Abstract
Congenital diaphragmatic hernia (CDH) is a common (1 in 3,000 live births) major congenital malformation that results in significant morbidity and mortality. The discovery of CDH loci using standard genetic approaches has been hindered by its genetic heterogeneity. We hypothesized that gene expression profiling of developing embryonic diaphragms would help identify genes likely to be associated with diaphragm defects. We generated a time series of whole-transcriptome expression profiles from laser captured embryonic mouse diaphragms at embryonic day (E)11.5 and E12.5 when experimental perturbations lead to CDH phenotypes, and E16.5 when the diaphragm is fully formed. Gene sets defining biologically relevant pathways and temporal expression trends were identified by using a series of bioinformatic algorithms. These developmental sets were then compared with a manually curated list of genes previously shown to cause diaphragm defects in humans and in mouse models. Our integrative filtering strategy identified 27 candidates for CDH. We examined the diaphragms of knockout mice for one of the candidate genes, pre–B-cell leukemia transcription factor 1 (Pbx1), and identified a range of previously undetected diaphragmatic defects. Our study demonstrates the utility of genetic characterization of normal development as an integral part of a disease gene identification and prioritization strategy for CDH, an approach that can be extended to other diseases and developmental anomalies.
Keywords: pleuroperitoneal folds, expression profiling, diaphragm development
Congenital diaphragmatic hernia (CDH) encompasses a broad spectrum of developmental defects including complete absence (diaphragmatic aplasia), well-circumscribed defects or “holes” (posterolateral and central CDH), and thinning and/or muscularization defects (eventration and sac type CDH). Despite advances in surgical techniques and neonatal management, limited improvements in survival and long-term morbidity have been realized over the past two decades (1–3). Among CDH cases, 40% have a phenotype that also includes other malformations or associations with genetic syndromes (complex CDH) (2, 4). Such extreme phenotypic variability, likely reflecting multiple etiologies, and limited knowledge of diaphragm developmental processes have slowed the pace of gene discovery (5). Mice, however, provide a surrogate system to elucidate the molecular mechanisms active during diaphragm morphogenesis and those perturbed in CDH (6, 7).
The diaphragm develops between embryonic day (E)10.5 to E15.5 in mice, corresponding to the 4th to 10th weeks of human gestation (6). Among the different components of the primordial diaphragm, the pleuroperitoneal folds (PPFs) are the anlagen of its lateral muscular component where ≈80% of human defects occur (4, 8). PPFs taper medially from the lateral cervical body wall and fuse ventrally with the septum transversum and the posthepatic mesodermal plate (9–12).
Previous whole-transcriptome studies in other organ systems, such as the mouse lung and heart, have demonstrated connections between transcriptional programs in normal development and those perturbed in disease processes (13–15). Similar correlations have been described between diaphragm morphogenesis and CDH only at the level of individual genes (8, 16–18). In this study, we analyzed global trends in gene expression in the PPFs (at E11.5 and E12.5) compared with in the mature diaphragm (at E16.5) by using Short Time-series Expression Miner (STEM) (19) to cluster genes by similarities in temporal expression patterns, as well as Gene Set Enrichment Analysis (GSEA) (20, 21) to assess the expression data in the context of biological processes and signaling pathways. Based on the assumption that CDH candidate genes would have similar temporal expression trends and would be part of the same biological pathways as those already implicated in CDH, we used known CDH-associated genes as “baits” to filter gene sets and to identify novel candidates. One such gene, pre–B-cell leukemia transcription factor 1 (Pbx1), validated this strategy by demonstrating previously undetected diaphragm defects in knockout mice.
Results
Generation of Expression Data from Laser-Captured Developing Diaphragms.
Transcriptional profiles were generated from laser-capture microdissection (LCM) in C57BL/6J mouse diaphragms at three timepoints during normal embryonic development. Gene expression trends were characterized within the PPF tissue at E11.5 and E12.5 (Fig. 1 A and B) and within the mature diaphragm at E16.5 (Fig. 1C).
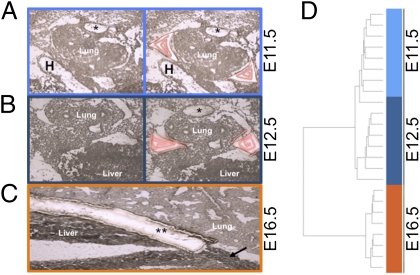
Fig. 1.
Expression study of microdissected tissue. (A and B) Unstained and unfixed transverse cryosections of E11.5 (A) and E12.5 (B) mouse embryo before (Left) and after (Right) laser capture microdissection (LCM) (H, heart; *, fused dorsal aorta). Captured tissue is outlined in red. (C) Sagittal cryosection of an E16.5 mouse embryo showing an anatomically mature diaphragm (arrow) (**, microdissected diaphragm). (D) Unsupervised hierarchical clustering of 871 differentially expressed probes (Welch modified t test) identifies two distinct clusters [the early (E11.5, E12.5) and late (E16.5)] Additionally, E11.5 and E12.5 separate in two subclusters, except for one outlier.
There were no statistically significant changes in gene expression between E11.5 and E12.5 (Welch's t test, P < 0.01). However, a comparison of early (combined E11.5 and E12.5) versus late (E16.5) diaphragm transcript levels revealed 871 probes corresponding to 770 genes listed in the Mouse Genome Informatics (MGI database; March 2011) that were differentially expressed (Welch's t test; P < 0.01) (Dataset S1). Unsupervised hierarchical clustering of the differentially expressed probes resulted in two distinct groups (1), genes up-regulated within the fully muscularized diaphragm and (2) genes up-regulated within the primordial diaphragm; these groups are further described in Fig. S1. Five genes within the second cluster were known to be associated with abnormal diaphragm development in murine models of gene inactivation (Slit3, Pdgfrα, Mmp2, Gli3, and Ilf3), and two others are orthologs of genes associated with syndromic forms of human CDH (STRA6 and GPC3) (22–30) (cumulative enrichment P = 2.9 × 10−4), suggesting that CDH-associated genes are active during early diaphragm formation.
Genes above the median of normalized hybridization intensities for intronic probes were interpreted as expressed (Dataset S2). It is, however, possible that genes below the thresholds are expressed, but below the detection capacity of the array.
Temporal Trends in Gene Expression and Molecular Pathways Active During Diaphragm Development.
To identify significant temporal expression trends (profiles) during diaphragm development and the genes associated with each profile, we used the STEM clustering system. STEM analysis revealed five gene expression profiles; three (8, 13, and 12) (Fig. 2A) showed upward trends in gene expression at E16.5 when the diaphragm is fully muscularized, whereas two (7 and 2) (Fig. 2A) showed highest expression during early development (E11.5 and E12.5). Their functional and phenotypic signatures were analyzed by using Visual Annotation Display (VLAD) to test for enrichment of terms in the Gene Ontology (GO) (31) and Mammalian Phenotype (MP) ontology (32). Genes clustering within STEM profiles 8, 13, and 12 were associated with muscle development and metabolism (GO), and with abnormal muscle and cardiovascular phenotypes (MP). Genes in STEM profiles 7 and 2 were associated with transcriptional regulation and cell proliferation (among others) (GO), and with embryonic lethality, as well as developmental abnormalities of the body wall (among others) (MP) (Dataset S3).
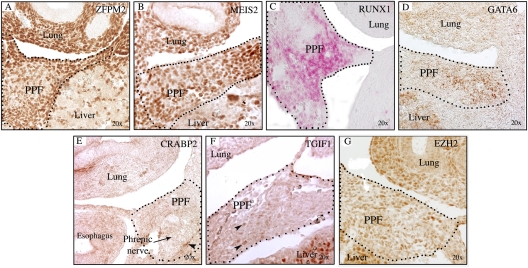
Fig. 2.
Expression of CDH candidate genes in E12.5 transverse sections. ZFPM2 (A) and MEIS2 (B) are expressed in diaphragmatic and lung bud mesenchyme. RUNX1 (C) and GATA6 (D) expression are restricted to the most lateral primordial diaphragmatic mesenchyme and are not expressed in the lung or body wall. CRABP2 (E), TGIF1 (F), and EZH2 (G) are expressed in the PPFs.
Because the biological significance of gene expression differences can often be more readily appreciated in the context of pathways, we performed GSEA. Seventy-five GO and 33 canonical pathway gene sets correlated with late diaphragmatic development, i.e., members of this list tended to occur among the genes up-regulated at E16.5. The highest ranking sets (with an acceptable false discovery rate of <25%) were associated with structural muscle proteins and mitochondria. One hundred thirteen GO gene sets and 16 canonical pathways correlated with the E11.5 and E12.5 timepoints; among them 12 GO gene sets and 15 canonical pathways had a false discovery rate of <25% (Dataset S4). These pathways include the WNT, TGFβ, ALK, and axonal guidance, all of which are important in mesodermal differentiation, a key component of the early developing diaphragm (33–35).
Gene Selection and Prioritization.
To prioritize candidate genes from the analysis of diaphragm development transcriptional profiles, we used a strategy based on the hypothesis that novel CDH candidate genes have similar temporal expression trends and participate in the same biological pathways as those already implicated in CDH. Accordingly, all five STEM gene sets were assessed (by hypergeometric distribution after correction for multiple testing) for enrichment of 48 genes previously shown to be associated with mouse models of CDH and/or in human genetic conditions that have CDH as part of their phenotype (Dataset S5). Similarly all 31 biological pathway gene sets obtained by GSEA were also examined for overrepresentation of the same 48 genes.
As a result, this filtering strategy revealed a single STEM profile (profile 7) (301 genes) and five GSEA gene sets (totaling 216 genes) (Table 1) that passed the significance threshold. Interestingly, all genes in these gene sets were expressed at highest levels in the primordial diaphragm compared with the mature diaphragm. The top priority CDH candidates were predicted to belong to the intersecting subset of genes that overlapped both biological (GSEA) and temporal (STEM) contexts (Fig. S2 B and C). This approach resulted in a list of 31 prioritized genes: Alx1, Casp8ap2, Chd3, Crabp2, Efna5, Epha3, Epha4, Epha7, Ephb2, Ezh2, Fzd2, Gata6, Gli3, Hells, Hmga2, Ilf3, Khdrbs1, Meil2, Pbx1, Phf21a, Robo1, Runx1, Sema3a, Smarca1, Smarcc1, Snai2, Tgif1, Twist1, Uhrf1, Unc5c, Zfpm2. Among these prioritized genes, 27 were considered candidates as four [Gli3, Ilf3, Zfpm2 (Fog2), and Robo1] had already been causally associated with diaphragm defects (Dataset S5).
Table 1.
Enrichment analysis
| Profiles and gene sets | Program (database) | P value | Overlap with CDH associated genes |
| Profile 7 | STEM | 4.46 × 10−7 | Gli3, Gpc3, Ilf3, Mmp2, Pdgfra, Robo1, Stra6, Wt1, Zfpm2 |
| Regulation of transcription, DNA dependent | GSEA (CP) | 1.21 × 10−5 | Gli2, Gata4, Zfpm2, Rarb, Nr2f2, Ilf3 |
| Regulation of RNA metabolic process | GSEA (CP) | 1.50 × 10−5 | Gli2, Gata4, Zfpm2, Rarb, Nr2f2, Ilf3 |
| Axon guidance | GSEA (CP) | 2.76 × 10−3 | Efnb1, Slit3, Robo1 |
| Transcription repressor activity | GSEA (CP) | 6 × 10−3 | Zfpm2, Nr2f2, Ilf3 |
| Basal cell carcinoma | GSEA (CP) | 0.035 | Gli2, Gli3 |
Expression of Prioritized Genes in Primordial Diaphragm Mesenchyme.
To delineate expression patterns and to validate the prioritized genes in the developing diaphragm, we performed immunohistochemistry (IHC) on E11.5 and/or E12.5 PPF tissue of C57BL/6J mouse embryos on seven candidates [Zfpm2, Meis2, Runx1, Gata6, Crabp2, Tgif1, Ezh2 (Fig. 2), and Pbx1 (Fig. 3A)] for which appropriate antibodies were available and hybridization was successful. ZFPM2 expression was detected throughout the midlung field, as well as the PPF mesenchyme. PBX1 was expressed in the PPFs and other mesenchymal tissues, as was MEIS2, a PBX1 binding partner. RUNX1 expression was detected in the primordial diaphragm just medial to the body wall and in the liver; no expression was seen in the lungs, body wall, or limb buds at this stage. GATA6 expression was restricted to the lateral PPF regions, whereas CRABP2, TGIF1, and EZH2 were expressed diffusely in the PPF. Additionally, expression of Pbx1 was confirmed by reverse transcription–quantitative PCR (RT-qPCR) (Fig. 3B).
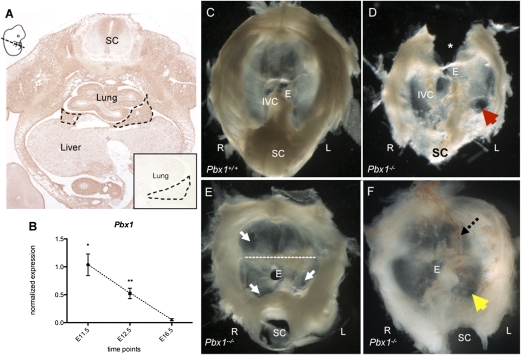
Fig. 3.
PBX1 expression and characterization of Pbx1 null mutant diaphragms. (A) IHC labeling of PBX1 shows expression in the E11.5 primordial diaphragm. The cartoon (Upper Left) indicates the plane of section. (Inset) PBX1 is not expressed in the mutant mouse PPF. (B) RT-qPCR validation of Pbx1 expression levels indicate significant down-regulation (±SEM) E11.5–E16.5 (*) and E12.5–E16.5 (**) (P < 0.05, two-tailed Welch t test). (C–F) Dissected diaphragms of E15.5 Pbx1 mutants (D–F) and control embryo (C) viewed from the abdomen. (D) Circumscribed muscularization defect of the left posterolateral aspect of the diaphragm (indicated by a red arrow) and diffuse decreased muscularization in the lateral regions bilaterally. *, Due to abnormal fusion of the heart to the anterior aspect of diaphragm and thoracic wall, a section of diaphragm was removed during heart dissection. (E) Multiple areas of abnormal muscle patterning in the anterior and posterior regions of the diaphragm (white arrows), with an abnormal band of tissue (dotted line). We observed reduced musculature in the crural region, without an independent opening for the IVC. (F) Herniated stomach fundus (yellow arrow). Abnormally increased vascularity (dotted arrow). E, esophagus; IVC, inferior vena cava; SC, spinal cord.
Pbx1 Knockout Mice Have Diaphragmatic Defects.
To determine whether the 27 genes were worthy candidates, Pbx1 was selected for functional validation of the prioritization strategy. The choice of Pbx1 was based purely on feasibility, namely embryonic survival up to E15.5; it was not biased by biological preconceptions. Whereas wild-type embryos expressed PBX1 in the mesenchymal tissues of the PPF, lungs, and body wall by IHC analysis (Fig. 3A), Pbx1−/− embryos (36) did not (Fig. 3A, Inset). These mice were generated by insertion of a PGK-neo cassette in exon 3, upstream of the homeodomain (36). Three knockout litters composed of four homozygote mutant embryos, eight heterozygotes, and nine wild types, were harvested at E15.5. Of these embryos, three homozygote mutants were suitable for examination of whole diaphragms; homozygotes, but not heterozygotes, showed diaphragmatic and muscle patterning defects (Fig. 4 D–F). Two mutants had left-sided posterolateral defects with an intact membrane but absent musculature. In one, the fundus of the stomach was herniated into the thorax. This mutant also had abnormal muscular patterning with a region of enhanced vascularity adjacent to the circumscribed posterolateral defect (Fig. 3F). Expression of muscle markers (PAX3, MYOD, MYOG) was abnormal in E11.5 Pbx1 mutants (details in Fig. S3).
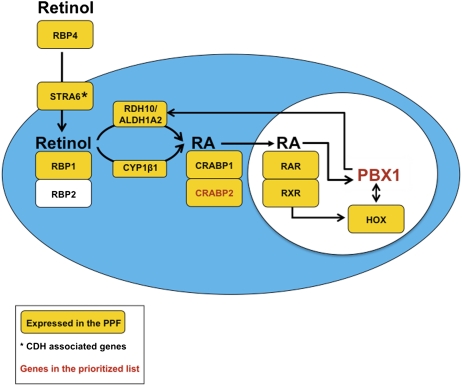
Fig. 4.
PBX1 transcriptional regulation in the PPFs. PBX1 may function downstream of the RA pathway and act via a feedback loop involving Aldh1a2 (46) (details in the text). Yellow boxes represent genes in significant STEM and/or GSEA gene sets; *, CDH-associated genes; red letters, members of the prioritized list of candidates.
Discussion
To determine whether the genes important for development are those perturbed in congenital anomalies affecting the diaphragm, we generated gene expression datasets that represent a global transcriptional survey of genes and biological pathways active during early (E11.5/E12.5) and late (E16.5) diaphragm embryogenesis. The molecular environment of the early diaphragm is characterized by elevated expression of embryonic “master regulator” genes involved in a variety of signaling pathways critical for early developmental processes (i.e., tissue specification and patterning) (37, 38), whereas in the late diaphragm, enhanced expression of genes associated with differentiated skeletal muscle system processes, such as muscle contraction, maintenance, and energy metabolism, was observed.
We identified 27 CDH candidate genes by using a prioritization strategy to filter the transcriptome datasets. Many of the 27 genes play cooperative roles in critical embryonic signaling pathways including retinoic acid (RA) signaling (Crabp2, Tgif1, Pbx1, Gata6, Ezh2), WNT canonical signaling (Fzd2, Pbx1, Gata6), TGFβ signaling (Tgfβ2, Tgif1, Pbx1), and Ephrin signaling/axon guidance (Epha3, Efna5, Epha4, Epha7, Ephb2, Sema3a, Unc5c) (39–47).
The human orthologs of these 27 genes do not map to previously identified CDH “hotspots” (i.e., recurring deletions or duplications that confer risk for CDH) (5), although a cytogenetic duplication that potentially involves the PBX1 locus may confer risk for CDH (48). Taken as a whole, these observations suggest our approach to gene discovery is complementary to genomic studies.
Among the 27 candidates, Pbx1 was selected to validate the prioritization strategy. Examination of Pbx1 knockout embryos at E15.5 revealed a range of diaphragmatic muscularization and tissue patterning defects (Fig. 4). Pbx1 encodes a three amino acid loop extension (TALE) homeodomain protein that functions as a transcriptional cofactor central to numerous regulatory networks that modulate cell specification, segmental patterning of gene expression, and mesenchymal precursor cell migration (49, 50).
PBX1 is known to interact with HOXA5 and HOXB5 (among others) (51, 52), which were identified as being expressed in the primordial diaphragm (as per STEM and GSEA). Because PBX1 directs local RA production by transcriptional regulation of Aldh1a2 in murine hindbrain mesoderm (46), we extrapolate that loss of PBX1 in the primordial diaphragm may lead to decreased RA. This decrease in RA, in turn, may affect transcription the RA regulated Hoxa5 and/or Hoxb5 genes (53, 54) (Fig. 4). Furthermore, rodents treated with the RA inhibitor nitrofen, a common model of CDH-associated pulmonary hypoplasia, showed abnormal expression patterns of HOXA5 in the developing lung buds (55).
In addition to RA, other signaling pathways, such as SHH, WNT, and TGFβ (49, 50, 56, 57), are considered important for proper diaphragm formation (42), because genes involved in the SHH pathway (e.g., Gli2, Gli3), the WNT pathway (e.g., Ctbp1, Ctbp2), and the TGFβ pathway (e.g., Fbn1) are associated with CDH phenotypes in knockout mice and/or in humans (24, 58, 59). PBX1 has been shown to influence SHH signaling during early embryonic patterning of the limb and neural tube with its MEIS and PKNOX1/HOX binding partners via Gli3 (60, 61). Furthermore, PBX1, in a complex with TGIF1/PKNOX1 and SMAD proteins, modulates transcription of TGFβ pathway target genes in pituitary cells (56), and also up-regulates genes involved in WNT signaling via interaction with PKNOX1/MEIS2 proteins in murine midface ectoderm (62). Expression of these PBX1 interactors was identified in the primordial diaphragm (as per STEM and GSEA). Therefore, we surmise that abnormal Pbx1-mediated regulation of transcription factors involved in one or several diaphragm-associated signaling pathway(s) (RA, SHH, WNT, and/or TGFβ) may lead to the tissue patterning and muscularization defects observed in diaphragms of E15.5 knockout mice. Abnormal expression patterns of MYOD1 in the developing diaphragm anlagen of Pbx1 mutants at E11.5 may result from impaired initiation of myogenesis. The homeodomain proteins PBX and MEIS have been shown to be necessary for MYOD1 binding of the Myog promoter and, possibly, other downstream targets, thus promoting the myogenic fate (63–65).
Among the 27 candidates, 8 are associated with human or mouse phenotypes, in which CDH was not among the spectrum of defects according to MGI or OMIM. The failure to detect CDH does not negate our prioritization approach, because CDH may be a subtle defect only visible upon careful scrutiny or occur as a low penetrant component of the overall phenotype. Additionally, mutations in some of the prioritized genes may not be compatible with life.
In conclusion, the unbiased transcriptome data generated herein have significantly expanded the number of high quality candidates for CDH. These candidates warrant follow-up in resequencing studies of human CDH cohorts, where risk alleles may be detected by mutational burden tests in case-control studies. The diaphragmatic defects identified in Pbx1 knockout mice provide proof of principle that prioritization strategies are capable of predicting CDH-associated genes and illustrate the power of integrating normal developmental expression data from the mouse with previous knowledge of mouse and human disease-causing mutations. Furthermore, genes involved in the causation of severe diaphragm anomalies, such as CDH, become candidates for causing less severe diaphragm-related phenotypes, for example gastroesophageal reflux. We predict that this approach can be generalized to a wide array of congenital anomalies that exhibit genetic heterogeneity.
Materials and Methods
Animals.
Tissue collections for LCM, IHC, and for diaphragm examination were performed as detailed in SI Materials and Methods.
LCM.
LCM, RNA extraction, purification, and amplification were performed as detailed in SI Materials and Methods.
Expression Arrays.
Gene expression experiments were performed on Affymetrix Mouse Gene 1.0_ST microarrays. The RNA from multiple laser captured tissues was pooled for each embryo, but each embryo was analyzed separately. Hybridization was performed according to the manufacturer's instructions.
Data Analyses.
The microarray data from E11.5, E12.5, and E16.5 embryos were normalized in the R statistical programming environment (version 2.9.2) by using Robust Multichip Averaging [RMA; ref. 66; implemented in the Affymetrix package for R (http://bioconductor.org/packages/2.1/bioc/html/affy.html)]. Differentially expressed genes were identified by using Welch's t test, assuming unequal variances, with a Bonferroni correction. Significance was set at P < 0.01. To determine genes expressed at each time-point, the median of the normalized hybridization intensities for 5,222 probes designed to correspond to introns was calculated by using an R script and used as a threshold (for E11.5 and E12.5, the median was 6.3; for E16.5, the median was 6.2). Groups of genes with similar temporal expression patterns were identified by using STEM (http://gene.ml.cmu.edu/stem/) (19), a modified k-means clustering algorithm (a method which partitions data in a predefined number of groups, based on their mean) (67). In GSEA, a continuous phenotype label and a Pearson metric was applied to rank genes and gene sets enriched at a nominal P < 0.05 and a false discovery rate <0.25, as discussed by Subramanian et al. (20).
Candidate Gene Prioritization.
A list of 48 known diaphragm defect genes (Dataset S5) was generated by manually curating data from several sources including PubMed; Online Mendelian Inheritance in Man, OMIM, McKusick-Nathans Institute of Genetic Medicine, Johns Hopkins University (Baltimore), and the National Center for Biotechnology Information, National Library of Medicine (Bethesda, MD) [last accession November 2010]; and the Mouse Genome Informatics (MGI) database, The Jackson Laboratory [last accession November 2010]. STEM and GSEA both partition the expression data into subsets of genes that participate in a common pathway (GSEA) or specific temporal expression profiles (STEM). To identify the subsets most likely to contain unique CDH genes, we reasoned that unique CDH causing genes are likely to belong to the pathways or expression profiles most enriched for known disease genes, used as “baits”. We measured the representation of CDH-associated genes in the different GSEA pathways, or STEM-derived expression datasets, using a hypergeometric distribution (a probability test describing the number of successes in a number of draws from a finite population) (68), and considered only gene sets that were significantly enriched (P < 0.05 after adjustment for multiple hypothesis testing). For example, if an enriched GSEA-defined pathway contained 10 genes of which 8 were known to cause CDH, the two remaining genes would be likely to be involved in CDH themselves. Although some genes occur in more then one pathway, each enrichment calculation is independent, thus preventing bias.
RT-qPCR.
qPCR experiments and data normalization were performed as detailed in SI Materials and Methods.
IHC.
IHC was performed as described (69). Sections were visualized with the ABComplex/HRP Vectastain Detection System according to manufacturer's protocols (Vector Laboratories) after 3,3′-diaminobenzidine (DAB) enhancement (Sigma) (CRABP2, EZH2, GATA6, MEIS2, MYOD1, MYOG, PAX3, PBX1b, TGIF1, ZFPM2), or by the VECTASTAIN ABC-AP KIT (AP-1000) (Vector Laboratories) (RUNX1). The combinations of primary and biotinylated secondary antibodies are listed in Table S1.
Note.
During the preparation of the manuscript, another gene among the novel candidates (Fzd2) was implicated as causal of CDH (47), further substantiating the effectiveness of our gene discovery approach.
Supplementary Material
Acknowledgments
We thank the Jackson Laboratory Scientific Services for valuable expertise and Dr. Jim Denegre, Dr. Eric Antoniou, Sonya Kamdar, Karen Moore, Ellen Akeson, and Dr. Leslie Bechtold. Pbx1 knockout embryos were provided by Dr. Michael Cleary and Carmencita Nicholas (Stanford University). We also thank Mark Daly and David MacLaughlin for helpful suggestions. This work was submitted by M.K.R. in partial fulfillment for her doctoral degree.
Footnotes
The authors declare no conflict of interest.
Data deposition: The array data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE35243).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1121621109/-/DCSupplemental.
References
- 1.Stege G, Fenton A, Jaffray B. Nihilism in the 1990s: The true mortality of congenital diaphragmatic hernia. Pediatrics. 2003;112:532–535. doi: 10.1542/peds.112.3.532. [DOI] [PubMed] [Google Scholar]
- 2.Pober BR. Overview of epidemiology, genetics, birth defects, and chromosome abnormalities associated with CDH. Am J Med Genet C Semin Med Genet. 2007;145C:158–171. doi: 10.1002/ajmg.c.30126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dott MM, Wong LY, Rasmussen SA. Population-based study of congenital diaphragmatic hernia: Risk factors and survival in Metropolitan Atlanta, 1968-1999. Birth Defects Res A Clin Mol Teratol. 2003;67:261–267. doi: 10.1002/bdra.10039. [DOI] [PubMed] [Google Scholar]
- 4.Pober BR, Russell MK, Ackerman KG. Congenital diaphragmatic hernia overview. In: Pagon RABT, Dolan CR, Stephens K, editors. GeneReviews. Seattle: University of Washington; 2010. [PubMed] [Google Scholar]
- 5.Holder AM, et al. Genetic factors in congenital diaphragmatic hernia. Am J Hum Genet. 2007;80:825–845. doi: 10.1086/513442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ackerman KG, Greer JJ. Development of the diaphragm and genetic mouse models of diaphragmatic defects. Am J Med Genet C Semin Med Genet. 2007;145C:109–116. doi: 10.1002/ajmg.c.30128. [DOI] [PubMed] [Google Scholar]
- 7.Fisher JC, Bodenstein L. Computer simulation analysis of normal and abnormal development of the mammalian diaphragm. Theor Biol Med Model. 2006;3:9. doi: 10.1186/1742-4682-3-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Babiuk RP, Zhang W, Clugston R, Allan DW, Greer JJ. Embryological origins and development of the rat diaphragm. J Comp Neurol. 2003;455:477–487. doi: 10.1002/cne.10503. [DOI] [PubMed] [Google Scholar]
- 9.Greer JJ, Allan DW, Martin-Caraballo M, Lemke RP. An overview of phrenic nerve and diaphragm muscle development in the perinatal rat. J Appl Physiol. 1999;86:779–786. doi: 10.1152/jappl.1999.86.3.779. [DOI] [PubMed] [Google Scholar]
- 10.Greer JJ, et al. Structure of the primordial diaphragm and defects associated with nitrofen-induced CDH. J Appl Physiol. 2000;89:2123–2129. doi: 10.1152/jappl.2000.89.6.2123. [DOI] [PubMed] [Google Scholar]
- 11.Mayer S, Metzger R, Kluth D. The embryology of the diaphragm. Semin Pediatr Surg. 2011;20:161–169. doi: 10.1053/j.sempedsurg.2011.03.006. [DOI] [PubMed] [Google Scholar]
- 12.Iritani I. Experimental study on embryogenesis of congenital diaphragmatic hernia. Anat Embryol (Berl) 1984;169:133–139. doi: 10.1007/BF00303142. [DOI] [PubMed] [Google Scholar]
- 13.Kho AT, et al. Conserved mechanisms across development and tumorigenesis revealed by a mouse development perspective of human cancers. Genes Dev. 2004;18:629–640. doi: 10.1101/gad.1182504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Naxerova K, et al. Analysis of gene expression in a developmental context emphasizes distinct biological leitmotifs in human cancers. Genome Biol. 2008;9:R108. doi: 10.1186/gb-2008-9-7-r108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lage K, et al. A large-scale analysis of tissue-specific pathology and gene expression of human disease genes and complexes. Proc Natl Acad Sci USA. 2008;105:20870–20875. doi: 10.1073/pnas.0810772105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ackerman KG, et al. Fog2 is required for normal diaphragm and lung development in mice and humans. PLoS Genet. 2005;1:58–65. doi: 10.1371/journal.pgen.0010010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Clugston RD, Zhang W, Greer JJ. Gene expression in the developing diaphragm: Significance for congenital diaphragmatic hernia. Am J Physiol Lung Cell Mol Physiol. 2008;294:L665–L675. doi: 10.1152/ajplung.00027.2008. [DOI] [PubMed] [Google Scholar]
- 18.Dingemann J, Doi T, Ruttenstock E, Puri P. The role of primary myogenic regulatory factors in the developing diaphragmatic muscle in the nitrofen-induced diaphragmatic hernia. Pediatr Surg Intl. 2011;27:579–582. doi: 10.1007/s00383-010-2834-8. [DOI] [PubMed] [Google Scholar]
- 19.Ernst J, Bar-Joseph Z. STEM: A tool for the analysis of short time series gene expression data. BMC Bioinformatics. 2006;7:191. doi: 10.1186/1471-2105-7-191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Subramanian A, et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Subramanian A, Kuehn H, Gould J, Tamayo P, Mesirov JP. GSEA-P: A desktop application for Gene Set Enrichment Analysis. Bioinformatics. 2007;23:3251–3253. doi: 10.1093/bioinformatics/btm369. [DOI] [PubMed] [Google Scholar]
- 22.Bleyl SB, et al. Candidate genes for congenital diaphragmatic hernia from animal models: Sequencing of FOG2 and PDGFRalpha reveals rare variants in diaphragmatic hernia patients. Eur J Hum Genet. 2007;15:950–958. doi: 10.1038/sj.ejhg.5201872. [DOI] [PubMed] [Google Scholar]
- 23.Golzio C, et al. Matthew-Wood syndrome is caused by truncating mutations in the retinol-binding protein receptor gene STRA6. Am J Hum Genet. 2007;80:1179–1187. doi: 10.1086/518177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim PC, Mo R, Hui Cc C. Murine models of VACTERL syndrome: Role of sonic hedgehog signaling pathway. J Pediatr Surg. 2001;36:381–384. doi: 10.1053/jpsu.2001.20722. [DOI] [PubMed] [Google Scholar]
- 25.Liu J, et al. Congenital diaphragmatic hernia, kidney agenesis and cardiac defects associated with Slit3-deficiency in mice. Mech Dev. 2003;120:1059–1070. doi: 10.1016/s0925-4773(03)00161-8. [DOI] [PubMed] [Google Scholar]
- 26.Neri G, Gurrieri F, Zanni G, Lin A. Clinical and molecular aspects of the Simpson-Golabi-Behmel syndrome. Am J Med Genet. 1998;79:279–283. doi: 10.1002/(sici)1096-8628(19981002)79:4<279::aid-ajmg9>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- 27.Oh J, et al. Mutations in two matrix metalloproteinase genes, MMP-2 and MT1-MMP, are synthetic lethal in mice. Oncogene. 2004;23:5041–5048. doi: 10.1038/sj.onc.1207688. [DOI] [PubMed] [Google Scholar]
- 28.Shi L, et al. NF90 regulates cell cycle exit and terminal myogenic differentiation by direct binding to the 3′-untranslated region of MyoD and p21WAF1/CIP1 mRNAs. J Biol Chem. 2005;280:18981–18989. doi: 10.1074/jbc.M411034200. [DOI] [PubMed] [Google Scholar]
- 29.Yano S, et al. Familial Simpson-Golabi-Behmel syndrome: Studies of X-chromosome inactivation and clinical phenotypes in two female individuals with GPC3 mutations. Clin Genet. 2010;80:466–471. doi: 10.1111/j.1399-0004.2010.01554.x. [DOI] [PubMed] [Google Scholar]
- 30.Pasutto F, et al. Mutations in STRA6 cause a broad spectrum of malformations including anophthalmia, congenital heart defects, diaphragmatic hernia, alveolar capillary dysplasia, lung hypoplasia, and mental retardation. Am J Hum Genet. 2007;80:550–560. doi: 10.1086/512203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ashburner M, et al. The Gene Ontology Consortium Gene ontology: Tool for the unification of biology. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Smith CL, Eppig JT. The mammalian phenotype ontology: Enabling robust annotation and comparative analysis. Wiley Interdiscip Rev Syst Biol Med. 2009;1:390–399. doi: 10.1002/wsbm.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Grigoryan T, Wend P, Klaus A, Birchmeier W. Deciphering the function of canonical Wnt signals in development and disease: conditional loss- and gain-of-function mutations of beta-catenin in mice. Genes Dev. 2008;22:2308–2341. doi: 10.1101/gad.1686208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Harland RM. The transforming growth factor beta family and induction of the vertebrate mesoderm: bone morphogenetic proteins are ventral inducers. Proc Natl Acad Sci USA. 1994;91:10243–10246. doi: 10.1073/pnas.91.22.10243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Winnier G, Blessing M, Labosky PA, Hogan BL. Bone morphogenetic protein-4 is required for mesoderm formation and patterning in the mouse. Genes Dev. 1995;9:2105–2116. doi: 10.1101/gad.9.17.2105. [DOI] [PubMed] [Google Scholar]
- 36.Selleri L, et al. Requirement for Pbx1 in skeletal patterning and programming chondrocyte proliferation and differentiation. Development. 2001;128:3543–3557. doi: 10.1242/dev.128.18.3543. [DOI] [PubMed] [Google Scholar]
- 37.Hunt P, Krumlauf R. Hox codes and positional specification in vertebrate embryonic axes. Annu Rev Cell Biol. 1992;8:227–256. doi: 10.1146/annurev.cb.08.110192.001303. [DOI] [PubMed] [Google Scholar]
- 38.Martínez L, et al. [The etiology of congenital diaphragmatic hernia and esophageal atresia: The Hox genes] Cir Pediatr. 2007;20:223–228. [PubMed] [Google Scholar]
- 39.Bertolino E, Reimund B, Wildt-Perinic D, Clerc RG. A novel homeobox protein which recognizes a TGT core and functionally interferes with a retinoid-responsive motif. J Biol Chem. 1995;270:31178–31188. doi: 10.1074/jbc.270.52.31178. [DOI] [PubMed] [Google Scholar]
- 40.Qin P, Cimildoro R, Kochhar DM, Soprano KJ, Soprano DR. PBX, MEIS, and IGF-I are potential mediators of retinoic acid-induced proximodistal limb reduction defects. Teratology. 2002;66:224–234. doi: 10.1002/tera.10082. [DOI] [PubMed] [Google Scholar]
- 41.Mey J, Babiuk RP, Clugston R, Zhang W, Greer JJ. Retinal dehydrogenase-2 is inhibited by compounds that induce congenital diaphragmatic hernias in rodents. Am J Pathol. 2003;162:673–679. doi: 10.1016/s0002-9440(10)63861-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bielinska M, et al. Molecular genetics of congenital diaphragmatic defects. Ann Med. 2007;39:261–274. doi: 10.1080/07853890701326883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kuijper S, Turner CJ, Adams RH. Regulation of angiogenesis by Eph-ephrin interactions. Trends Cardiovasc Med. 2007;17:145–151. doi: 10.1016/j.tcm.2007.03.003. [DOI] [PubMed] [Google Scholar]
- 44.Clugston RD, Zhang W, Greer JJ. Early development of the primordial mammalian diaphragm and cellular mechanisms of nitrofen-induced congenital diaphragmatic hernia. Birth Defects Res A Clin Mol Teratol. 2010;88:15–24. doi: 10.1002/bdra.20613. [DOI] [PubMed] [Google Scholar]
- 45.Hogue J, et al. A novel EFNB1 mutation (c.712delG) in a family with craniofrontonasal syndrome and diaphragmatic hernia. Am J Med Genet A. 2010;152A:2574–2577. doi: 10.1002/ajmg.a.33596. [DOI] [PubMed] [Google Scholar]
- 46.Vitobello A, et al. Hox and Pbx factors control retinoic acid synthesis during hindbrain segmentation. Dev Cell. 2011;20:469–482. doi: 10.1016/j.devcel.2011.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wat MJ, et al. Genomic alterations that contribute to the development of isolated and non-isolated congenital diaphragmatic hernia. J Med Genet. 2011;48:299–307. doi: 10.1136/jmg.2011.089680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Otake K, et al. Congenital diaphragmatic hernia with a pure duplication of chromosome 1q: Report of the first surviving case. Pediatr Surg Int. 2009;25:827–831. doi: 10.1007/s00383-009-2421-z. [DOI] [PubMed] [Google Scholar]
- 49.Capellini TD, et al. Pbx1/Pbx2 requirement for distal limb patterning is mediated by the hierarchical control of Hox gene spatial distribution and Shh expression. Development. 2006;133:2263–2273. doi: 10.1242/dev.02395. [DOI] [PubMed] [Google Scholar]
- 50.Laurent A, Bihan R, Omilli F, Deschamps S, Pellerin I. PBX proteins: much more than Hox cofactors. Int J Dev Biol. 2008;52:9–20. doi: 10.1387/ijdb.072304al. [DOI] [PubMed] [Google Scholar]
- 51.Lu Q, Kamps MP. Structural determinants within Pbx1 that mediate cooperative DNA binding with pentapeptide-containing Hox proteins: Proposal for a model of a Pbx1-Hox-DNA complex. Mol Cell Biol. 1996;16:1632–1640. doi: 10.1128/mcb.16.4.1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shen WF, et al. Hox homeodomain proteins exhibit selective complex stabilities with Pbx and DNA. Nucleic Acids Res. 1996;24:898–906. doi: 10.1093/nar/24.5.898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kim C, Nielsen HC. Hoxa-5 in mouse developing lung: Cell-specific expression and retinoic acid regulation. Am J Physiol Lung Cell Mol Physiol. 2000;279:L863–L871. doi: 10.1152/ajplung.2000.279.5.L863. [DOI] [PubMed] [Google Scholar]
- 54.Mahony S, et al. Ligand-dependent dynamics of retinoic acid receptor binding during early neurogenesis. Genome Biol. 2011;12:R2. doi: 10.1186/gb-2011-12-1-r2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Volpe MV, Wang KT, Nielsen HC, Chinoy MR. Unique spatial and cellular expression patterns of Hoxa5, Hoxb4, and Hoxb6 proteins in normal developing murine lung are modified in pulmonary hypoplasia. Birth Defects Res A Clin Mol Teratol. 2008;82:571–584. doi: 10.1002/bdra.20481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bailey JS, Rave-Harel N, McGillivray SM, Coss D, Mellon PL. Activin regulation of the follicle-stimulating hormone beta-subunit gene involves Smads and the TALE homeodomain proteins Pbx1 and Prep1. Mol Endocrinol. 2004;18:1158–1170. doi: 10.1210/me.2003-0442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Capellini TD, et al. Pbx1/Pbx2 govern axial skeletal development by controlling Polycomb and Hox in mesoderm and Pax1/Pax9 in sclerotome. Dev Biol. 2008;321:500–514. doi: 10.1016/j.ydbio.2008.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hildebrand JD, Soriano P. Overlapping and unique roles for C-terminal binding protein 1 (CtBP1) and CtBP2 during mouse development. Mol Cell Biol. 2002;22:5296–5307. doi: 10.1128/MCB.22.15.5296-5307.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pereira L, et al. Pathogenetic sequence for aneurysm revealed in mice underexpressing fibrillin-1. Proc Natl Acad Sci USA. 1999;96:3819–3823. doi: 10.1073/pnas.96.7.3819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Vokes SA, Ji H, Wong WH, McMahon AP. A genome-scale analysis of the cis-regulatory circuitry underlying sonic hedgehog-mediated patterning of the mammalian limb. Genes Dev. 2008;22:2651–2663. doi: 10.1101/gad.1693008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Coy S, Caamaño JH, Carvajal J, Cleary ML, Borycki AG. A novel Gli3 enhancer controls the Gli3 spatiotemporal expression pattern through a TALE homeodomain protein binding site. Mol Cell Biol. 2011;31:1432–1443. doi: 10.1128/MCB.00451-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ferretti E, et al. A conserved Pbx-Wnt-p63-Irf6 regulatory module controls face morphogenesis by promoting epithelial apoptosis. Dev Cell. 2011;21:627–641. doi: 10.1016/j.devcel.2011.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Berkes CA, et al. Pbx marks genes for activation by MyoD indicating a role for a homeodomain protein in establishing myogenic potential. Mol Cell. 2004;14:465–477. doi: 10.1016/s1097-2765(04)00260-6. [DOI] [PubMed] [Google Scholar]
- 64.Maves L, et al. Pbx homeodomain proteins direct Myod activity to promote fast-muscle differentiation. Development. 2007;134:3371–3382. doi: 10.1242/dev.003905. [DOI] [PubMed] [Google Scholar]
- 65.Heidt AB, Rojas A, Harris IS, Black BL. Determinants of myogenic specificity within MyoD are required for noncanonical E box binding. Mol Cell Biol. 2007;27:5910–5920. doi: 10.1128/MCB.01700-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Irizarry RA, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 67.MacQueen J. Some methods for classification and analysis of multivariate observations. In: Le Cam LM, Neyman J, editors. Fifth Berkeley Symposium on Mathematical Statistics and Probability. Univ of California Press, Berkeley, CA; 1967. pp. 281–297. [Google Scholar]
- 68.Rice JA. Mathematical Statistics and Data Analysis. Belmont, CA: Duxbury; 2007. [Google Scholar]
- 69.Loscertales M, Mikels AJ, Hu JK, Donahoe PK, Roberts DJ. Chick pulmonary Wnt5a directs airway and vascular tubulogenesis. Development. 2008;135:1365–1376. doi: 10.1242/dev.010504. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.