Abstract
Estrogen receptor (ER) binds to distal enhancers within the genome and requires additional factors, such as the Forkhead protein FoxA1, for mediating chromatin interactions. We now show that the human Groucho protein, Transducin-like enhancer protein 1 (TLE1), positively assists some ER-chromatin interactions, a role that is distinct from its general role as a transcriptional repressor. We show that specific silencing of TLE1 inhibits the ability of ER to bind to a subset of ER binding sites within the genome, a phenomenon that results in perturbations in phospho-RNA Pol II recruitment. Furthermore, TLE1 is essential for effective ER-mediated cell division. We have discovered a distinct role for TLE1, as a necessary transcriptional component of the ER complex, where it facilitates ER-chromatin interactions.
Estrogen receptor (ER) is the defining transcription factor of luminal breast tumors (1) and is the target of most endocrine-based breast cancer therapies, including tamoxifen and aromatase inhibitors (2). Understanding how estrogen–ER initiates transcription events is paramount in understanding how endocrine therapies work and what happens if they fail.
ER transcriptional activity involves numerous proteins, and a significant amount of work has shown an essential role for ER-associated cofactors in regulating chromatin. Recently a previously undescribed class of regulatory proteins called pioneer factors have been implicated in maintaining transcription factor interactions with the chromatin (3, 4). FoxA1 is an ER pioneer factor that promotes ER binding to chromatin, a requirement for estrogen-mediated cell growth (5, 6). Furthermore, genome-wide mapping of FoxA1 and ER revealed that half of all ER binding sites co-occur at FoxA1 binding regions in the genome (7, 8). Preliminary data suggests that pioneer factors function differently to known ER cofactors; rather than enzymatically modulating chromatin structure or functioning as adapter proteins for other cofactors, they are directly required to maintain interactions between ER and the chromatin (5, 6).
The Groucho/transducin-like enhancer of split (TLE) proteins also possesses similar properties to pioneer factors, namely that they can bind to histones (9) within condensed chromatin, independently of other proteins (10). TLE proteins have a well-characterized role as repressor proteins for various transcriptional regulators including Wnt and Notch pathways (reviewed in ref. 11). Interestingly, using chromatin reconstitution assays, TLE and FoxA1 proteins were shown to interact, independently of other proteins (10), and TLE proteins were shown to interact with and modulate Pax2 transcriptional activity (12), a protein that was recently implicated as an ER-associated repressor of transcription (13). Furthermore, estrogen receptor related-γ (ERR-γ) interacts with TLE1, where it was subsequently shown to be a transcriptional activator (14); this is in contrast to the general role for TLE1 as a repressor of transcription (15).
Results
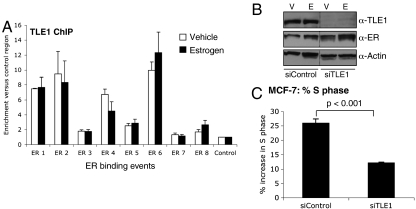
We explored the possibility that TLE proteins may play a role in ER-mediated transcriptional activation. Because TLE1 was previously shown to interact with a nuclear receptor (14), we focused on this family member. To determine if TLE1 could bind to regions of the genome known to be distal ER interacting cis-regulatory elements, we performed ChIP of TLE1 in ER positive MCF-7 breast cancer cells. Cells were hormone deprived for 3 d to induce cell cycle arrest and treated with vehicle or 100 nM estrogen for 45 min, a time period known to result in ER binding to chromatin (5, 16). TLE1 ChIP was performed, followed by quantitative PCR of a number of previously identified ER binding sites (17). Real-time PCR data suggested that TLE1 bound to approximately half of the tested ER binding sites and that importantly, TLE1 binding occurred prior to estrogen treatment (Fig. 1A), in an ER independent manner. In fact, estrogen treatment did not influence TLE1 binding, suggesting that TLE1-chromatin interactions are an estrogen-ER independent event. This is reminiscent of what was shown for FoxA1 (5) and suggests that TLE1 may be able to interact directly with condensed chromatin, potentially mediating ER binding events. In support of this, the Zaret lab has shown that the mouse homologs of TLE proteins directly associate with chromatin via the histone tails, independently of other factors (10). TLE1 binding to chromatin may not be influenced by estrogen–ER, but TLE1 bound regions may exist as docking sites for the ER complex following ligand treatment. Following estrogen treatment, we could show by Re-ChIP, a method to show cooccupancy of two proteins on the same region of the chromatin (Fig. S1), that ER/TLE1 form complexes together on the chromatin.
Fig. 1.
TLE1 binds to chromatin at ER binding events and is not influenced by estrogen. (A) TLE1 ChIP after vehicle (white bars) or estrogen (black bars) treatment followed by real-time PCR of ER binding sites. TLE1 binds to a subset of regions tested independently of ligand. The data are the average of three replicate experiments ± Std Dev. (B) siRNA to TLE1 (or control siRNA) was transfected into cells and total protein was immunoblotted. The uncropped figure is in Fig. S1. C. siControl or siTLE1 transfected MCF-7 cells were treated with estrogen for 24 h and %S phase was determined by PI staining and flow cytometry. The %S phase was decreased when TLE1 was silenced. The data are the average of three independent replicates ± Std Dev and example histograms are shown in Fig. S1.
To assess if TLE1 is required for estrogen-mediated proliferation, we specifically silenced TLE1 using siRNA (Western blot showing effective silencing is shown in Fig. 1B), treated cells with estrogen for 24 h, and assessed proliferation by measuring %S phase using flow cytometry. Specific silencing of TLE1 resulted in a significant (p < 0.001) inhibition of MCF-7 cell growth, confirming a requirement for TLE1 in the estrogen growth response (Fig. 1C). Examples of the flow cytometry histograms are provided in Fig. S1. This was validated using two independent siRNAs o TLE1 and using an independent measure of cell growth (cell confluence) (Fig. S1). We could also show a TLE1-dependent estrogen proliferation response in ZR75-1 cells, another ER positive cell line (Fig. S1).
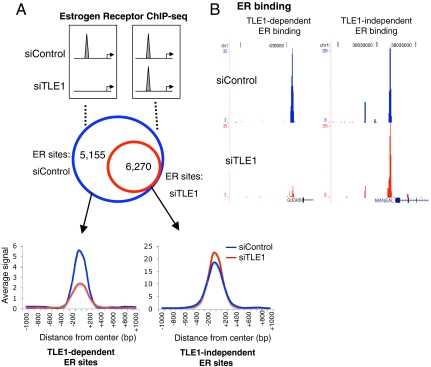
We attempted to map TLE1 binding events by ChIP sequencing (ChIP-seq), but numerous efforts with all commercially available antibodies failed, suggesting that the enrichment we get by ChIP is not sufficient to translate to global sequencing data. However, to glean global insight into whether TLE1 plays a role in modulating ER-chromatin interactions, as had previously been seen for FoxA1 (8), we explored whether silencing of TLE1 impacted ER binding. Importantly, silencing of TLE1 did not affect ER protein levels (Fig. 1B). To obtain a global insight into the potential role of TLE1 in regulating ER binding, we transfected cells with siControl or siTLE1, treated cells with estrogen for 45 min, and performed genome-wide ER ChIP-seq. High throughput Illumina sequencing of ER ChIP in siControl or siTLE1 conditions resulted in approximately 16 million aligned reads for both conditions. A second independent replicate was performed. Peak calling was performed using Model-based analysis of ChIP-seq (MACS) (18), and we considered peaks that occurred only in both biological replicates. We found distinct differences in ER binding profiles in the presence and absence of TLE1. We found 11,425 ER binding events that occurred in both replicates of the siControl-transfected cells, but only 6,270 ER peaks that remained in both replicates following silencing of TLE1. As such, ER binding at 5,155 regions was substantially reduced in multiple experiments, in the absence of TLE1 (Fig. 2A). This suggests that approximately 45% of all ER binding events are dependent on the presence of TLE1 for maintaining chromatin interactions (Fig. 2A). Examples of a TLE1-dependent and a TLE1-independent ER binding event are shown in Fig. 2B. Interestingly, when assessing the global signal intensity of the two categories (TLE1-dependent and TLE1-independent ER binding events), those regions that were decreased when TLE1 was silenced tended to be the weaker ER binding events (Fig. 2A). The TLE1-independent ER binding events were generally stronger and actually gained a subtle increase in binding intensity in the absence of TLE1 (Fig. 2A). A third independent ER ChIP-seq experiment was performed, and again global ER binding signal intensity was decreased when TLE1 was silenced (Fig. S2). A number of these TLE1 dependent sites were validated by ER ChIP and quantitative PCR using an independent siRNA to TLE1. As a separate measure of TLE1-mediated ER binding to chromatin, we transfected cells with siControl or siTLE1, performed chromatin fractionation (19), and Western blotted total chromatin bound ER. The data confirmed that specific silencing of TLE1 resulted in a global decrease in chromatin associated ER (Fig. S2).
Fig. 2.
Silencing of TLE1 inhibits a subset of ER binding sites. (A) Cells were transfected with control or TLE1 siRNA and subsequently treated with estrogen for 45 min. ER ChIP-sequencing was performed. The data represent the overlap in ER binding peaks in the presence and absence of TLE1 from two independent replicates. We compared the ER binding signal intensity in the two categories (the TLE1-independent and TLE1-dependent ER binding events). The signal intensity is shown as an average of all the binding events in that category, in a window of ± 1 kb from the center of the binding event. (B) An example of an ER binding event that is lost upon silencing TLE1 (TLE1-dependent) and one example where ER binding remains (TLE1-independent).
Given our recent finding that ER binding to chromatin requires FoxA1, even under conditions when cells are treated with the antiproliferative drug tamoxifen (8), we assessed whether TLE1 played a similar role. We transfected hormone-depleted MCF-7 cells with siControl or siTLE1, treated cells with tamoxifen for 45 min, and performed an ER ChIP, followed by real-time PCR. Specific silencing of TLE1 inhibited tamoxifen-mediated recruitment of ER to the sites tested (Fig. S2), confirming that TLE1 is required for ER-chromatin interactions even when exposed to additional ligands, such as tamoxifen.
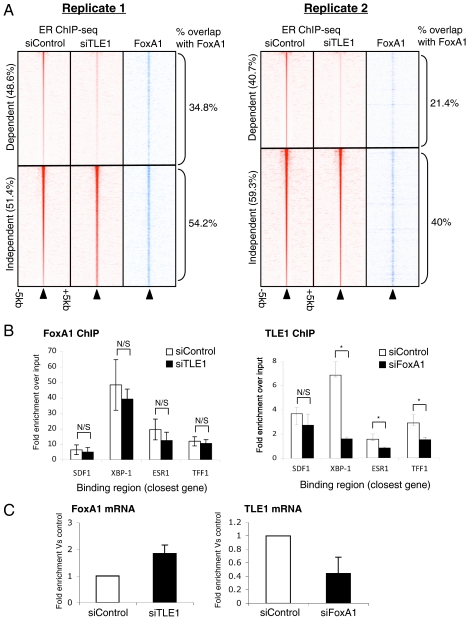
Previous data had suggested that FoxA1 binding occurs at a significant percentage of ER binding sites, where it was proposed to function as a putative pioneer factor for ER (7). To identify if FoxA1 and TLE1 cooperate at shared cis-regulatory elements or function independently of each other, we compared the ER binding sites mapped in the presence or absence of TLE1, with the FoxA1 binding sites generated by ChIP-seq (8). Each ER ChIP-seq replicate was analyzed independently. For replicate 1, TLE1 was required for 48.6% of all ER binding events. Of all the ER-dependent binding events in this replicate, 34.8% were cobound by FoxA1 versus 54.2% of the TLE1-independent ER binding events (Fig. 3A). For replicate two, 40.7% of all ER binding events were TLE1 dependent. Within replicate two, 21.4% of the TLE1-dependent ER binding events overlapped with a FoxA1 binding event versus 40% for the TLE1-independent ER binding events. These data would suggest that TLE1 is required for a substantial fraction of ER binding events, but those that are also bound by FoxA1 are less likely to be affected when TLE1 is silenced. The ER binding events that are regulated by TLE1, but not FoxA1, are more likely to be affected when TLE1 is specifically silenced. As expected, specific silencing of TLE1 did not alter FoxA1 binding, as measured by ChIP, at any of the tested sites (Fig. 3B), yet silencing of FoxA1 reduced TLE1 mRNA levels (Fig. 3C), possibly explaining the decreased TLE1 binding to the tested sites (Fig. 3B).
Fig. 3.
TLE1 dependent ER binding events are less likely to be cobound by FoxA1. (A) Heat map showing ER binding signal intensity for each individual ER ChIP-seq replicate following transfection of siTLE1 or control siRNA. The heat map represents binding signal intensity with binding events ranked from the strongest to the weakest binding events. Each line represents an individual binding event, and the data are shown in a window of ± 5 kb around the center of the binding event. Also shown is the FoxA1 binding signal intensity in each category at the corresponding region and the fraction of each category (TLE1-independent or TLE1-dependent ER binding events) that overlap with a FoxA1 binding event. (B) Hormone-deprived MCF-7 cells were transfected with siRNA to TLE1 (or control siRNA) or FoxA1. ChIP of the reciprocal factor was performed followed by real-time PCR of known ER binding events. FoxA1 binding was not influenced by silencing of TLE1, but TLE1 binding was decreased when FoxA1 was silenced. N/S denotes not significant and * denotes p < 0.05. (C) Changes in mRNA levels of FoxA1 or TLE1 when the other gene was silenced by siRNA.
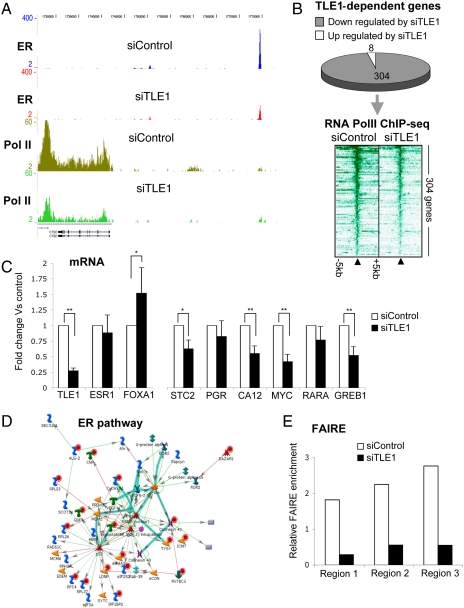
To gain global insight into the role that TLE1 plays in regulating gene transcription events, we transfected MCF-7 cells with siControl or siTLE1 and performed phospho-RNA Pol II ChIP-seq. Duplicate biological replicates were performed and were assessed for phosphorylated RNA Pol II binding at promoters and within gene bodies. When considering genes that had a 1.5-fold (or more) decrease in phospho-RNA Pol II sequencing tags in both replicates (after normalization) following silencing of TLE1, an example is shown in Fig. 4A. We found 304 genes that were dependent on TLE1 for recruitment of phospho-RNA Pol II occupancy. This represented the significant majority of genes that had reproducible changes in phospho-RNA Pol II occupancy (Fig. 4B). As such, in MCF-7 breast cancer cells, TLE1 primarily appears to function as a positive regulator of ER-mediated transcription. To determine whether TLE1 was required for transcript production of estrogen-regulated genes, we assessed mRNA levels of several genes following silencing of TLE1 and could confirm a requirement for a fraction of the tested genes (Fig. 4C).
Fig. 4.
ER-mediated gene expression requires TLE1. Cells were hormone deprived, transfected with siControl or siTLE1, treated with estrogen for 1 h and phospho-RNA Pol II ChIP-seq was performed. We considered differentially regulated genes those that changed by more that 1.5-fold (between siTLE1 and siControl treatments) in both independent replicates. (A) An example of an estrogen-regulated gene (CTSD) and the changes in phospho-RNA Pol II occupancy and ER binding in the presence (siControl) or absence (siTLE1) of TLE1. Following silencing of TLE1, ER binding, and phospho-RNA Pol II occupancy is decreased. (B) The 312 genes contained reproducible changes in phospho-RNA Pol II occupancy following silencing of TLE1, of which 304 are genes that show decreased phospho-RNA Pol II occupancy in the absence of TLE1. Also shown is the phospho-RNA PolII binding intensity around the transcription start sites of the TLE1 dependent genes, in a window of ± 5 kb. Phospho-RNA Pol lI occupancy is decreased following silencing of TLE1. (C) Several genes were assessed using quantitative RT-PCR following silencing of TLE1 in proliferating cells. Also included are changes in TLE1, ESR1 (ER) and FoxA1 mRNA levels as controls. * denotes p < 0.05 and ** denotes p < 0.01. (D) The 304 TLE1 dependent genes were analyzed for enriched biological pathways. We found the gene signatures for the ER pathway to be overrepresented. (E) FAIRE was conducted in MCF-7 cells transfected with siControl or siTLE1. Real-time PCR of several ER binding region were assessed. The experiment was repeated in quadruplicate and one representative experiment is shown.
The list of 304 TLE1-dependent genes included well-known ER target genes such as cyclin D1, STC2, and progesterone receptor. CCND1 (cyclin D1) has been shown to be sufficient to induce cell cycle progression in MCF-7 cells (20), and, therefore, the decrease in phospho-RNA Pol II recruitment to CCND1 likely contributes to the altered proliferative status following silencing of TLE1. By mining the 304 TLE1 dependent genes for enriched biological pathways, we find both the ER (Fig. 4D) and MYC pathways are significantly overrepresented. MYC is a well-established ER target gene and is thought to be responsible for roughly half of the genes regulated by ER (21). Therefore, the changes in phospho-RNA Pol II recruitment following silencing of TLE1 confirm that TLE1 is required for functional gene expression of genes that reflect ER-regulated pathways. Interestingly, this involves some of the weaker ER binding events, confirming that these weaker ER binding regions are still transcriptionally important.
To confirm these findings, we performed gene expression microarray analysis of hormone-depleted cells transfected with siTLE1 (or siControl) and treated with vehicle or estrogen treatment for 6 h. Microarrays measure total steady state transcript levels, as compared to phospho-RNA Pol II occupancy that assesses active transcription. Although total transcript levels represent additional variables (such as transcript stability), we sought to confirm a TLE1 dependency on ER target genes using a separate and independent measure of transcriptional output. Six biological replicates were performed and the changes in estrogen-mediated gene expression following silencing of TLE1 were identified using Illumina beadchip microarrays. As shown in Fig. S3, silencing of TLE1 had a significant inhibitory effect on a large fraction of estrogen-regulated genes, including classic target genes such as CA12, GREB-1, TFF-1, HEY2, and CXCL12. Although the microarray analysis measures different endpoints from phospho-RNA Pol II mapping, it confirmed a dependency on TLE1 for effective estrogen-mediated transcription.
To assess whether TLE1 played a direct role in mediating chromatin structure at ER binding regions, we conducted FAIRE (formaldehyde-assisted isolation of regulatory elements) analysis, a method for enriching nucleosome-depleted regions of the genome (22). We transfected MCF-7 cells with siControl or siTLE1 and collected chromatin for FAIRE. Real-time PCR of three ER binding regions were assessed, and the normalised data confirm that TLE1 is required for optimal euchromatic conditions at the tested regions (Fig. 4E). As such, TLE1 is required for optimal chromatin accessibility, potentially explaining why ER binding is perturbed at specific regions in the absence of TLE1.
Discussion
A significant amount of work over the past decade has focused on the role that ER cofactors play in regulating chromatin. More recently a previously undescribed class of ER regulators, pioneer factors, have been identified as critical regulators of ER function (5, 6). Much of the initial work identifying and characterizing pioneer factor binding together with regulation of condensed chromatin comes from the Zaret lab (3, 4, 10). We, and others, have subsequently found a role for these pioneer factors in ER biology, where they have been shown to be critical mediators of ER-chromatin interactions. They therefore constitute a previously undescribed level of ER regulation: They play a role in maintaining ER-cofactor interactions with the chromatin during transcriptional activity. These proteins that assist other transcription factors in binding to chromatin appear to be able to associate with condensed chromatin via distinct mechanisms. FoxA proteins can mimic linker histone (3) and therefore associate via consensus DNA motifs that exist in regions between nucleosomes. ER binding to condensed chromatin has been shown to be more dependent on FoxA1, as compared to nucleosome-depleted “open” chromatin (8). However, the presence of the Forkhead motif is generally required for FoxA1 binding, providing some specificity for FoxA1 binding.
TLE proteins can associate with condensed chromatin by binding to histone tails of the nucleosomes, rather than via specific DNA consensus motifs (10). Therefore, FoxA and TLE proteins possess the unique abilities to bind condensed chromatin, but this is achieved using distinct mechanisms. Our data confirm that nucleosome-free chromatin (representative of active regulatory regions) is affected when TLE1 is modulated (Fig. 4E). Interestingly, our data would suggest that ER binding can simultaneously be recruited by different proteins. TLE1 is less required for ER binding if FoxA1 is cobound with ER, but ER binding is more dependent on TLE1 if FoxA1 is absent. Our data suggest that TLE1 mRNA levels are influenced by FoxA1, suggesting that FoxA1 may be upstream of TLE1. Silencing of FoxA1 results in global inhibition of ER binding (8). This is likely to be explained via two mechanisms, the first being a direct requirement for FoxA1 to mediate ER-chromatin interactions. The second mechanism is that FoxA1 is required for expression of TLE1 (and possibly other pioneer factors) and therefore a decrease in TLE1 levels (resulting from decreased FoxA1 levels) influences ER-chromatin interactions at genomic regions that are not direct FoxA1 binding domains. As expected, silencing of FoxA1 also results in decreased proliferation of MCF-7 cells (6), similar to our data for TLE1. A model describing the interplay between ER, FoxA1 and TLE1 is provided in Fig. S4.
We now show that ER binding to chromatin involves TLE1, which is critical for maintaining ER binding to a subset of chromatin regions. The downstream consequences of decreased TLE1 levels (and decreased ER binding) are changes in gene transcription and inhibition of estrogen-ER-mediated proliferation in breast cancer cell lines. However, it is currently unclear what influences TLE1 binding in breast cancer cells. The Zaret lab has previously shown that TLE proteins can bind to chromatin via histone tails (10) and as such, specific histone modifications and chromatin structure may be a critical determinant of TLE1 binding events. Previous work has suggested that H3K4me1 and H3K4me2 marks occur at distal ER binding enhancers and that these specific marks influence FoxA1 binding (7). It is possible that the same situation exists for TLE1 binding capacity.
TLE proteins have generally been characterized as repressor factors, although examples exist where they function as activators (15). Whether TLE factors impart repressive or active effects may be dictated by the associated transcription factors. In breast cancer cells, TLE1 appears to be a critical factor required for active ER binding and transcriptional activity in breast cancer cells. TLE1 binding to chromatin is independent of estrogen treatment and can occur even when ER is not associated with the chromatin. Following estrogen treatment, ER can associate with the chromatin, but the specific regions of the genome that can be bound by ER are dictated by additional proteins, such as TLE1 and FoxA1. Changes in ER binding dynamics in different contexts may be explained by alterations in TLE1 and/or FoxA1 binding profiles or potentially by additional factors that may function in similar chromatin-tethering roles. Alternatively, changes in ER phosphorylation state, cofactor stoichiometry or chromatin structure may “reveal” TLE1 and/or FoxA1 binding regions that can subsequently be bound by ER.
Understanding the factors involved in the mechanism of ER binding to DNA provides a unique opportunity for inhibition of ER action in breast cancer, by physically impeding ER-DNA interactions and subsequently blocking ER-mediated transcription.
Methods and Materials
Cell Lines.
MCF-7 cells were grown for 3 d in hormone deprived media (phenol red-free D-MEM, 5% charcoal/dextran-treated fetal bovine serum), changing the media every day. Synthetic 17β-Estradiol (Sigma E2758) was added to the media at a final concentration of 100 nM and 4-hydroxtamoxifen (Sigma H7904) was used at 1 μM for the indicated times. The ZR-75 cell line was maintain in RPMI, 10% FBS with penicillin and streptomycin. Before experiments they were grown for 3 d in hormone deprived media (phenol red-free RPMI, 5% charcoal/dextran-treated fetal bovine serum), changing the media every day.
Chromatin Immunoprecipitation.
ChIP experiments were performed as previously described (5). Antibodies used were ERα (HC-20), from Santa Cruz (Santa Cruz Biotechnologies) and TLE1 (ab15587), RNA Pol II (ab5131) and FoxA1 (ab5089) from Abcam. Primer sequences are provided in Fig. S5.
ChIP-Sequencing Experiments.
The ChIP DNA was verified by real-time PCR, and the DNA was processed for Illumina sequencing as previously described (23), using 36-bp reads on a GAIIx. Sequences generated by the Illumina genome analyzer were aligned against NCBI Build 36.3 of the human genome using MAQ: Mapping and assembly with qualities (http://maq.sourceforge.net/) with default parameters. The aligned reads were converted to BED format using a custom script. Peaks were called using MACS (18).
For analysis of RNA Pol II binding, we considered entire gene bodies of all Refseq genes, plus an additional 1,000 bases upstream (respecting ± strand). The threshold was based on read density (the mean read density minus 1/4 of the standard deviation), as well as a minimum number of reads (at least 80 reads in one of the libraries). We considered genes to be differentially regulated if normalized tag count changed by at least 1.5-fold in both replicate experiments.
Small Interfering RNA.
siRNA experiments were performed using Lipofectamine2000 (Invitrogen); 25 μL was used in a total of 5 mL of Optimem (Invitrogen) together with 50 nM of each siRNA for 6 h. The sequence of the siRNAs were siTLE1 (D-015528-01): GAACAAGCCUGACAAGUAC; siTLE1 (D-015528-17): GGAAAAUGGAAUCGACAAA; siTLE1 siGenome SMARTpool (M-015528-01) and siFoxA1: GAGAGAAAAAAUCAACAGC, from Dharmacon (Thermo Scientific Dharmacon RNAi Technologies). After 48 h of transfection, cells were treated with control ethanol vehicle or 17β-estradiol.
Western Blotting.
Nuclear lysate was extracted using lysis buffer (20 mM Hepes, 10 mM NaCl, 1.5 mM MgCl, 20% glycerol, 0.1% triton and 0.1 mM DTT) on ice for 5 min then spun at 13,000 rpm (supernatant contains the cytoplasmic fraction), the pellet was then resuspended in lysis buffer with 600 mM NaCl and incubated at 4 °C for 1 h then spun for 10 min at 13,000 rpm and 30 μg of the nuclear protein lysate was resolved by 10% SDS PAGE. Antibodies used were ERα (HC-20) diluted at 1∶500, from Santa Cruz (Santa Cruz Biotechnologies), TLE1 (sc9121) diluted at 1∶500 and β-actin (ab6276) diluted at 1∶20,000 from Abcam. Secondary antibodies obtained from Dako were used at 1∶2,000 dilution.
Microarray Analysis.
MCF-7 cells were hormone deprived, transfected with siTLE1 or siControl and treated with estrogen or vehicle for 6 h. RNA was collected from six biological replicates. The Illumina BeadChip (HumanWG-12 version 4) bead-level data were preprocessed, log2-transformed, and quantile-normalized using the bead array package (24, 25) in Bioconductor (26). Differential expression analysis was performed using limma eBayes (Smyth 2004 with a Benjamini and Hochberg multiple test correction procedure (27) to identify statistically significant differentially expressed genes (adjusted P value < 0.05).
Flow Cytometry.
Cells were plated at equal confluence, transfected as previously described (17) and total cells were harvested for flow cytometry analysis, using propidium iodide staining.
Cell Growth Assay.
Cells were plated at equal confluence, grown in hormone-depleted DMEM media and treated with vehicle or 100 nM 17β-estradiol. Confluence of cells was analyzed using the live-cell imaging Incucyte™ Analyzer (Bucher Biotec AG).
Formaldehyde-Assisted Isolation of Regulatory Elements.
FAIRE was performed as described by ref. 22. Primers are included in Fig. S5.
Statistical Analyses.
Statistical analyses were performed using two tailed paired T tests and only values lower than p value < 0.05 were considered statistical. In all figures, the data are the average of a minimum of three independent replicates ± Std Dev.
Supplementary Material
Acknowledgments.
The authors thank Stewart MacArthur, Ros Russell, Sarah Vowler, Rory Stark, and Matt Eldridge for discussions and bioinformatics support. We acknowledge the support of the University of Cambridge, Cancer Research UK, and Hutchison Whampoa Limited. K.A.H. is supported by a Breast Cancer Campaign project grant, and D.T.O. and J.S.C. are supported by ERC (European Research Council) Starting Grants.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. K.P. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1018863108/-/DCSupplemental.
References
- 1.Sorlie T, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci USA. 2001;98:10869–10874. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ali S, Coombes RC. Endocrine-responsive breast cancer and strategies for combating resistance. Nat Rev Cancer. 2002;2:101–112. doi: 10.1038/nrc721. [DOI] [PubMed] [Google Scholar]
- 3.Cirillo LA, et al. Binding of the winged-helix transcription factor HNF3 to a linker histone site on the nucleosome. EMBO J. 1998;17:244–254. doi: 10.1093/emboj/17.1.244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cirillo LA, et al. Opening of compacted chromatin by early developmental transcription factors HNF3 (FoxA) and GATA-4. Mol Cell. 2002;9:279–289. doi: 10.1016/s1097-2765(02)00459-8. [DOI] [PubMed] [Google Scholar]
- 5.Carroll JS, et al. Chromosome-wide mapping of estrogen receptor binding reveals long-range regulation requiring the forkhead protein FoxA1. Cell. 2005;122:33–43. doi: 10.1016/j.cell.2005.05.008. [DOI] [PubMed] [Google Scholar]
- 6.Laganiere J, et al. Location analysis of estrogen receptor alpha target promoters reveals that FOXA1 defines a domain of the estrogen response. Proc Natl Acad Sci USA. 2005;102:11651–11656. doi: 10.1073/pnas.0505575102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lupien M, et al. FoxA1 translates epigenetic signatures into enhancer-driven lineage-specific transcription. Cell. 2008;132:958–970. doi: 10.1016/j.cell.2008.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hurtado A, Holmes KA, Ross-Innes CS, Schmidt D, Carroll JS. FOXA1 is a key determinant of estrogen receptor function and endocrine response. Nat Genet. 2011;43:27–33. doi: 10.1038/ng.730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Palaparti A, Baratz A, Stifani S. The Groucho/transducin-like enhancer of split transcriptional repressors interact with the genetically defined amino-terminal silencing domain of histone H3. J Biol Chem. 1997;272:26604–26610. doi: 10.1074/jbc.272.42.26604. [DOI] [PubMed] [Google Scholar]
- 10.Sekiya T, Zaret KS. Repression by Groucho/TLE/Grg proteins: genomic site recruitment generates compacted chromatin in vitro and impairs activator binding in vivo. Mol Cell. 2007;28:291–303. doi: 10.1016/j.molcel.2007.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cinnamon E, Paroush Z. Context-dependent regulation of Groucho/TLE-mediated repression. Curr Opin Genet Dev. 2008;18:435–440. doi: 10.1016/j.gde.2008.07.010. [DOI] [PubMed] [Google Scholar]
- 12.Cai Y, Brophy PD, Levitan I, Stifani S, Dressler GR. Groucho suppresses Pax2 transactivation by inhibition of JNK-mediated phosphorylation. EMBO J. 2003;22:5522–5529. doi: 10.1093/emboj/cdg536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hurtado A, et al. Regulation of ERBB2 by oestrogen receptor-PAX2 determines response to tamoxifen. Nature. 2008;456:663–666. doi: 10.1038/nature07483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hentschke M, Borgmeyer U. Identification of PNRC2 and TLE1 as activation function-1 cofactors of the orphan nuclear receptor ERRgamma. Biochem Biophys Res Commun. 2003;312:975–982. doi: 10.1016/j.bbrc.2003.11.025. [DOI] [PubMed] [Google Scholar]
- 15.Fisher AL, Caudy M. Groucho proteins: Transcriptional corepressors for specific subsets of DNA-binding transcription factors in vertebrates and invertebrates. Genes Dev. 1998;12:1931–1940. doi: 10.1101/gad.12.13.1931. [DOI] [PubMed] [Google Scholar]
- 16.Shang Y, Hu X, DiRenzo J, Lazar MA, Brown M. Cofactor dynamics and sufficiency in estrogen receptor-regulated transcription. Cell. 2000;103:843–852. doi: 10.1016/s0092-8674(00)00188-4. [DOI] [PubMed] [Google Scholar]
- 17.Carroll JS, et al. Genome-wide analysis of estrogen receptor binding sites. Nat Genet. 2006;38:1289–1297. doi: 10.1038/ng1901. [DOI] [PubMed] [Google Scholar]
- 18.Zhang Y, et al. Model-based Analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137–R137.9. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Narita M, et al. A novel role for high-mobility group a proteins in cellular senescence and heterochromatin formation. Cell. 2006;126:503–514. doi: 10.1016/j.cell.2006.05.052. [DOI] [PubMed] [Google Scholar]
- 20.Prall OW, Rogan EM, Musgrove EA, Watts CK, Sutherland RL. c-Myc or cyclin D1 mimics estrogen effects on cyclin E-Cdk2 activation and cell cycle reentry. Mol Cell Biol. 1998;18:4499–4508. doi: 10.1128/mcb.18.8.4499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Musgrove EA, et al. Identification of functional networks of estrogen- and c-Myc-responsive genes and their relationship to response to tamoxifen therapy in breast cancer. PLoS One. 2008;3:e2987. doi: 10.1371/journal.pone.0002987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Eeckhoute J, et al. Cell-type selective chromatin remodeling defines the active subset of FOXA1-bound enhancers. Genome Res. 2009;19:372–380. doi: 10.1101/gr.084582.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schmidt D, et al. ChIP-seq: Using high-throughput sequencing to discover protein-DNA interactions. Methods. 2009;48:240–248. doi: 10.1016/j.ymeth.2009.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dunning MJ, Smith ML, Ritchie ME, Tavare S. beadarray: R classes and methods for Illumina bead-based data. Bioinformatics. 2007;23:2183–2184. doi: 10.1093/bioinformatics/btm311. [DOI] [PubMed] [Google Scholar]
- 25.Cairns JM, Dunning MJ, Ritchie ME, Russell R, Lynch AG. BASH: A tool for managing BeadArray spatial artefacts. Bioinformatics. 2008;24:2921–2922. doi: 10.1093/bioinformatics/btn557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gentleman RC, et al. Bioconductor: Open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80–R80.16. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Benjamini Y, Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J R Stat Soc Series B Stat Methodol. 1995;57:289–300. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.