Abstract
Gonadal sex determination in vertebrates generally follows a sequence of genetically programmed events. In what is seemingly becoming a pattern, all confirmed or current candidate “master” sex-determining genes reported in this group, e.g., SRY in eutherian mammals, DMY/dmrt1bY in medaka, DM-W in the African clawed frog, and DMRT1 in chicken encode transcription factors. In contrast, here we show that a male-specific, duplicated copy of the anti-Müllerian hormone (amh) is implicated in testicular development of the teleost fish Patagonian pejerrey (Odontesthes hatcheri). The gene, termed amhy because it is found in a single metacentric/submetacentric chromosome of XY individuals, is expressed much earlier than the autosomal amh (6 d after fertilization vs. 12 wk after fertilization) and is localized to presumptive Sertoli cells of XY males during testicular differentiation. Moreover, amhy knockdown in XY embryos resulted in the up-regulation of foxl2 and cyp19a1a mRNAs and the development of ovaries. These results are evidence of a functional amh duplication in vertebrates and suggest that amhy may be the master sex-determining gene in this species. If confirmed, this would be a unique instance of a hormone-related gene, a member of the TGF-β superfamily, in such a role.
The sexual fate of the differentiating gonads in vertebrates is under the control of specific genes that initiate and direct the developmental pathway. A few genes have been already identified as master sex determiners, and they all encode transcription factors, e.g., SRY in eutherian mammals (1), DMY/dmrt1bY in medaka (2, 3), DM-W in the African clawed frog (4), and DMRT1 in chicken (5). These findings might be construed as evidence that transcription factors always trigger gonadal sex determination in vertebrates. However, the molecular pathway of sex determination has been studied in relatively few nonmammalian species, and most of the details of this process remain elusive.
We have recently identified a sex-linked locus in Odontesthes hatcheri (Atherinopsidae), a South American gonochoristic fish with an XX-XY sex determination system (6, 7). The existence of a sex-linked single nucleotide polymorphism (SNP) marker associated with this locus has allowed us to profile the expression of a series of genes involved in early sex differentiation of putative females (XX genotype) and males (XY genotype). Analyses performed during early stages of embryonic and larval development revealed a comparatively early mRNA expression of an anti-Müllerian hormone homolog [(amh); also known as Müllerian inhibitory substance/factor, or mis/mif (8)] in relation to other teleosts (9, 10) and showed that this unique feature was due to the up-regulation of a duplicated copy of this gene. AMH, a member of the TGF-β superfamily, is secreted by Sertoli cells and is responsible for the regression of Müllerian ducts during male fetal development in mammals, birds, and reptiles (11–13). Fish have amh even though they lack Müllerian ducts. However, as with mammals and birds, fish amh is generally considered to be autosomal and is placed with other hormones or steroidogenic enzymes downstream from the molecular cascade of sex differentiation in relation to transcription factors (14, 15).
This report describes a unique case of an amh paralogue in vertebrates. More importantly, this study shows that this gene is restricted to the male genome and that it is required for testis determination in O. hatcheri. These findings establish a hormone-related gene in such a role and an alternative mechanism for transcriptional control of sex determination in vertebrates.
Results
Males Carry a Duplicated Copy of the Anti-Müllerian Hormone Gene.
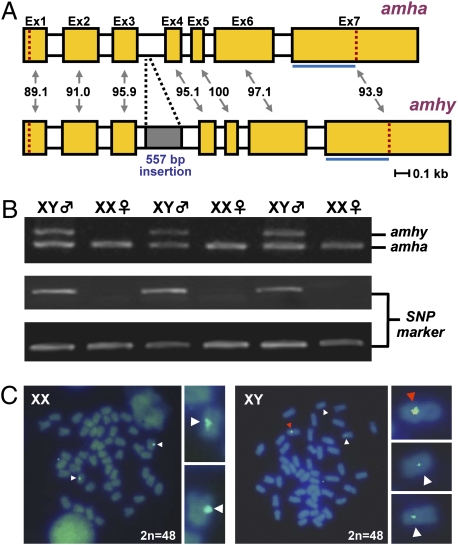
To clarify the reason for the unusual expression profile of amh in O. hatcheri, extensive sequencing was conducted with mRNAs expressed in larval and adult males. Such analysis revealed the presence of two different amh transcripts originated from two different loci. We also determined that one of these loci was present only in the male genome and was responsible for the early transcription of amh in XY gonads; this copy was therefore named amhy (Y chromosome-specific amh). RACE PCR was performed, and full cDNA sequence (2,059 bp) was obtained from mRNA of a 3-wah (weeks after hatching) XY larva. The nucleotide identity values between corresponding exons of amhy and the autosomal amh (amha) ranged from 89.1% to 100% (Fig. 1A). The deduced protein comprised 514 amino acids, which includes the characteristic TGF-β domain (amino acids 421–514) with seven canonical cysteine residues. Amino acid identity values of Amhy in relation to Amha were 92.2% for the entire protein and 91.4% for the TGF-β domain. Intron sequences were also characterized by PCR using the respective genomic DNA and revealing a 557-bp amhy-specific insertion in the third intron as the main structural difference within untranscribed intragenic regions (Fig. 1A). Primers flanking this intron were then designed and used for PCR-based sex genotyping. The comparison of genotypic sex with the histological sex of the gonads in 112 individuals derived from four crosses resulted in 100% matching (Fig. 1B) whereas sex genotyping using the previously reported sex-linked SNP marker (6) showed disagreement in two animals. From these results, the genetic distance between amhy and the SNP marker was estimated as 1.78 cM. The presence of amhy was also confirmed in the genomes of males from another cultivated stock from Japan (Kanagawa; n = 24) and natural populations from Argentina (Piedra del Aguila and Mari Menuco; n = 12 for each location). The 5′ flanking region of amhy was amplified by genome walking, and, together with the amhy gene sequence, was used as a probe (7.3 kb) for physical mapping on metaphase chromosomes. Signals were detected in a pair of acrocentric/telocentric chromosomes of XX and XY genotypes, probably due to the high homology of amha and amhy (∼90%), and in a single metacentric/submetacentric chromosome of XY fish (Fig. 1C). The presence of a single and relatively stronger signal in only one metacentric/submetacentric chromosome in XY fish indicates that this is the Y chromosome of O. hatcheri.
Fig. 1.
amhy gene structure, relation with SNP marker and phenotypic sex, and physical mapping. (A) Schematic representation of amhy and amha gene structure in O. hatcheri. Exons (Ex1 to Ex7) are represented by filled boxes, and the percentage identity between corresponding exons is indicated. Open segments represent introns, and the gray-shaded area shows the location of the insertion in the amhy third intron. The location of the TGF-β domain (blue line) and of the start and stop codons (red dotted lines in the first and seventh exons, respectively) are indicated. (B) Pattern of amha (1,057 bp; Top panel, lower band) and amhy (1,614 bp; Top panel, upper band) amplification using primers flanking the third intron and of the sex-linked SNP marker in sex genotyping. Results show complete matching between genotypic sex (inferred by both amhy amplification and SNP marker) and phenotypic sex (only six animals are shown). (Middle) The SNP marker shows the amplification of the male-specific PCR fragment, and the Bottom lane the respective control. (C) Fluorescence in situ hybridization using as probe the amhy gene and its 5′ flanking region (7.3 kb) on metaphase spreads from XX and XY fish. The fragment was based on the amhy sequence but the probe hybridizes also to amha. XX individuals show two signals in a pair of acrocentric/telocentric chromosomes whereas XY individuals show, in addition to this pair of signals (presumptive amha), a strong signal (presumptive amhy) in a single metacentric/submetacentric chromosome.
amhy mRNA Expression Encompasses Temporal Gonadal Differentiation.
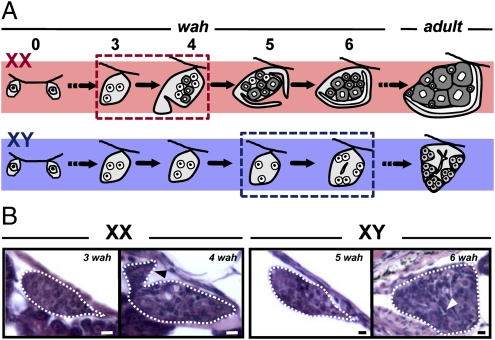
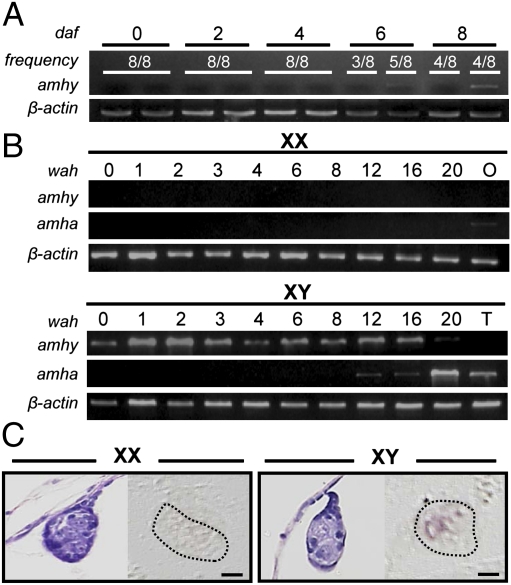
Gonadal sex differentiation in O. hatcheri began at 4 wah in females (XX individuals) and 6 wah in males (XY individuals) (Fig. 2 A and B). The first signs of ovarian and testicular differentiation were the appearance of a somatic cell outgrowth in the ventral portion of the gonad, which leads to the formation of the ovarian cavity and of the rudiments of the main sperm duct in the medullar region of the gonad, respectively. amhy transcripts were detected by RT-PCR specifically in putative XY males from 6 d after fertilization (4 d before hatching) through hatching. It was also present in XY males during testicular differentiation (juvenile stage) but was not detected after 20 wah and in adult testes (Fig. 3 A and B). In contrast, amha expression was absent during embryonic and early larval development of both sexes and was detected only after 12 wah in XY genotypes as well as in adult testes. Weak expression was observed also in adult ovaries. In situ hybridization using an antisense probe that hybridizes to both amhy and amha showed that the transcripts were located in somatic cells, presumably Sertoli cells, surrounding germ cells in XY gonads of 3-wah larvae (Fig. 3C). The sense probe did not produce any signal in both XX and XY gonads. Because at this stage only amhy expression is detected (by RT-PCR; Fig. 3 A and B), it is surmised that the signals correspond exclusively to amhy.
Fig. 2.
Time course of gonadal sex differentiation in XX and XY fish. (A) Schematic representation of the morphological changes in female and male gonads during sex differentiation (wah: weeks after hatching). (B) Light histology of the critical developmental stages indicated by dotted boxes in A. Black (XX; 4 wah) and white (XY; 6 wah) arrowheads indicate the appearance of somatic cell outgrowths (ovarian differentiation) and rudiments of the main sperm duct (testicular differentiation), respectively.
Fig. 3.
XY-specific expression of amhy mRNA during embryogenesis and gonadal sex differentiation. (A and B) Temporal expression of amhy (1,699 bp) and amha (1,716 bp) mRNAs by RT-PCR in (A) embryos (daf: days after fertilization) and in (B) XX and XY larval trunks (wah: weeks after hatching). The β-actin (457-bp) gene was used as endogenous control for all samples. T: testis; O: ovary. (C) Localization of amhy mRNA (Right) in gonadal sections of XX and XY larvae at 3 wk after hatching (in situ hybridization with an antisense probe). (Left) H&E-stained sections of the same individuals. (Scale bars, 10 μm.)
Morpholino-Mediated amhy Knockdown Inhibits Testicular Differentiation, Resulting in Ovarian Development in XY Fish.
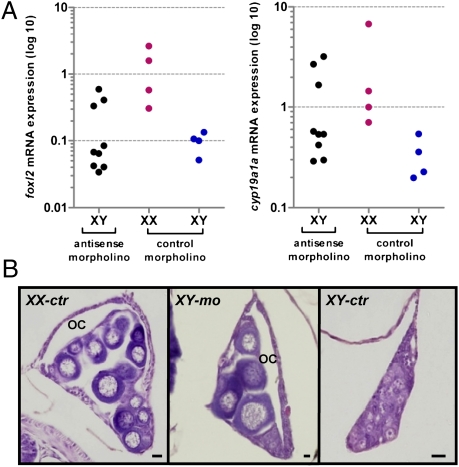
The injection of an amh antisense morpholino (MO) in XY embryos resulted in up-regulation of foxl2 and cyp19a1a mRNA expression in 3 of 9 larvae, producing values similar to those of XX control larvae at 4 wah (Fig. 4A) and development of ovaries in 11 of 50 larvae. In contrast, control XY larvae showed normal (low) foxl2 and cyp19a1a expression and none developed ovaries (n = 36). The difference in the frequency of XY females between the experimental groups was statistically significant (P < 0.01). Gonadal sex differentiation in the 11 sex-reversed males was characterized by the formation of the ovarian cavity and the appearance of previtellogenic oocytes and was histologically indistinguishable from that in control XX fish (Fig. 4B).
Fig. 4.
Feminization in amhy knockdown XY fish. (A) Relative expression of foxl2 and cyp19a1a mRNAs in morpholino-injected fish at 4 weeks after hatching (wah). foxl2 and cyp19a1a expression was normalized by the respective β-actin values. The three XY individuals in the antisense morpholino group with female-like, high foxl2 values also show high values for cyp19a1a. (B) Histological appearance of the gonads in mispaired morpholino-injected XX control fish (XX-ctr), antisense morpholino-injected XY fish (XY-mo), and mispaired morpholino-injected XY fish (XY-ctr) at about 12 wah. OC: ovarian cavity. (Scale bars, 20 μm.)
Discussion
This study describes a possible mechanism of genotypic sex determination in vertebrates whereby the main trigger would not be a transcription factor. In the mechanism reported here, a duplicated copy of the anti-Müllerian hormone (amh) gene, a TGF-β superfamily member and a well-characterized hormone in mammals, plays the key role in primary sex determination in the gonochoristic teleost O. hatcheri. The gene was termed amhy because it is present only in a single chromosome (Y chromosome) of XY (male) individuals. The evidences that support amhy as the master sex determiner in O. hatcheri, in addition to its tight linkage with the male sex, include its expression in putative Sertoli cells before the onset of and during gonadal differentiation and the fact that its knockdown resulted in up-regulation of foxl2 and cyp19a1a mRNA expression and ovarian development in XY genotypes.
Homologs of the mammalian amh gene have been described in several nonmammalian vertebrates including birds, reptiles, and even teleost fishes, a group devoid of Müllerian ducts. However, all previous studies have described single-copy homologs (16). This is a unique case of a duplicated amh locus and, most importantly, of a male-specific paralogue. The two loci, amhy and the autosomic amha, shared high homology in nucleotide and amino acid sequences. Like the amh of other teleosts, both loci are codified by seven exons (17), compared with five exons in mammals (18) and birds (19). The high homology between the two amhs also spans the TGF-β domain and the seven cystein residues necessary for protein homodimerization that characterize this family of growth factors. In the untranscribed intragenic region, the most evident structural difference between them was an amhy-specific insertion of about 0.5 kbp within the third intron. Ongoing studies are now looking into the promoter regions of both genes, which seem to differ considerably compared with exons and introns and which may explain the different expression profiles of both loci.
The mRNA expression of the two amhs colocalized spatially but not temporally. amhy transcription was first detected in putative Sertoli cells surrounding the germ cells of genotypic males during middle embryonic development. These features are very similar to the mRNA expression of medaka DMY/dmrt1bY (2, 3) and clearly precede the first signs of morphological differentiation of ovaries and testes in O. hatcheri (this study; see also ref. 20). amhy expression could not be detected in juveniles after 20 wah or in adult testes. In contrast, amha transcripts were detected only from later stages of testicular differentiation (12 wah onward) and in adult gonads of both sexes. In other teleosts, amh has been implicated in the regulation of germ cell proliferation and spermatogenesis (21, 22). These findings suggest that amhy has been inserted upstream of amha in the molecular cascade of sexual development in O. hatcheri, perhaps leading to a subfunctionalization of amhs. Thus, amhy may have been encharged of sex determination and amha of testicular maturation and/or spermatogenesis.
Morpholino-mediated amhy knockdown inhibited testicular development in genotypic males; increased foxl2 and cyp19a1a mRNA expression, which are critical genes for ovarian development in teleosts (10); and led to the formation of histologically normal ovaries in these fish. Although the morpholinos used in this study could presumably interfere with the expression of both amhy and amha genes, because of their high identity, the different expression dynamics of both genes in relation to the timing of gonadal sex differentiation described above and the timing of injection leaves little doubt that only amhy was actually blocked to a significant extent. Mutations of AMH and its receptor AMHRII result in premature reduction of the follicle pool in females and the persistent Müllerian duct syndrome in mice and humans, but not in functional XY sex reversal (23, 24). amh signaling and amhrII seem to be involved in the regulation of primordial germ-cell proliferation before gonadal sex differentiation in Japanese medaka but are located downstream of DMY/dmrt1bY (22). The case of O. hatcheri is different from previous reports, however, because amhy appears to be the master switch for sex. Further studies including gain-of-function analysis will be conducted to clarify whether amhy alone suffices for testicular determination. We also aim to clarify whether control of germ-cell proliferation is the target of amhy and a determinant of sex in O. hatcheri. We are also trying to clarify if both amhs share the same receptor.
In conclusion, our findings suggest that pivotal control of sex determination may not be restricted to DNA-binding factors (1–5), and, in this context, amhy might represent evidence of a sex-determining hormone in vertebrates. The present data also illustrate that even genes that do not originally participate in primary sex determination can override the genetic hierarchy in the differentiation cascade and become established as master switches of sexual fate by gene duplication events. The presence of amhy has been confirmed in captive-reared and wild stocks of O. hatcheri from Japan and Argentina, respectively, and is currently under investigation in phylogenetically related species to trace its evolution.
Materials and Methods
Experimental Animals and Rearing Procedures.
Fertilized eggs of O. hatcheri were obtained by artificial insemination using gametes from broodstock fish kept at the Aquatic Animal Rearing Facilities, Tokyo University of Marine Science and Technology. Fish used in this study were approximately the 11th generation after introduction from Argentina into Japan and were obtained from the Ehime Prefecture Fisheries Experimental Station. Egg incubation and rearing procedures followed previous studies (7) and were performed at 17–21 °C to avoid the possibility of temperature-induced sex reversal (25). Samples from another captive-reared stock from Japan (Kanagawa Prefecture Fisheries Experimental Station) and wild stocks from Argentina (Lakes Piedra del Aguila and Mari Menuco) were used to probe the occurrence of amhy in other populations.
DNA Extraction and Sex Genotyping.
DNA was extracted from caudal-fin tissue in all samples by the phenol:chloroform protocol and subjected to PCR using a set of primers designed to amplify both amha and amhy (MisREFw2: CTGAAGCACTGAGGCGGAAC; MisRERv2: CTCGCTGGAGGATAAGCCGA) for determination of genotypic sex. The amplification conditions consisted of 3 min at 94 °C, followed by 30 cycles of 30 s at 94 °C, 30 s at 60 °C, and 1 min at 72 °C, and a final elongation for 5 min at 72 °C. To estimate the distance between the sex-linked SNP marker and the amhy gene, 112 fishes derived from four different crosses were also analyzed by the SNP marker as previously described (6).
Cloning and Sequencing.
For isolation of O. hatcheri amha and amhy ORFs, total RNA was extracted from adult testes and trunks of 3-wk-old larvae using TRIzol (Invitrogen). The first-strand cDNA was synthesized according to previous studies (9). PCR products were electrophoresed in 1% agarose gel, purified, and sequenced in an ABI PRISM 3100 capillary sequencer (Applied Biosystems) using the BigDye Terminator method. Sequences were analyzed with GENETYX version 9.0.
Genome Walking.
The amhy 5′-flanking region (National Center for Biotechnology Information accession code HM_153804) was amplified using the Universal Genome Walker Kit (Clontech Laboratories), gene-specific reverse primers, and the AP1 and AP2 primers following the manufacturer's protocol. PCR products were processed and analyzed as described in the previous section.
Fluorescence In Situ Hybridization.
Metaphases were obtained by the air-drying method (26). A 7.3-kb DNA fragment containing the full sequence of amhy gene and its promoter was labeled with Alexa Fluor 488 using a FISH Tag DNA Kit (Molecular Probes), and hybridization was performed following the manufacturer's protocol. Signal amplification was performed using anti-Alexa Fluor 488 rabbit IgG and Alexa Fluor 488 goat anti-rabbit IgG (Molecular Probes). Chromosomes were counterstained using DAPI (1 μg/mL), and the metaphases were observed under a Nikon fluorescence microscope (Eclipse E600) using the filters B-2A and UV-2A (Olympus). The images were acquired with a CCD camera (Pixera CL600) and digitally merged using GIMP software (Version 2.2).
Expression Analysis by RT-PCR.
Total RNA extraction, cDNA synthesis, and expression analysis by RT-PCR in tissues, embryos, and larvae trunks followed methods from previous studies (9, 27). Specific primers for analyzing transcription of amhy and amha by RT-PCR were designed on the basis of regions with lowest homology between the two loci. The forward primers AmhyFw (GGAGGTCGCAGTTTCGAG) and AmhaFw (ACGCGGGTCACACAGGCGTTTC) were designed in the first exon within the 5′ untranslated region, whereas the respective reverse primers AmhyRv (GCATAAATACTGCACACAAC) and AmhaRv (CCGTACTGCATAAAACAAAC) were designed in the seventh exon. The β-actin gene was used as positive control (β-actinFw: CATCACACCTTGTACAATGAGCTGA; β-actinRv: AGCTCTTTTCCAATGATGAAGAGGA).
In Situ Hybridization and Light Histology.
In situ hybridization and light histology using H&E counterstaining and staining, respectively, were performed according to protocols described previously (9, 27). For RNA probe synthesis, T7 and T3 phage promoter sequences were appended to the forward (GTAATACGACTCACTATAGGGCGGGACAGTGATGAGCAGAAATGGA) and the reverse (GCAATTAACCCTCACTAAAGGGCTCACCTGCTGCTGTTTCCTTCC) primers, respectively.
Morpholino Microinjection.
Morpholinos were purchased from Gene Tools. Sequences were amh-MO GAATATCACCAGGACGGCCAACAT (underline shows the start codon) and ctr-amh-MO GTACAACCGGCAGGACCACTATAAG (mispaired control morpholino). Each morpholino was resuspended in distilled water to form a 1-mM stock solution. For microinjection, eggs were inseminated in a solution of glutathione 10 mM (pH 10.0) to prevent the hardening of the chorion and incubated at 17–20 °C. Morpholinos were further diluted (0.0285–0.05 mM) in 10 mM Tris-HCl/1mM EDTA buffer (pH 7.0) and coinjected with phenol red (20–50 nL) as a tracer into the yolk of embryos at the 1- to 32-cell stage using a microinjector (Narishige). Embryos and larvae were maintained at 17–21 °C for expression analysis of foxl2, a marker of ovarian development (10), at 4 wah and for determination of phenotypic sex by light histology at ∼3 mo after hatching. Fisher's exact test was used to analyze whether the difference between the frequency of XY females in the antisense morpholino and control groups was statistically significant.
Relative Quantification of foxl2 and cyp19a1a mRNAs.
The quantification of foxl2 and cyp19a1a expression in morpholino-injected larvae at 4 wah was performed by real-time RT-PCR using the primers RT-Foxl2Fw (TCATGAACAACTCCTGGTCGTT), RT-Foxl2Rv (GGCCATCTGACAGGACGTGTA), RT-AromGFw2 (GCGAGCTGTCTGGCTGAGAA), and RT-AromGRv2 (AGGAGCAGCAGCATGAAGAAGA). Both primer sets were designed on the basis of the O. hatcheri foxl2 and cyp19a1a sequences (National Center for Biotechnology Information accession codes FJ548572 and EF051123, respectively) and were used under the conditions described previously (9). β-actin (Obb-actinFw: GCTGTCCCTGTACGCCTCTGG; Obb-actinRv: GCTCGGCTGTGGTGGTGAAGC) was used as the endogenous control gene.
Acknowledgments
We thank Mr. P. Hualde for providing samples of wild Patagonian pejerrey from Argentina; the Ehime and Kanagawa Prefectural Fisheries Experimental Stations and Ms. E. Koshimizu for samples from Japan; and Dr. N. Mise for technical advice. We are also indebted to Dr. L. Orban and Dr. V. L. Trudeau for critically reviewing the manuscript. This work was supported in part by grants-in-aid from the Ministry of Education, Culture, Sports, Science and Technology of Japan (to C.A.S.) and Tokyo University of Marine Science and Technology (to R.S.H.).
Footnotes
References
- 1.Sinclair AH, et al. A gene from the human sex-determining region encodes a protein with homology to a conserved DNA-binding motif. Nature. 1990;346:240–244. doi: 10.1038/346240a0. [DOI] [PubMed] [Google Scholar]
- 2.Matsuda M, et al. DMY is a Y-specific DM-domain gene required for male development in the medaka fish. Nature. 2002;417:559–563. doi: 10.1038/nature751. [DOI] [PubMed] [Google Scholar]
- 3.Nanda I, et al. A duplicated copy of DMRT1 in the sex-determining region of the Y chromosome of the medaka, Oryzias latipes. Proc Natl Acad Sci USA. 2002;99:11778–11783. doi: 10.1073/pnas.182314699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yoshimoto S, et al. A W-linked DM-domain gene, DM-W, participates in primary ovary development in Xenopus laevis. Proc Natl Acad Sci USA. 2008;105:2469–2474. doi: 10.1073/pnas.0712244105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Smith CA, et al. The avian Z-linked gene DMRT1 is required for male sex determination in the chicken. Nature. 2009;461:267–271. doi: 10.1038/nature08298. [DOI] [PubMed] [Google Scholar]
- 6.Koshimizu E, Strüssmann CA, Okamoto N, Fukuda H, Sakamoto T. Construction of a genetic map and development of DNA markers linked to the sex-determining locus in the Patagonian pejerrey (Odontesthes hatcheri) Mar Biotechnol (NY) 2010;12:8–13. doi: 10.1007/s10126-009-9194-1. [DOI] [PubMed] [Google Scholar]
- 7.Hattori RS, et al. Establishment of a strain inheriting a sex-linked SNP marker in Patagonian pejerrey (Odontesthes hatcheri), a species with both genotypic and temperature-dependent sex determination. Anim Genet. 2010;41:81–84. doi: 10.1111/j.1365-2052.2009.01948.x. [DOI] [PubMed] [Google Scholar]
- 8.Hattori RS, et al. Characterization and expression profiles of DMRT1, AMH, SF1 and P450aro genes during gonadal sex differentiation in Patagonian pejerrey Odontesthes hatcheri. Cybium. 2008;32:95–96. [Google Scholar]
- 9.Fernandino JI, Hattori RS, Kimura H, Strüssmann CA, Somoza GM. Expression profile of anti-Müllerian hormone during gonadal development in pejerrey Odontesthes bonariensis, a teleost fish with strong temperature-dependent sex determination. Dev Dyn. 2008;237:3192–3199. doi: 10.1002/dvdy.21731. [DOI] [PubMed] [Google Scholar]
- 10.Piferrer F, Guiguen Y. Fish gonadogenesis part II: Molecular biology and genomics of sex differentiation. Rev Fish Sci. 2008;16:35–55. [Google Scholar]
- 11.Josso N, di Clemente N, Gouédard L. Anti-Müllerian hormone and its receptors. Mol Cell Endocrinol. 2001;179:25–32. doi: 10.1016/s0303-7207(01)00467-1. [DOI] [PubMed] [Google Scholar]
- 12.Teixeira J, Maheswaran S, Donahoe PK. Müllerian inhibiting substance: An instructive developmental hormone with diagnostic and possible therapeutic applications. Endocr Rev. 2001;22:657–674. doi: 10.1210/edrv.22.5.0445. [DOI] [PubMed] [Google Scholar]
- 13.Rey R, Lukas-Croisier C, Lasala C, Bedecarrás P. AMH/MIS: What we know already about the gene, the protein and its regulation. Mol Cell Endocrinol. 2003;211:21–31. doi: 10.1016/j.mce.2003.09.007. [DOI] [PubMed] [Google Scholar]
- 14.DeFalco T, Capel B. Gonad morphogenesis in vertebrates: Divergent means to a convergent end. Annu Rev Cell Dev Biol. 2009;25:457–482. doi: 10.1146/annurev.cellbio.042308.13350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Williams TM, Carroll SB. Genetic and molecular insights into the development and evolution of sexual dimorphism. Nat Rev Genet. 2009;10:797–804. doi: 10.1038/nrg2687. [DOI] [PubMed] [Google Scholar]
- 16.Paibomesai MI, Moghadam HK, Ferguson MM, Danzmann RG. Clock genes and their genomic distributions in three species of salmonid fishes: Associations with genes regulating sexual maturation and cell cycling. BMC Res Notes. 2010;3:215. doi: 10.1186/1756-0500-3-215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Halm S, Rocha A, Miura T, Prat F, Zanuy S. Anti-Müllerian hormone (AMH/AMH) in the European sea bass: Its gene structure, regulatory elements, and the expression of alternatively-spliced isoforms. Gene. 2007;388:148–158. doi: 10.1016/j.gene.2006.10.018. [DOI] [PubMed] [Google Scholar]
- 18.Cate RL, et al. Isolation of the bovine and human genes for Müllerian inhibiting substance and expression of the human gene in animal cells. Cell. 1986;45:685–698. doi: 10.1016/0092-8674(86)90783-x. [DOI] [PubMed] [Google Scholar]
- 19.Eusèbe D, et al. Cloning and expression of the chick anti-Müllerian hormone gene. J Biol Chem. 1996;271:4798–4804. doi: 10.1074/jbc.271.9.4798. [DOI] [PubMed] [Google Scholar]
- 20.Strüssmann CA, Cota JCC, Pronlor G, Higuchi H, Takashima F. Temperature effects on sex differentiation of two South American atherinids, Odontesthes argentinensis and Patagonina hatcheri. Environ Biol Fishes. 1996;47:143–154. [Google Scholar]
- 21.Miura T, Miura C, Konda Y, Yamauchi K. Spermatogenesis-preventing substance in Japanese eel. Development. 2002;129:2689–2697. doi: 10.1242/dev.129.11.2689. [DOI] [PubMed] [Google Scholar]
- 22.Morinaga C, et al. The hotei mutation of medaka in the anti-Mullerian hormone receptor causes the dysregulation of germ cell and sexual development. Proc Natl Acad Sci USA. 2007;104:9691–9696. doi: 10.1073/pnas.0611379104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.di Clemente N, Belville C. Anti-Müllerian hormone receptor defect. Best Pract Res Clin Endocrinol Metab. 2006;20:599–610. doi: 10.1016/j.beem.2006.09.004. [DOI] [PubMed] [Google Scholar]
- 24.Behringer RR, Cate RL, Froelick GJ, Palmiter RD, Brinster RL. Abnormal sexual development in transgenic mice chronically expressing Müllerian inhibiting substance. Nature. 1990;345:167–170. doi: 10.1038/345167a0. [DOI] [PubMed] [Google Scholar]
- 25.Strüssmann CA, Saito T, Usui M, Yamada H, Takashima F. Thermal thresholds and critical period of thermolabile sex determination in two atherinid fishes, Odontesthes bonariensis and Patagonina hatcheri. J Exp Zool. 1997;278:167–177. [Google Scholar]
- 26.Almeida-Toledo LF, Foresti F, Daniel MF, Toledo-Filho SA. Nucleolar chromosome variants in Sternopygus macrurus (Pisces, Sternopygidae) from three Brazilian river basins. Caryologia. 1993;46:53–61. [Google Scholar]
- 27.Hattori RS, et al. Thermolabile sex determination in Hd-rR medaka Oryzias latipes: Gender sensitivity, thermal threshold, critical period, and DMRT1 expression profile. Sex Dev. 2007;1:138–146. doi: 10.1159/000100035. [DOI] [PubMed] [Google Scholar]