Abstract
Large-scale immunization has profoundly impacted control of many infectious diseases such as measles and smallpox because of the ability of vaccination campaigns to maintain long-term herd immunity and, hence, indirect protection of the unvaccinated. In the case of human influenza, such potential benefits of mass vaccination have so far proved elusive. The central difficulty is a considerable viral capacity for immune escape; new pandemic variants, as well as viral escape mutants in seasonal influenza, compromise the buildup of herd immunity from natural infection or deployment of current vaccines. Consequently, most current influenza vaccination programs focus mainly on protection of specific risk groups, rather than mass prophylactic protection. Here, we use epidemiological models to show that emerging vaccine technologies, aimed at broad-spectrum protection, could qualitatively alter this picture. We demonstrate that sustained immunization with such vaccines could—through potentially lowering transmission rates and improving herd immunity—significantly moderate both influenza pandemic and seasonal epidemics. More subtly, phylodynamic models indicate that widespread cross-protective immunization could slow the antigenic evolution of seasonal influenza; these effects have profound implications for a transition to mass vaccination strategies against human influenza, and for the management of antigenically variable viruses in general.
Influenza is a major disease of humans and animals (1). Current influenza vaccines induce immunity primarily against the variable viral surface antigen hemagglutinin (HA). Owing to ongoing evolution of HA—manifested on the population level as antigenic “drift” (2)—current vaccines must be reviewed semiannually, in anticipation of the upcoming winter influenza season in the Northern and Southern hemispheres (3). There is evidence that current vaccines can elicit a degree of herd immunity (4–7), even when imperfectly matched (8); however, ongoing antigenic drift renders this effect too short-lived for any lasting epidemiological impact. Similarly, vaccination for pandemic response is limited to a largely reactive function that can only be fully initiated once a pandemic virus has emerged (9).
There is therefore increasing interest in developing vaccines targeting viral proteins more conserved than currently targeted HA epitopes (10–14). By inducing immunity against different subtypes of influenza, and different strains of the same subtype, such vaccines do not require knowledge of what strain is emerging; they could thus provide better pandemic and epidemic mitigation than antiviral drugs or social distancing (15), by permitting long-term suppression of transmission. In doing so, these vaccines may afford qualitatively unique opportunities concerning the epidemiology and evolution of influenza, which we explore here.
There are many different cross-protective vaccine candidates under study (10–14). Such vaccines do not necessarily prevent infection, but can be effective in reducing viral shedding, and can greatly reduce morbidity and mortality in animal models (10, 11). Fig. 1A shows recent experimental results (11) of a vaccine based on two conserved viral components, the matrix protein M2 and the nucleoprotein NP derived from an H1N1 strain. Vaccinated ferrets challenged with H5N1 exhibited a striking (order-of-magnitude) drop in nasal viral shedding over the course of infection. Further studies reporting comparable results are shown in Fig. S1 in SI Materials and Methods. If reducing viral shedding also reduces infectiousness (Table S1), then studies such as these suggest a potential impact of cross-protective vaccines on limiting onward transmission.
Fig. 1.
Reduction in viral shedding and potential effects on transmission and epidemiology. (A) Results of cross-protective vaccination and lethal H5N1 challenge in ferrets. Adult ferrets, 6 per group, were immunized with three doses of DNA vaccine encoding influenza A matrix 2 and nucleoprotein (NP+M2) or influenza B nucleoprotein (B/NP) given intramuscularly at 2-wk intervals, followed by intranasal boosting with recombinant adenovirus vectors (rAd) expressing the same antigen(s) 1 mo later. Animals were challenged with 5 LD50 of A/Vietnam/1203/04 (H5N1) 6 wk after boosting, and virus titers in nasal wash samples from days 1, 3, 5, 7, and 9 after challenge determined by 50% egg infectious dose (EID50) assay. Mean virus titers are shown ±SEM. The NP+M2 group differs significantly from the B/NP group at all times, (P < 0.05 by one-way ANOVA). Note that the ferret model shows susceptibility to influenza infection, similar symptoms to those in humans, and is transmission-competent. For additional details, see ref. 11. (Reprinted from ref. 11, Copyright 2009, with permission from Elsevier.). (B) Simulated outcome of pandemic emergence in a vaccinated population, taking R0 = 2, consistent with previous pandemics, and assuming that vaccinated individuals have transmission potential reduced by a proportion c.
Accordingly we refer to “cross-protection” as broadly including: (i) protection of vaccinated individuals by reducing viral shedding and/or morbidity and mortality, but not necessarily preventing infection, and (ii) offering this protection against different subtypes (e.g., H3N2 and H5N1), and against divergent strains of the same subtype (e.g., drift variants of H3N2). Most important in our context is the reduction in transmission that could result from vaccination.
Results
A central concept in infection control is the basic reproduction number R0, the mean number of infections arising from a single infected case in an otherwise susceptible population (16). For human influenza R0 is typically 1.5–3 (17, 18), significantly less than some other respiratory infections such as measles (19, 20). Where prior HA immunity exists, we refer instead to the “effective” reproduction number, Reff.
In light of the comparatively low R0 of influenza, well-established concepts of herd immunity (21) would suggest that, in principle, influenza pandemics could be substantially mitigated by persistently lowering transmission. Such effects could be achieved by deployment of cross-protective vaccines in advance of a pandemic in a way that would otherwise be impracticable with conventional, strain-matched vaccines. Fig. 1B illustrates implications for control, obtained by incorporating cross-protective vaccination in a standard susceptible-infectious-recovered (SIR) framework (Materials and Methods). In contrast with most other models of vaccination (22, 23), here we assume conservatively that vaccination lowers infectiousness (see e.g., refs. 24 and 25) but not susceptibility to infection. In particular, Fig. 1B illustrates that even incomplete vaccination coverage (modestly >50% of the population) has the potential for substantial reductions in viral spread, owing to the comparatively low R0 of influenza. As stated above, these results relate to well-established concepts of indirect protection; uniquely, however, novel vaccines raise the prospect of implementing such powerful concepts for pandemic control.
Evolutionary dynamics of seasonal influenza pose more subtle questions, primarily: how might mass, cross-protective vaccination programs, sustained over several years, shape influenza virus evolution? (Although we focus here on HA drift, our overall findings would apply generically to other variable viral components such as neuraminidase.)
Notable features of recent A/H3N2 evolution include its trunk-like phylogeny (2), in which a single lineage dominates at any given time, and punctuations in its antigenic evolution (26); the resulting sequential antigenic “clusters” often necessitate major reformulation of existing vaccines. (By contrast, a conserved antigen vaccine might be able to control all these variants uniformly.) Two prevailing paradigms capturing these and other aspects of influenza evolution, are the “epochal evolution” (27) and the “strain transcending immunity” models (28). Here, we present results of incorporating vaccination in the former; importantly, however, we indicate that our qualitative results apply equally to the latter and across the range of candidate explanations.
Briefly, the epochal evolution model proposes that influenza HA evolves along a neutral antigenic network (27); thus, whereas most substitutions do not result in significant immune escape, over time, they accumulate a genetic background in which certain context-specific mutations can ultimately effect a disproportionate antigenic change. (This process can be viewed, for example, in terms of conformational changes in HA epitopes.) Here, we include cross-protective vaccination in a recent, phenomenological formulation of this model (29), a framework incorporating stochasticity and annual seasonality in viral transmission. Fig. 2 illustrates schematically the model structure in the case of a single circulating strain, with further details summarized in Materials and Methods. Essentially, as in Fig. 1B, we assume a simple vaccination program in which a proportion q of the population has cross-protective immunity, reducing their transmission potential by a proportion c.
Fig. 2.
Compartmental formulation of infection and vaccination dynamics in the epochal evolution model, shown for a single antigenic variant. The lower row represents vaccinated individuals; α is the per capita rate of vaccination, whereas ω is the rate of waning of vaccine-derived immunity. Immunity within a single antigenic cluster wanes at a per-capita rate γ; infected individuals recover at a rate ν; there is a constant per-capita birth and death rate μ (not shown on figure for clarity), and the force of infection λ is given by λ = β (I + (1 − c)I(v)), where β is the rate of infection and c is the proportion by which transmission is reduced in vaccinated individuals.
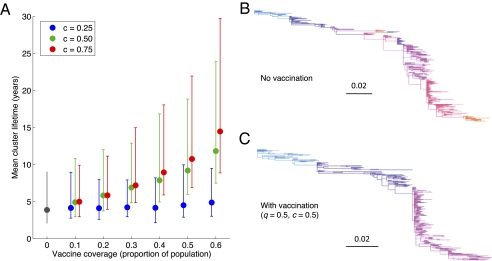
Results (Fig. 3A) show that cross-protective vaccination substantially slows the process of natural HA drift, in particular lengthening the mean “residence time” of each antigenic cluster. Specifically, as in Fig. 1B, cross-protective vaccination does not target HA, but acts indirectly through suppressing transmission, thus lowering epidemic sizes. Indeed, the significance of affecting transmission is apparent in the case of a low-efficacy vaccine (c = 0.25 in Fig. 3A), which has little evolutionary impact even at high coverages (q = 0.6). Fig. 1B illustrates how even at such coverages, such a vaccine effects only a limited reduction in epidemic sizes. Nonetheless, given the comparatively low R0 of influenza, Fig. 3A demonstrates that even partial protection (c = 0.5 or greater) offers the potential for major vaccine-induced modulation of influenza evolution. Moreover vaccination conserves influenza's trunk-like phylogeny (Fig. 3 B and C). We note that the alternative framework of strain-transcending immunity shows behavior in conceptual agreement with Fig. 3: Although the shape of the phylogeny is insensitive to R0 (28), this model predicts a lengthening strain lifetime with lowering R0 (30).
Fig. 3.
Results from the “epochal evolution” model of influenza surface antigenic evolution (see Materials and Methods, and Table S2 for parameters used). (A) Antigenic cluster lifetime, over 150 repetitions. Points indicate means, and error bars span 95% of simulation outcomes. (B) Simulated HA phylogeny over 30 y in the absence of vaccination, with changes in color representing punctuations in antigenic evolution. (C) As for B but in the presence of vaccination, with vaccine parameters shown. Although demonstrated here for a particular choice of q and c, the overall ”trunk-like” phylogeny is similarly conserved for other parameter values plotted in A.
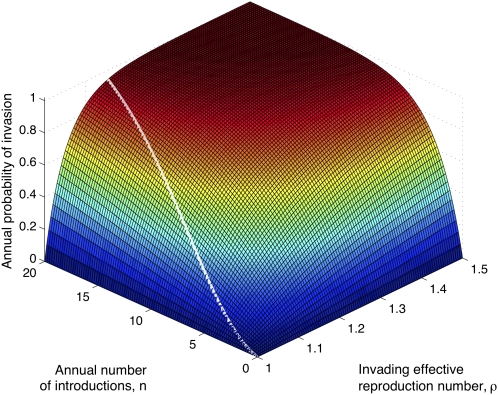
The epochal and strain-transcending immunity models capture drift by very different biological mechanisms. However, a simple analogy illustrates that vaccination essentially operates via a common dynamical mechanism in both these scenarios. Conceptually, sustained transmission dampening has a twofold evolutionary impact: (i) by lowering infection prevalence, mutants are generated at a reduced rate in the population. (ii) Reducing cumulative incidence implies fewer individuals with HA immunity, thus eroding the transmission advantage for any antigenic novelty that does arise (equivalently, lowering Reff for novel variants and, thereby, increasing the probability of their stochastic loss). These effects are analogous to control of an invading zoonotic pathogen, which might aim to reduce the annual rate n of introductions in the human population (akin to effect i), and lowering the reproduction number ρ in humans (akin to effect ii). Specifically, the annual probability of emergence of such a pathogen is P = 1–1/ρn. Fig. 4 illustrates this analogy and its correspondence with cross-protective vaccination. Another potential explanation for influenza dynamics invokes a network of immune responses against a limited set of antigenic types (31). In this formulation too, cross-protective vaccination would control all antigenic types uniformly, thereby potentially slowing the rate of antigenic change.
Fig. 4.
A simple schematic model illustrating the effect of cross-protective vaccination. The surface plots the annual probability of emergence of a zoonotic pathogen, one undergoing n introductions per year (x axis) with reproduction number ρ (y axis). For illustration, the white line indicates a subset of this surface relevant to influenza strain replacement, a downward trajectory denoting the effect of vaccination. Any single point on the line corresponds to a choice of RV, the reproduction number in the presence of vaccination and in the absence of HA immunity (see Eq. S3, and the comment thereafter). In the context of the epochal model, effects associated with both n and ρ make comparable contributions to slowing HA drift (SI Materials and Methods).
Discussion
Our work illustrates how novel vaccines could qualitatively change the nature of influenza control, adding rationale for large-scale immunization programs that would otherwise be prohibitively compromised by antigenic drift and shift. Moreover, slowing antigenic drift could reduce the frequency of reformulation of existing vaccines to match surface antigens of circulating viruses, and this effect would be of great importance for certain risk groups of the population.
An instructive thought experiment is to take the potential effect of mass vaccination to its conceptual extreme; consider a scenario where, whatever the details of the cross-protective vaccination program, transmission is reduced and drift is slowed to such an extent that most individuals do not live sufficiently long to acquire repeat infections. Consequently there is little or no selective advantage for antigenic novelty, and the paradigm of control is reduced to that of limiting SIR-like dynamics: in other words, a measles-like scenario (32). A crucial distinction, however, is that influenza shows much less transmission, and a shorter infectious period, than measles (16); hence, our results that even modest levels of vaccination could significantly affect pandemic spread and seasonal influenza evolution.
Although our models entail several simplifying assumptions, fundamental mechanisms illustrated in Fig. 4 indicate that our essential conclusion on the evolution-slowing effects of cross-protective vaccination is likely to be robust to these. Nonetheless, as with any modeling approach, there are some important caveats to our work. For simplicity we have assumed a homogeneous population and, in doing so, neglect any heterogeneity in transmission potential among hosts. However, this assumption is likely to be conservative. If the majority of transmission is attributable to a subgroup of the population, such as schoolchildren, then concentration of vaccine in this subgroup may achieve as much impact as random vaccination, but with lower overall vaccination effort (16). Large-scale spatial structure is another potentially significant heterogeneity, with several studies (e.g., refs. 33 and 34) indicating a dominant role for East/South-East Asia in seeding temperate regions with new seasonal variants each year. Here too, a concentration of vaccination effort at the geographic sources of antigenic novelty could achieve more efficient control than a more evenly distributed coverage.
Another important consideration is the possibility of vaccine program failure. Here, we have assumed a constant vaccination effort α; the effects of temporary shortfalls in vaccine supply could be mitigated by sufficiently long-lasting, vaccine-induced immunity. An important corollary of our work is thus the need to quantify the strength, breadth, and duration of transmission blocking for novel vaccines (see Table S1 for relevant sources). A different scenario, however, is the emergence of a fully transmissible vaccine escape mutant, an event that would expose an accumulated pool of susceptibles. The antigens targeted by cross-protective vaccines are, of course, relatively conserved among viral strains and subtypes and are far less variable than HA (see Table S1 for relevant studies). Moreover, targeting a combination of antigens, such as in the NP+M2 studies described here (Fig. 1A), may well limit the likelihood of emergence of such escape mutants. The degree to which viral escape could be blocked by such combination strategies will be an important question for future research. Overall, any adverse outcomes of vaccine program failure would be mitigated by the use or reintroduction of strain-matched vaccines (not modeled here).
Finally, a crucial question arising from this work is how transmission potential may be estimated from viral shedding data. Indeed, studies summarized in Table S1 offer empirical support for a positive correlation between shedding and infectiousness. However, explicit quantitative relationships between shedding and transmission potential remain unclear. Such a relationship need not be linear (see ref. 35 for a discussion): For example a “threshold” effect is possible, yielding transmission only above a certain level of cumulative shedding. Elucidating this relationship will be an important task for future work.
In summary, the advent of cross-protective vaccines could open important strategic options in the control of pandemic and seasonal influenza by prophylactic mass vaccination. More generally, this analysis illustrates the potential for cross-protective, transmission-reducing vaccines to control the dynamics of any antigenically variable virus. Such vaccines could potentially reduce the complex question of predicting and managing viral evolution to the conceptually simpler task of maximizing and maintaining cross protective herd immunity.
Materials and Methods
Basic Model of Transmission Dynamics.
We take a population of size N; for a single wave of infection with a virus having basic reproduction number R0, write S and I for the numbers of susceptible and infected, respectively. Assume conservatively that a cross-protective vaccine does not protect against infection, nor does it reduce the infectious period. However, it moderates viral shedding and the clinical course of infection, which reduces infectiousness (and thus onward transmission) by a factor c, where 0 < c < 1, and c = 1 corresponds to abolishment of transmission. We assume that, at the time of pandemic emergence, a random proportion q of the population has vaccine-induced immunity, taking this to remain constant over the timescale of the pandemic. Those having vaccine-induced immunity are designated with a superscript v (e.g., S(v)).
Neglecting births and deaths over the epidemic time course and assuming homogeneous mixing, basic model equations are:
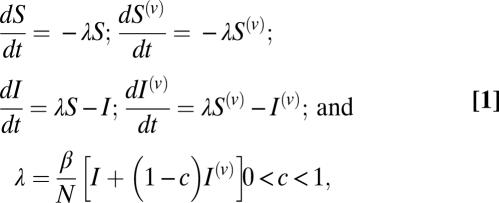 |
where β is the rate of infection and time has been normalized with respect to the mean infectious period. Note that the force of infection λ is the same for both vaccinated and unvaccinated classes, owing to equal susceptibility.
The disease-free equilibrium has: S = (1 – q) N; S(v) = q N; and all other quantities zero. Infection is then initiated by a perturbation to I. A proportion (1 – φ) of the population is ultimately infected (Fig. 1B), where φ is the nontrivial solution of
Such a nontrivial solution exists as long as R0(1-qc) > 1, that is as long as sustained transmission occurs.
Epochal Evolution Model.
Overview of the model without vaccination.
We use the stochastic, phenomenological implementation of this model developed in ref. 29, because this lends itself more readily to repeated simulations than the original, mechanistic model presented in ref. 27. Here, we give an overview of the model structure, with further details given in ref. 29.
The phylodynamic model incorporates two structural “tiers.” The first tier models the epidemiological dynamics of antigenic clusters, including their stochastic emergence, competition, and loss (Fig. 3A), whereas the second tier simulates HA sequence evolution within a given antigenic cluster, yielding genetic data that can be used in reconstructing phylogenies (Fig. 3 B and C).
Tier 1.
Although implemented as a stochastic process (see “Numerical simulations” below), the dynamics of antigenic cluster i, in the absence of vaccination, can be summarized by the deterministic equations:
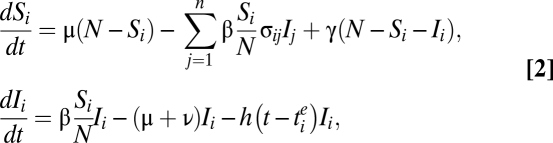 |
where Si and Ii, are, respectively, the numbers susceptible and infected, with reference to a strain from antigenic cluster i. Here, γ summarizes antigenic evolution within a single cluster; β is the rate of infection (assumed to be seasonally varying according to a sinusoidal form, as in ref. 29); 1/ν the mean recovery period; 1/μ the mean host lifetime; n the total number of antigenic clusters that have emerged by time t; and N the population size.
We take σij, the antigenic cross-reactivity between strains from clusters i and j, as described in ref. 29. An implication of this assumption is a constant cross-reactivity between any cluster and its descendant; however, because cross-protective vaccination is assumed to control all such variants uniformly, we do not expect our precise choice of cross-reactivity to qualitatively affect our evolutionary results.
Finally, the function h in Eq. 2 represents the key evolutionary component of Tier 1, the rate at which an individual infected with a strain from cluster i gives rise to a novel antigenic cluster. This rate depends on the age of cluster i, as follows: defining tie as the time at which cluster i emerged in the population, we write
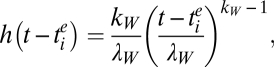 |
that is, a Weibull hazard function parameterized by a shape parameter kW and scale parameter λW. Taking kW > 1 captures the phenomenology of antigenic jumps becoming more likely with increasing age of the incumbent antigenic cluster (that is, (t − tie)), as described in detail in ref. 29. In a stochastic formulation, the magnitude of h(t − tie) determines the probability that a strain from cluster i that is infecting an individual mutates into a strain belonging to a new antigenic cluster. Such an event increments n by 1 in Eq. 2; this and other transitions are described in detail in table 1 of ref. 29.
Tier 2.
Taking as input the antigenic cluster changes simulated in tier 1 (as driven by the function h), this tier simulates molecular evolution within a given antigenic cluster to simulate a phylogenetic tree. Note that this tier provides no feedback to tier 1. Details of tier 2 implementation are as given in ref. 29.
For this model, neglecting the small contribution from per capita antigenic transition rate h, we have:
 |
Incorporation of cross-protective vaccination.
We now develop this model to include new terms for vaccination and the loss of vaccine-derived immunity. We assume for simplicity that HA immunity and conserved-antigen immunity are independent, identifying the former with infection and the latter with cross-protective vaccination. Thus, for example, we neglect CTL-based immunity naturally elicited by infection, and the role of anti-HA vaccination: we do not expect either of these features to alter qualitative results. (Incorporating the former would compound the transmission-reducing effects of cross-protective vaccination, and its omission is thus a conservative assumption.)
As described in the main text, we assume a vaccination program such that a proportion q of the population have cross-protective immunity, acting equally across all “HA clusters.” As in Eq. 1 above, such individuals have the same susceptibility to infection as others, but have a reduced rate of transmission β(1 – c), where 0 < c < 1.
Writing 1/ω for the mean duration of vaccine-derived immunity, we also define α as the per capita vaccination rate required to maintain immunity in a proportion q of the population: we discuss in the SI Materials and Methods how q, α, and ω are related.
The modified model is summarized with a compartmental flow diagram for a single antigenic variant in Fig. 2. It is straightforward to show that the vaccination-derived reproduction number RV, (i.e., in the presence of vaccination and in the absence of HA immunity), is
Numerical simulations.
We implement a stochastic simulation of this model in the presence of vaccination, mapping all deterministic rates described above to transition rates in a Markov process, and using a Binomial leap algorithm (36). The simulation therefore incorporates host demographic stochasticity (i.e., in N), as well as stochasticity in the infection and evolution process. Definitions and default values for parameters used here are provided in Table S2. All parameter values not related to vaccination are drawn from ref. 29, and sensitivity analyses are conducted on the remaining (SI Materials and Methods and Figs. S2–S4).
Supplementary Material
Acknowledgments
We thank Prof. Adrian Hill and Dr. Julia Gog for helpful discussion. This research was supported by National Institutes of Health Grant R01 GM083983-01. B.T.G. was also supported by the Bill and Melinda Gates Foundation; National Science Foundation Grant EF-0742373; the Research and Policy for Infectious Disease Dynamics program of the Science and Technology Directorate, Department of Homeland Security; and the Fogarty International Center, National Institutes of Health. K.K. and O.R. were supported by National Science Foundation Grant EF-08-27416; K.K. was further supported by the James S. McDonnell Foundation and the Research and Policy for Infectious Disease Dynamics program of the Science and Technology Directorate, Department of Homeland Security; and O.R. was further supported by Sir Henry Wellcome Fellowship Grant WT092311MF. S.L.E. and G.E.P. were supported by funds from the Center for Biologics Evaluation and Research, Food and Drug Administration.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1113342109/-/DCSupplemental.
References
- 1.Palese P. Influenza: Old and new threats. Nat Med. 2004;10(12 Suppl):S82–S87. doi: 10.1038/nm1141. [DOI] [PubMed] [Google Scholar]
- 2.Fitch WM, Bush RM, Bender CA, Cox NJ. Long term trends in the evolution of H(3) HA1 human influenza type A. Proc Natl Acad Sci USA. 1997;94:7712–7718. doi: 10.1073/pnas.94.15.7712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carrat F, Flahault A. Influenza vaccine: The challenge of antigenic drift. Vaccine. 2007;25:6852–6862. doi: 10.1016/j.vaccine.2007.07.027. [DOI] [PubMed] [Google Scholar]
- 4.Monto AS, Davenport FM, Napier JA, Francis TJ., Jr Effect of vaccination of a school-age population upon the course of an A2-Hong Kong influenza epidemic. Bull World Health Organ. 1969;41:537–542. [PMC free article] [PubMed] [Google Scholar]
- 5.Piedra PA, et al. Herd immunity in adults against influenza-related illnesses with use of the trivalent-live attenuated influenza vaccine (CAIV-T) in children. Vaccine. 2005;23:1540–1548. doi: 10.1016/j.vaccine.2004.09.025. [DOI] [PubMed] [Google Scholar]
- 6.Reichert TA, et al. The Japanese experience with vaccinating schoolchildren against influenza. N Engl J Med. 2001;344:889–896. doi: 10.1056/NEJM200103223441204. [DOI] [PubMed] [Google Scholar]
- 7.Rudenko LG, et al. Efficacy of live attenuated and inactivated influenza vaccines in schoolchildren and their unvaccinated contacts in Novgorod, Russia. J Infect Dis. 1993;168:881–887. doi: 10.1093/infdis/168.4.881. [DOI] [PubMed] [Google Scholar]
- 8.Hurwitz ES, et al. Effectiveness of influenza vaccination of day care children in reducing influenza-related morbidity among household contacts. JAMA. 2000;284:1677–1682. doi: 10.1001/jama.284.13.1677. [DOI] [PubMed] [Google Scholar]
- 9.Fedson DS. Pandemic influenza and the global vaccine supply. Clin Infect Dis. 2003;36:1552–1561. doi: 10.1086/375056. [DOI] [PubMed] [Google Scholar]
- 10.Epstein SL, Price GE. Cross-protective immunity to influenza A viruses. Expert Rev Vaccines. 2010;9:1325–1341. doi: 10.1586/erv.10.123. [DOI] [PubMed] [Google Scholar]
- 11.Price GE, et al. Vaccination focusing immunity on conserved antigens protects mice and ferrets against virulent H1N1 and H5N1 influenza A viruses. Vaccine. 2009;27:6512–6521. doi: 10.1016/j.vaccine.2009.08.053. [DOI] [PubMed] [Google Scholar]
- 12.Berthoud TK, et al. Potent CD8+ T-cell immunogenicity in humans of a novel heterosubtypic influenza A vaccine, MVA-NP+M1. Clin Infect Dis. 2011;52:1–7. doi: 10.1093/cid/ciq015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wei CJ, et al. Induction of broadly neutralizing H1N1 influenza antibodies by vaccination. Science. 2010;329:1060–1064. doi: 10.1126/science.1192517. [DOI] [PubMed] [Google Scholar]
- 14.Corti D, et al. A neutralizing antibody selected from plasma cells that binds to group 1 and group 2 influenza A hemagglutinins. Science. 2011;333:850–856. doi: 10.1126/science.1205669. [DOI] [PubMed] [Google Scholar]
- 15.Cauchemez S, et al. Closure of schools during an influenza pandemic. Lancet Infect Dis. 2009;9:473–481. doi: 10.1016/S1473-3099(09)70176-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Anderson RM, May RM. Infectious Diseases of Humans: Dynamics and Control (Oxford Science Publications) Oxford, UK: Oxford Univ Press; 1992. [Google Scholar]
- 17.Yang Y, et al. The transmissibility and control of pandemic influenza A (H1N1) virus. Science. 2009;326:729–733. doi: 10.1126/science.1177373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mills CE, Robins JM, Lipsitch M. Transmissibility of 1918 pandemic influenza. Nature. 2004;432:904–906. doi: 10.1038/nature03063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Edmunds WJ, Gay NJ, Kretzschmar M, Pebody RG, Wachmann H. ESEN Project. European Sero-epidemiology Network The pre-vaccination epidemiology of measles, mumps and rubella in Europe: Implications for modelling studies. Epidemiol Infect. 2000;125:635–650. doi: 10.1017/s0950268800004672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Metcalf CJ, Bjørnstad ON, Grenfell BT, Andreasen V. Seasonality and comparative dynamics of six childhood infections in pre-vaccination Copenhagen. Proc Biol Sci. 2009;276:4111–4118. doi: 10.1098/rspb.2009.1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Anderson RM, May RM. Vaccination and herd immunity to infectious diseases. Nature. 1985;318:323–329. doi: 10.1038/318323a0. [DOI] [PubMed] [Google Scholar]
- 22.McLean AR, Blower SM. Imperfect vaccines and herd immunity to HIV. Proc Biol Sci. 1993;253:9–13. doi: 10.1098/rspb.1993.0075. [DOI] [PubMed] [Google Scholar]
- 23.Medlock J, Galvani AP. Optimizing influenza vaccine distribution. Science. 2009;325:1705–1708. doi: 10.1126/science.1175570. [DOI] [PubMed] [Google Scholar]
- 24.Basta NE, Halloran ME, Matrajt L, Longini IMJ., Jr Estimating influenza vaccine efficacy from challenge and community-based study data. Am J Epidemiol. 2008;168:1343–1352. doi: 10.1093/aje/kwn259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McLean AR, Blower SM. Modelling HIV vaccination. Trends Microbiol. 1995;3:458–462. doi: 10.1016/s0966-842x(00)89010-1. [DOI] [PubMed] [Google Scholar]
- 26.Smith DJ, et al. Mapping the antigenic and genetic evolution of influenza virus. Science. 2004;305:371–376. doi: 10.1126/science.1097211. [DOI] [PubMed] [Google Scholar]
- 27.Koelle K, Cobey S, Grenfell B, Pascual M. Epochal evolution shapes the phylodynamics of interpandemic influenza A (H3N2) in humans. Science. 2006;314:1898–1903. doi: 10.1126/science.1132745. [DOI] [PubMed] [Google Scholar]
- 28.Ferguson NM, Galvani AP, Bush RM. Ecological and immunological determinants of influenza evolution. Nature. 2003;422:428–433. doi: 10.1038/nature01509. [DOI] [PubMed] [Google Scholar]
- 29.Koelle K, Khatri P, Kamradt M, Kepler TB. A two-tiered model for simulating the ecological and evolutionary dynamics of rapidly evolving viruses, with an application to influenza. J R Soc Interface. 2010;7:1257–1274. doi: 10.1098/rsif.2010.0007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Minayev P, Ferguson N. Incorporating demographic stochasticity into multi-strain epidemic models: Application to influenza A. J R Soc Interface. 2009;6:989–996. doi: 10.1098/rsif.2008.0467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Recker M, Pybus OG, Nee S, Gupta S. The generation of influenza outbreaks by a network of host immune responses against a limited set of antigenic types. Proc Natl Acad Sci USA. 2007;104:7711–7716. doi: 10.1073/pnas.0702154104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Grenfell BT, Bjørnstad ON, Kappey J. Travelling waves and spatial hierarchies in measles epidemics. Nature. 2001;414:716–723. doi: 10.1038/414716a. [DOI] [PubMed] [Google Scholar]
- 33.Bedford T, Cobey S, Beerli P, Pascual M. Global migration dynamics underlie evolution and persistence of human influenza A (H3N2) PLoS Pathog. 2010;6:e1000918. doi: 10.1371/journal.ppat.1000918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Russell CA, et al. The global circulation of seasonal influenza A (H3N2) viruses. Science. 2008;320:340–346. doi: 10.1126/science.1154137. [DOI] [PubMed] [Google Scholar]
- 35.King AA, Shrestha S, Harvill ET, Bjørnstad ON. Evolution of acute infections and the invasion-persistence trade-off. Am Nat. 2009;173:446–455. doi: 10.1086/597217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tian T, Burrage K. Binomial leap methods for simulating stochastic chemical kinetics. J Chem Phys. 2004;121:10356–10364. doi: 10.1063/1.1810475. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.