos DA. Immunohistochemical study of ras and myc oncoproteins in apocrine breast lesions with and without papillomatosis. Eur J Gynaecol Oncol. 1992;13:309–315. [PubMed] [Google Scholar]
- 13.Bièche I, et al. Quantitation of MYC gene expression in sporadic breast tumors with a real-time reverse transcription-PCR assay. Cancer Res. 1999;59:2759–2765. [PubMed] [Google Scholar]
- 14.Sears RC. The life cycle of C-myc: From synthesis to degradation. Cell Cycle. 2004;3:1133–1137. [PubMed] [Google Scholar]
- 15.Welcker M, et al. The Fbw7 tumor suppressor regulates glycogen synthase kinase 3 phosphorylation-dependent c-Myc protein degradation. Proc Natl Acad Sci USA. 2004;101:9085–9090. doi: 10.1073/pnas.0402770101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yeh E, et al. A signalling pathway controlling c-Myc degradation that impacts oncogenic transformation of human cells. Nat Cell Biol. 2004;6:308–318. doi: 10.1038/ncb1110. [DOI] [PubMed] [Google Scholar]
- 17.Hemann MT, et al. Evasion of the p53 tumour surveillance network by tumour-derived MYC mutants. Nature. 2005;436:807–811. doi: 10.1038/nature03845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thibodeaux CA, et al. Immortalization and transformation of human mammary epithelial cells by a tumor-derived Myc mutant. Breast Cancer Res Treat. 2009;116:281–294. doi: 10.1007/s10549-008-0127-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang X, et al. Phosphorylation regulates c-Myc's oncogenic activity in the mammary gland. Cancer Res. 2011;71:925–936. doi: 10.1158/0008-5472.CAN-10-1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zeng L, et al. The mouse Fused locus encodes Axin, an inhibitor of the Wnt signaling pathway that regulates embryonic axis formation. Cell. 1997;90:181–192. doi: 10.1016/s0092-8674(00)80324-4. [DOI] [PubMed] [Google Scholar]
- 21.Zhang Y, Neo SY, Wang X, Han J, Lin SC. Axin forms a complex with MEKK1 and activates c-Jun NH(2)-terminal kinase/stress-activated protein kinase through domains distinct from Wnt signaling. J Biol Chem. 1999;274:35247–35254. doi: 10.1074/jbc.274.49.35247. [DOI] [PubMed] [Google Scholar]
- 22.Rui Y, et al. Axin stimulates p53 functions by activation of HIPK2 kinase through multimeric complex formation. EMBO J. 2004;23:4583–4594. doi: 10.1038/sj.emboj.7600475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu W, et al. Axin is a scaffold protein in TGF-beta signaling that promotes degradation of Smad7 by Arkadia. EMBO J. 2006;25:1646–1658. doi: 10.1038/sj.emboj.7601057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Arnold HK, et al. The Axin1 scaffold protein promotes formation of a degradation complex for c-Myc. EMBO J. 2009;28:500–512. doi: 10.1038/emboj.2008.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Malempati S, et al. Aberrant stabilization of c-Myc protein in some lymphoblastic leukemias. Leukemia. 2006;20:1572–1581. doi: 10.1038/sj.leu.2404317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sears R, Leone G, DeGregori J, Nevins JR. Ras enhances Myc protein stability. Mol Cell. 1999;3:169–179. doi: 10.1016/s1097-2765(00)80308-1. [DOI] [PubMed] [Google Scholar]
- 27.Escamilla-Powers JR, Sears RC. A conserved pathway that controls c-Myc protein stability through opposing phosphorylation events occurs in yeast. J Biol Chem. 2007;282:5432–5442. doi: 10.1074/jbc.M611437200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Salahshor S, Woodgett JR. The links between axin and carcinogenesis. J Clin Pathol. 2005;58:225–236. doi: 10.1136/jcp.2003.009506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Satoh S, et al. AXIN1 mutations in hepatocellular carcinomas, and growth suppression in cancer cells by virus-mediated transfer of AXIN1. Nat Genet. 2000;24:245–250. doi: 10.1038/73448. [DOI] [PubMed] [Google Scholar]
- 30.Webster MT, et al. Sequence variants of the axin gene in breast, colon, and other cancers: an analysis of mutations that interfere with GSK3 binding. Genes Chromosomes Cancer. 2000;28:443–453. [PubMed] [Google Scholar]
- 31.Chen B, et al. Small molecule-mediated disruption of Wnt-dependent signaling in tissue regeneration and cancer. Nat Chem Biol. 2009;5:100–107. doi: 10.1038/nchembio.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Penn LJ, Brooks MW, Laufer EM, Land H. Negative autoregulation of c-myc transcription. EMBO J. 1990;9:1113–1121. doi: 10.1002/j.1460-2075.1990.tb08217.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hsu W, Zeng L, Costantini F. Identification of a domain of Axin that binds to the serine/threonine protein phosphatase 2A and a self-binding domain. J Biol Chem. 1999;274:3439–3445. doi: 10.1074/jbc.274.6.3439. [DOI] [PubMed] [Google Scholar]
- 34.Benassi B, et al. c-Myc phosphorylation is required for cellular response to oxidative stress. Mol Cell. 2006;21:509–519. doi: 10.1016/j.molcel.2006.01.009. [DOI] [PubMed] [Google Scholar]
- 35.Hydbring P, et al. Phosphorylation by Cdk2 is required for Myc to repress Ras-induced senescence in cotransformation. Proc Natl Acad Sci USA. 2010;107:58–63. doi: 10.1073/pnas.0900121106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Murphy DJ, et al. Distinct thresholds govern Myc's biological output in vivo. Cancer Cell. 2008;14:447–457. doi: 10.1016/j.ccr.2008.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Smith DP, Bath ML, Metcalf D, Harris AW, Cory S. MYC levels govern hematopoietic tumor type and latency in transgenic mice. Blood. 2006;108:653–661. doi: 10.1182/blood-2006-01-0172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hsu W, Shakya R, Costantini F. Impaired mammary gland and lymphoid development caused by inducible expression of Axin in transgenic mice. J Cell Biol. 2001;155:1055–1064. doi: 10.1083/jcb.200107066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Soucek L, et al. Modelling Myc inhibition as a cancer therapy. Nature. 2008;455:679–683. doi: 10.1038/nature07260. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.