Abstract
The generation and maintenance of a plethora of neuronal subtypes is essential for normal brain function. Nevertheless, little is known about the molecular mechanisms that maintain the defining characteristics of neurons following their initial postmitotic specification. Using conditional gene ablation in mice, we demonstrate here that the homeodomain protein LIM homeobox (Lhx)7 is essential for maintaining the morphological and molecular characteristics of cholinergic interneurons of the striatum. Lhx7-depleted cholinergic interneurons extinguish expression of several subtype-specific markers, including choline acetyl transferase and Isl1, and are respecified into Lhx6-expressing mature GABAergic interneurons. Additional expression studies support a model where Lhx7 controls the choice between cholinergic or GABAergic identity by gating a cross inhibitory regulation between Isl1 and Lhx6. By demonstrating that the switch between alternative striatal interneuron fates depends on persistent activity of a single transcription factor, we provide evidence that the intrinsic plasticity of mammalian forebrain neuronal subtypes is maintained after the initial specification and lineage commitment and possibly throughout life.
Keywords: neuronal subtype specification, medial ganglionic eminence-derived interneurons
The information-processing ability of the nervous system depends on the formation of functional neuronal circuits composed of a highly heterogeneous population of neurons. Recent progress has identified molecular cascades that control the generation of distinct neuronal subtypes during development (1–3) but the mechanisms that maintain the identity of neurons following lineage commitment and differentiation are currently unclear. Identifying such mechanisms is critical for understanding phenotypic plasticity in the nervous system and, ultimately, influencing its ability to adjust during normal development, disease, or injury.
The striatum is a subcortical structure that integrates multiple inputs from the cortex, thalamus, and midbrain and relays information to the output domains of the basal ganglia via its principal population of projection neurons (4, 5). Balanced striatal output, which is critical for motor and cognitive activity, depends on local circuits controlled by distinct subpopulations of GABAergic and cholinergic interneurons (5). Both GABAergic and cholinergic striatal interneurons (GSIs and CSIs, respectively) are derived from Nkx2.1-expressing progenitors of the medial ganglionic eminence (MGE) (6, 7), which, upon exit from the cell cycle, express the LIM homeodomain (LIM HD) transcription factors LIM homeobox 6 (Lhx6) and LIM homeobox 7 (Lhx7) (8). A subset of these precursors maintains expression of Lhx6 and differentiate into mature GABAergic interneurons expressing the neuropeptide somatostatin (Sst) or the calcium-binding protein parvalbumin (Pv) (9, 10). The remaining precursors induce expression of the LIM HD protein Isl1, down-regulate Lhx6, and differentiate into CSIs (8). In adult animals, all GSIs are Lhx6+ and a fraction of these also coexpresses Lhx7. In contrast, Lhx6 is undetectable in CSIs, which invariably coexpress Isl1 and Lhx7. Genetic studies have established the requirement of Lhx7 and Isl1 for the specification of forebrain cholinergic neurons (11–14), but the potential role of these factors in maintaining the identity of CSIs following their specification and differentiation is currently unknown.
By combining in vivo fate mapping and phenotypic analysis of CSIs in which Lhx7 was conditionally ablated, we demonstrate here that Lhx7 is required subsequently to cholinergic subtype specification to actively maintain the phenotypic characteristics of these interneurons and prevent them from adopting GABAergic identity. Gain-of-function studies also suggest that Lhx7 is a molecular node that resolves the lineage choices of striatal interneuron precursors by modulating antagonistic expression of Isl1 and Lhx6. Our experiments reveal a remarkable degree of plasticity of postmitotic neurons and raise the possibility that cell differentiation choices can be readjusted at all developmental stages by manipulating the levels of key transcriptional regulators.
Results
Deletion of Lhx7 from Committed Postmitotic Cholinergic Neurons of the Forebrain.
To examine the role of Lhx7 in maintaining the subtype identity of CSIs following their initial specification, we generated a conditional allele of the locus (Lhx7fl), which allows cell type- and stage-specific deletion of the gene (Fig. S1 and Materials and Methods). The fate of Lhx7-depleted cells was followed by combining Lhx7fl with Lhx7LacZ, a null allele generated previously by inserting the LacZ reporter into the Lhx7 locus (12). To confirm that Lhx7fl is converted into a null allele upon Cre-mediated recombination, we analyzed brain sections from Lhx7+/LacZ, Lhx7fl/LacZ, and β-actin::Cre;Lhx7fl/LacZ animals (15). Consistent with Lhx7 deletion in β-actin::Cre;Lhx7fl/LacZ mice, Lhx7-positive cells were absent (Fig. S1D) and CSIs were dramatically reduced in the striatum (Fig. S1 E–G) (8, 12–14). Interestingly, reduction of CSIs was associated with increased number of Lhx6-expressing striatal interneurons (Fig. S1F), a characteristic feature of Lhx7-null mice (8, 12–14). Therefore, upon Cre-mediated recombination, Lhx7fl is converted into a null allele.
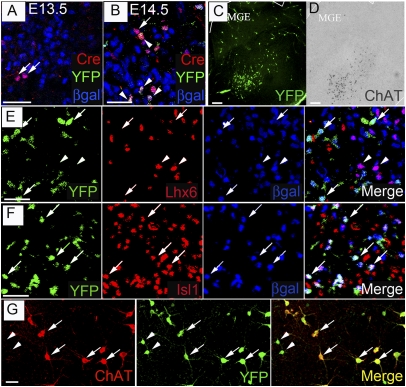
Because choline acetyltransferase (ChAT) mRNA is first detected in committed, immature cholinergic neurons of the striatum and basal forebrain at embryonic stages (Fig. 1C), we reasoned that deletion of Lhx7 by a transgene expressing Cre under the control of ChAT regulatory sequences (ChAT::Cre) would bypass the early requirement of the gene for cholinergic specification and allow us to address its role in maintaining the identity of the cholinergic sublineage. To validate our strategy, we fate-mapped cholinergic precursors in vivo by combining a ChAT::Cre transgene (16) with the Cre-dependent R26ReYFP lineage reporter (17) and the Lhx7LacZ allele (to mark Lhx7-expressing cells with β-gal). Animals were analyzed during embryogenesis for expression of Cre and YFP in β-gal+ cells of the MGE (where striatal interneurons are born) and LGE (the striatal anlage). At embryonic day (E)13.5, some β-gal+ cells expressed Cre (but not YFP) (Fig. 1A), and at E14.5, many β-gal+ cells coexpressed Cre and YFP (Fig. 1B). β-gal+YFP+ cells did not express Ki67, indicating that they were postmitotic (Fig. S2) (8). Induction of Cre and YFP in the striatal anlage followed a caudal to rostral temporal order (similar to the wave of birth and maturation of cholinergic neurons in the basal forebrain) (18), and the overall pattern of expression of YFP in the ventral forebrain was comparable to that of ChAT RNA (compare Fig. 1 C and D). Importantly, the vast majority of YFP+ cells in the basal forebrain of ChAT::Cre;Lhx7+/LacZ;R26ReYFP animals coexpressed β-gal, and many expressed the cholinergic interneuron marker Isl1 (Fig. 1F), but all were negative for the GABAergic marker Lhx6 (Fig. 1E). Furthermore, in the striatum of adult ChAT::Cre;Lhx7+/LacZ;R26YFP mice, virtually all β-gal+YFP+ neurons coexpressed ChAT (Fig. 1G, arrows). Although, at this stage, we observed a small number of YFP+ChAT− cells (Fig. 1G, arrowheads), these cells did not express β-gal and, therefore, did not interfere with our subsequent analysis. Taken together, these experiments demonstrate that the ChAT::Cre transgene is activated during embryogenesis in postmitotic CSIs after the divergence of the GABAergic and cholinergic lineages of the striatum.
Fig. 1.
The ChAT::Cre transgene is specifically expressed by Lhx7+ forebrain cholinergic neurons. (A–D) Ventral forebrain sections from E13.5 (A) or E14.5 (B–D) ChAT::Cre;Lhx7+/LacZ;R26ReYFP embryos immunostained for Cre, YFP, and β-gal (A and B) or YFP (C) or processed for ISH using a ChAT-specific riboprobe (D). Arrows in A and B indicate Cre+YFP− cells, and arrowheads in B indicate Cre+YFP+ cells. All Cre+ cells express β-gal. In the serial sections of ventral forebrain shown in C and D, the distribution of YFP signal is similar to that of ChAT mRNA. (E and F) Sections from the MGE-LGE boundary sections of E14.5 ChAT::Cre;Lhx7+/LacZ;R26ReYFP embryos triple-immunostained for YFP, Lhx6, and β-gal (E) or YFP, Isl1, and β-gal (F). Lhx6-expressing cells (arrowheads) are negative for YFP, and YFP+ cells (arrows) do not express Lhx6 (E). In contrast, YFP is detected in β-gal+ neurons expressing Isl1 (F). (G) Adult striatum sections immunostained for ChAT and YFP. YFP is expressed in virtually all cholinergic (ChAT+) neurons (arrows); some noncholinergic cells also express YFP (arrowheads). [Scale bars: 50 μm (A, B, E, and F); 100 μm (C and D).]
Lhx7 Is Required in Committed CSIs for Terminal Differentiation.
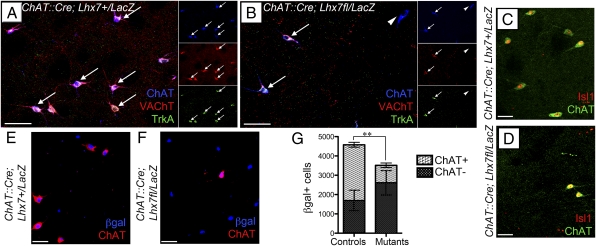
To examine whether Lhx7 is required in the mammalian forebrain to maintain the unique identity of CSIs, we compared the expression of the cholinergic markers ChAT, vesicular acetylcholine transporter (VAChT), and TrkA (19, 20) in the striatum of adult ChAT::Cre;Lhx7+/LacZ (control) and ChAT::Cre;Lhx7fl/LacZ (conditional mutant) mice. As expected, CSIs coexpressing ChAT, VAChT, and TrkA were readily detected in control animals (Fig. 2A, arrows). However, in the striatum of conditional mutants, the number of cells expressing these markers was dramatically reduced (compare Fig. 2 A and B). Most residual CSIs in these animals expressed all three markers, but, occasionally, we observed cells that expressed ChAT but were negative for TrkA and VAChT (Fig. 2B, arrowhead). The forebrain of ChAT::Cre;Lhx7fl/LacZ animals was also analyzed for expression of Isl1, a gene expressed in mature CSIs and required for their specification (8, 11). Consistent with the loss of CSIs, the number of cells expressing Isl1 was severely reduced (compare Fig. 2 C and D), with expression maintained only in residual cholinergic interneurons (Fig. 2D). Together, these findings suggest that deletion of Lhx7 from committed cholinergic precursors results in depletion of cholinergic neurons in the adult striatum.
Fig. 2.
Lhx7 activity is necessary for maintaining the cholinergic identity of striatal interneurons. (A–F) Sections from the striatum of adult ChaT::Cre;Lhx7+/LacZ (A, C, and E) and ChaT::Cre;Lhx7fl/LacZ (B, D, and F) animals immunostained for ChAT, VAChT, and TrkA (A and B), ChAT and Isl1 (C and D), or ChAT and β-gal (E and F). The number of cholinergic cells in conditional mutants is reduced and some ChAT+ neurons are negative for VAChT or TrkA (arrowhead in B). (G) Quantification of the cholinergic and noncholinergic subpopulations of β-gal+ cells in control and conditional mutant animals. The dramatic loss of cholinergic neurons in the striatum of ChAT::Cre;Lhx7fl/LacZ animals was associated with an increase in the number of noncholinergic β-gal+ cells (E–G). Bars represent means ± SD. [Scale bars: 100 μm (A and B); 50 μm (C–F).]
To examine whether the decrease of CSIs observed in ChAT::Cre;Lhx7fl/LacZ mice results from cell death, we compared the total number of β-gal+ cells in control and conditional mutant striatum. We observed that in conditional mutants, the absolute number of β-gal+ cells was reduced (Fig. 2G), suggesting a requirement of Lhx7 activity for the survival of a subset of these cells. However, twice as many cholinergic neurons were lost in the mutant striatum compared with β-gal+ cells (Fig. 2G), suggesting that upon deletion of Lhx7, a significant fraction of β-gal+ cholinergic neurons survive but extinguish expression of cholinergic markers. In support of this view, we recorded an increased number of β-gal+ChAT− cells in the striatum of ChAT::Cre;Lhx7fl/LacZ animals (Fig. 2 F and G). Together, our experiments indicate that Lhx7 is required to maintain the cholinergic identity of immature CSIs.
Upon Deletion of Lhx7 CSIs Switch Identity and Acquire Molecular and Morphological Features of GSIs.
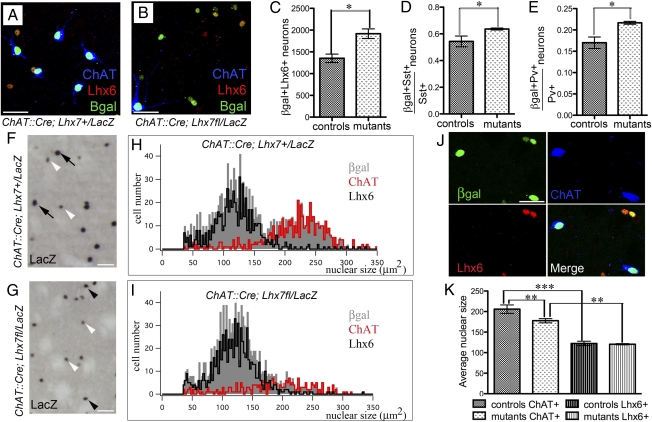
Because GSIs and CSIs share common Lhx6-expressing precursors, we investigated the possibility that deletion of Lhx7 from committed cholinergic interneurons is associated with reactivation of Lhx6 and acquisition of GABAergic characteristics. To explore this idea, we double immunostained forebrain sections from control and conditional mutant animals with antibodies for β-gal (to identify all striatal interneurons derived from Lhx7-expressing lineages) and Lhx6. Relative to controls, the total number of β-gal+ cells coexpressing Lhx6 (β-gal+Lhx6+) in the striatum of ChAT::Cre;Lhx7fl/LacZ animals increased by approx. 46% (Fig. 3 A–C), with Lhx6 never detected in residual ChAT+ neurons. Moreover, the number of Sst+ and Pv+ interneurons coexpressing LacZ increased in ChAT::Cre;Lhx7fl/LacZ animals relative to controls [16.7% increase of Sst+β-gal+ interneurons and 27% increase of Pv+β-gal+ interneurons (Fig. 3 D and E)]. These findings, together with the reduced representation of cholinergic neurons in the total β-gal+ subpopulation of the striatum, suggest that Lhx7-depleted CSIs switch identity and acquire molecular characteristic of GABAergic interneurons. Interestingly, β-gal+ cells were not detected in the cortex of conditional mutants indicating that Lhx7-depleted precursors do not adopt a cortical fate.
Fig. 3.
Lhx7-depleted CSI precursors acquire molecular and morphological characteristics of GSIs. (A and B) Sections from the striatum of ChAT::Cre;Lhx7+/LacZ (A) and ChAT::Cre;Lhx7fl/LacZ (B) animals triple-immunostained for ChAT, Lhx6, and β-gal. (C) Quantification of β-gal+Lhx6+ cells in the striatum of adult animals from the two genotypes. Note the increased representation of Lhx6+ cells among β-gal+ neurons in conditional mutants (A–C). (D and E) Quantification of the β-gal+ fraction of the Sst+ (D) or Pv+ (E) GSIs of adult conditional mutants and controls. (F and G) Sections from the striatum of ChAT::Cre;Lhx7+/LacZ (F) and ChAT::Cre;Lhx7fl/LacZ (G) adult animals immunostained for β-gal. (H and I) Distribution of nuclear size of all β-gal-expressing cells (gray), β-gal+Lhx6+ (black), and β-gal+ChAT+ (red) subpopulations in the striatum of ChAT::Cre;Lhx7+/LacZ (H) and ChAT::Cre;Lhx7fl/LacZ (I) mice. (J) Example of triple-immunostaining (β-gal, Lhx6, ChAT) used for the quantification shown in H and I. (K) Quantification of average nuclear size in the cholinergic (ChAT+) and GABAergic (Lhx6+) subpopulations in the striatum of ChAT::Cre;Lhx7+/LacZ and ChAT::Cre;Lhx7fl/LacZ animals. [Scale bars: 50 μm (A, B, F, G, and J).]
CSIs can be distinguished from GSIs because of their distinct cellular morphology and relatively large nuclear size (21, 22). Consistent with this, striatal interneurons in ChAT::Cre;Lhx7+/LacZ mice, which express the nuclear localized reporter β-gal, include neurons with large and intensely labeled nuclei, as well as neurons with relatively small and weakly labeled nuclei (Fig. 3F, black and white arrowheads, respectively). Interestingly, in ChAT::Cre;Lhx7fl/LacZ animals, the number of large and intensely stained nuclei was considerably smaller, whereas the number of small nuclei had increased (Fig. 3G). Triple immunostaining for β-gal, ChAT, and Lhx6 confirmed that large nuclei corresponded to CSIs, whereas the cells with small nuclei were GSIs (Fig. 3J).
To quantify the effects of Lhx7 deletion on nuclear morphology, the nuclear surface area and marker expression of striatal interneurons was compared in forebrain sections from control and conditional mutants triple immunostained for β-gal, ChAT, and Lhx6. In control sections, β-gal+ neurons segregated into two largely nonoverlapping groups; the first was characterized by relatively small nuclear size (peak around 120 μm2) and expression of Lhx6 (Fig. 3H, black), and the second by significantly larger nuclear size (peak around 230 μm2) and expression of ChAT (Fig. 3H, red). Similar analysis from ChAT::Cre;Lhx7fl/LacZ animals showed a dramatic reduction in the fraction of β-gal+ neurons with large nuclei, a concomitant decrease in the representation of cholinergic neurons and an increase in β-gal+ neurons belonging to the Lhx6+ group (Fig. 3I). The average nuclear size of the Lhx6+ population was indistinguishable in control and conditional mutant animals, consistent with the idea that cholinergic precursor-derived Lhx6+ interneurons acquire the nuclear size of GABAergic interneurons (Fig. 3K). Interestingly, we observed a relatively small but statistically significant reduction in the nuclear size of ChAT+ neurons in the striatum of Lhx7fl/LacZ;ChAT::Cre mice (Fig. 3K), indicating that the residual CSIs of these animals may have subtle phenotypic alterations. We conclude that deletion of Lhx7 from committed cholinergic interneurons results in molecular and morphological reprogramming of these cells toward the GABAergic identity.
Reprogramming of CSIs Occurs Shortly After Lhx7 Deletion and Does Not Require Cell Division.
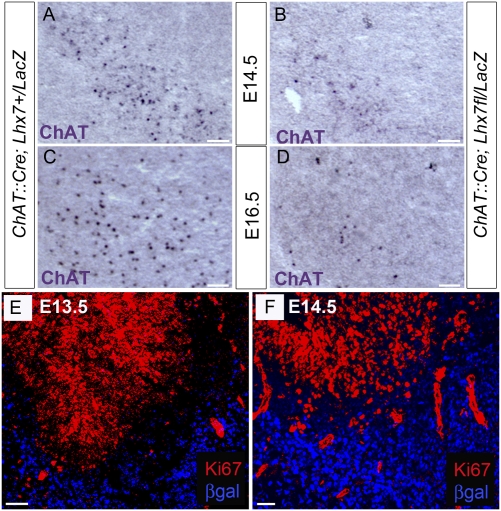
To define the stage at which Lhx7-depleted CSIs acquire GABAergic characteristics, we analyzed expression of ChAT mRNA in the basal forebrain of ChAT::Cre;Lhx7+/LacZ and ChAT::Cre;Lhx7fl/LacZ embryos. Reduction in the number of ChAT-expressing cells was evident already at E14.5 (compare Fig. 4 A and B) and was more pronounced by E16.5 (compare Fig. 4 C and D). Therefore, Lhx7 is required throughout the period of maturation of committed cholinergic precursors to actively maintain cholinergic identity, and deletion of the locus is followed shortly afterward by acquisition of GABAergic character.
Fig. 4.
Loss of cholinergic identity of Lhx7-depleted CSIs is not associated with reentry into the cell cycle. (A–D) ISH with a ChAT-specific riboprobe of ventral forebrain sections from E14.5 (A and B) and E16.5 (C and D) ChAT::Cre; Lhx7+/LacZ (A and C) and ChAT::Cre; Lhx7fl/LacZ (B and D) animals. (E and F) Ventral forebrain sections from E13.5 (E) and E14.5 (F) ChAT::Cre;Lhx7fl/LacZ embryos immunostained for β-gal and Ki67. No double-positive cells were detected at any of the stages analyzed. (Scale bars: 50 μm.)
To examine whether acquisition of GABAergic characteristics by Lhx7-depleted cholinergic interneurons requires reentry into the cell cycle, we immunostained forebrain sections of E13.5, E14.5, E15.5, and E16.5 ChAT::Cre;Lhx7fl/LacZ embryos for β-gal and Ki67 (Fig. 4 E and F). At no stage did we observe colocalization of the two markers, indicating that the Lhx7-expressing lineage remains postmitotic even after deletion of the gene and that the switch from a cholinergic to a GABAergic fate does not require cell division.
Lhx7 Maintains Cholinergic Identity by Regulating a Switch in Expression of Lhx6 and Isl1.
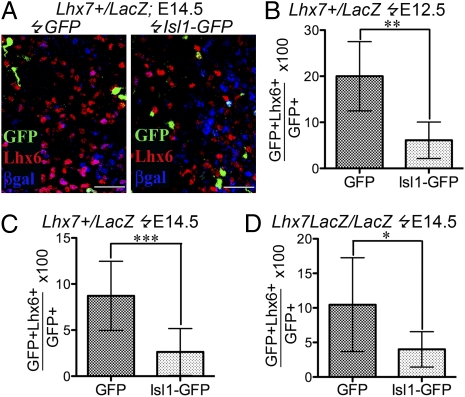
Given the critical roles of Lhx6 and Isl1 for GABAergic and cholinergic differentiation, respectively (10, 11), we posited that cross-repressive interactions between the two genes may control the segregation of the two striatal interneuron sublineages and that Lhx7 could maintain the cholinergic phenotype by sustaining the expression of Isl1 and, consequently, the down-regulation of Lhx6. To provide support for this hypothesis, we constitutively expressed Isl1 in the MGE of embryonic brain slices from wild-type embryos and analyzed its effect on Lhx6 expression. Vectors expressing either GFP (control) or Isl1-GFP were electroporated into the ventral region of forebrain slices from E12.5 (Fig. 5B) and E14.5 (Fig. 5C) Lhx7+/LacZ embryos. Forty-eight hours later, we quantified the percentage of GFP+ cells coexpressing Lhx6 (Fig. 5A and Materials and Methods). In control E12.5 slices, ∼20% of GFP+ cells coexpressed Lhx6. In contrast, expression of Isl1 led to a 70% reduction of the fraction of Lhx6+GFP+ cells (6.1%) (Fig. 5B). A similar effect was observed upon electroporation of Isl1 in E14.5 slices (reduction from 8.7% in GFP-electroporated to 2.6% in Isl1-electroporated slices; Fig. 5C). These studies suggest that Isl1 is capable of downregulating Lhx6 in the ventral forebrain of mouse embryos.
Fig. 5.
Isl1 down-regulates Lhx6 in the ventral forebrain. (A) Sections from ventral forebrain slices of E14.5 Lhx7+/LacZ embryos electroporated with GFP-only vector [left hemisphere (Left)] and Isl1-GFP vector [right hemisphere (Right)] were immunostained for GFP, Lhx6, and β-gal. (B and C) Quantification of the fraction of Lhx6-expressing GFP+ cells in ventral forebrain slices from E12.5 (B) or E14.5 (C) Lhx7+/LacZ embryos following electroporation of GFP-only or Isl1-GFP-expressing vectors. (D) Similar quantification following electroporation of E14.5 Lhx7LacZ/LacZ slices.
To further explore the role of Lhx7 in the regulation of Lhx6 expression by Isl1, we electroporated E14.5 forebrain slices from Lhx7-null (Lhx7LacZ/LacZ) animals with Isl1-GFP or GFP-only constructs (Fig. 5D). We observed that ∼10.5% of the GFP-electroporated cells in the MGE of Lhx7-null slices coexpressed Lhx6. In contrast, only 4% of the Isl1 electroporated cells coexpressed Lhx6, indicating that the ability of Isl1 to inhibit Lhx6 expression is largely independent of Lhx7. These findings provide support to the idea that cross repressive interactions between Isl1 and Lhx6 control sublineage segregation of striatal interneurons.
Discussion
All neuronal and glial subtypes in the CNS are derived from multipotential neuroepithelial progenitors that, under the influence of extracellular signals and intracellular factors, restrict their developmental potential and choose between alternative cell identities (2). “Terminal selector genes” encode transcription factors that coordinately regulate the expression of diverse molecular, morphological and functional characteristics that constitute the terminal phenotype of neurons (23). Lhx7 and Lhx6 belong to this class of genes because, respectively, they regulate multiple aspects of cholinergic and GABAergic interneuron differentiation in the mammalian forebrain (8, 10, 12–14). In vertebrates terminal selector genes have been studied mostly in the context of lineage restriction and forward neuronal differentiation, but their subsequent role in maintaining neuronal subtype identity is largely unknown. For example, constitutive germ-line deletion of Tlx1/3, Helt, Lbx1, and Sox6 resulted in the acquisition by the relevant cell lineages of alternative default cell fates (24–28). However, the potential plasticity of the neuronal identities promoted by these transcription factors has not been addressed. Using a conditional gene ablation strategy that deletes Lhx7 from committed forebrain cholinergic neurons, we demonstrate here that, in addition to its role in commitment and differentiation, Lhx7 activity is also critical for maintaining the lineage identity of CSIs. Several lines of evidence suggest that our experimental strategy bypasses the requirement of Lhx7 for the initial specification of striatal cholinergic precursors and allows us to explore its role in lineage-committed cholinergic neurons. First, Cre recombinase and its reporter YFP were induced exclusively in the nonproliferative mantle zone of the MGE, where the late cholinergic marker ChAT is expressed. Second, during embryogenesis, the Cre-dependent YFP reporter was specifically localized in cholinergic precursors marked by β-gal and Isl1 and was excluded from cells expressing Lhx6, which marks unspecified common precursors of striatal interneurons or specified GABAergic interneurons. Third, fate-mapping analysis of ChAT::Cre-expressing cells shows that in the adult striatum YFP was specifically expressed by β-gal+ cholinergic interneurons.
Striatal cholinergic interneurons are derived from Lhx6-expressing postmitotic precursors, which upon cholinergic commitment, induce Isl1, down-regulate Lhx6, and progressively acquire mature cholinergic markers and characteristic magnocellular morphology (8, 18, 29). In contrast, the lineally related GSIs maintain expression of Lhx6 and induce subtype-appropriate markers, such as Sst and Pv (Fig. 6). We demonstrate here that deletion of Lhx7 from committed cholinergic interneurons results not only in up-regulation of Lhx6 but also in expression of mature GABAergic-subtype markers and adoption of morphological features that are characteristic of GABAergic interneurons. Therefore, Lhx7-depleted cholinergic interneurons do not simply revert to an Lhx6+ common precursor stage but are capable of differentiating as Pv+ or Sst+ interneurons. Together, our studies argue that interneurons of the mammalian forebrain maintain remarkable phenotypic plasticity beyond cell cycle exit and commitment to terminal identity. Future studies could address the exciting possibility that GABAergic neurons originating from the Lhx7-depleted cholinergic lineage integrate into functional neuronal circuits of the basal forebrain.
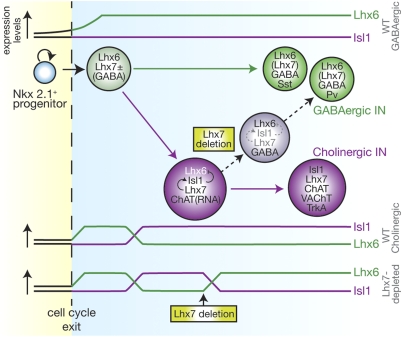
Fig. 6.
Model for Lhx7-dependent segregation and maintenance of alternative striatal interneuron subtypes. CSIs and GSIs are specified from common Nkx2.1+ MGE-derived progenitors, which, upon exit from the cell cycle, express Lhx6, GABA, and (at least some of them) Lhx7. A subset of these postmitotic precursors induce Isl1, which results in down-regulation of Lhx6, and differentiate into magnocellular cholinergic neurons expressing ChAT, VAChT, and TrkA (Cholinergic IN). Cells that do not induce Isl1 continue to express high levels of Lhx6 and differentiate into Pv+ or Sst+ GABAergic interneurons (GABAergic IN). Constitutive deletion of Lhx7 results in transient induction of Isl1 and the default differentiation of postmitotic precursors into mature GABAergic interneurons (8), suggesting that Lhx7 is required for maintaining Isl1 expression. The ability to adopt a GABAergic identity is not lost after cholinergic specification, because Lhx7 deletion in committed cholinergic neurons results in loss of Isl1, up-regulation of Lhx6, and acquisition of molecular and morphological features of GABAergic neurons. The changes in the relative levels of Isl1 and Lhx6 during normal differentiation of GSIs and CSIs are shown in the graphs labeled “WT GABAergic” and “WT Cholinergic,” respectively. The changes in the relative levels of Lhx6 and Isl1 as a result of Lhx7 deletion are depicted in the graph labeled “Lhx7-depleted.”
Our findings raise the possibility that Lhx7 activity is necessary to maintain the identity of fully differentiated CSIs during postnatal and adult stages. Interestingly, experiments involving withdrawal of trophic support in adult animals suggest that the cholinergic phenotype must be actively maintained throughout life. Thus, transection of septal cholinergic neuron projections to the hippocampus prevents target-derived nerve growth factor (NGF) signaling and leads to neuronal atrophy and loss of characteristic markers of cholinergic phenotype without cell death (30, 31). On the other hand, NGF infusion in the striatum of adult animals can protect or stimulate the cholinergic phenotype of interneurons (32–34). However, these studies have not determined whether upon withdrawal of neurotrophin signaling cholinergic neurons adopt an alternative neuronal identity. It would be interesting to investigate whether the effect of neurotrophins on cholinergic neurons is mediated by Lhx7 and Isl1 and determine whether variations in trophic signaling could trigger physiological changes in cholinergic identity in the mammalian forebrain.
What are the molecular mechanisms that ultimately control the choice and maintain the identity of striatal interneuron subtypes? The mutually exclusive expression of Isl1 and Lhx6 at all stages of striatal interneuron differentiation and the ability of Lhx6 to inhibit Lhx7/Isl1-induced cholinergic differentiation in chicken embryo spinal cord (8) suggest an antagonistic role of these transcription factors in striatal interneuron specification. Our present findings are consistent with this idea and further suggest that the antagonistic interaction between Isl1 and Lhx6 is also required to maintain the identity of CSIs beyond their initial specification (model in Fig. 6). By demonstrating here that overexpression of Isl1 in the ventral forebrain is sufficient to down-regulate Lhx6, we provide direct evidence for an inhibitory cross-regulatory interaction between these factors. However, what is the role of Lhx7 in such cross regulatory loop between Lhx6 and Isl1? We propose that Lhx7 activity is required for the sustained expression of Isl1 and the consequent down-regulation of Lhx6. This idea is consistent with the observation that in Lhx7-deficient embryos, Isl1 is only transiently induced in cholinergic interneurons precursors (8) and is supported by our present studies indicating that ablation of Lhx7 in cholinergic neurons results in down-regulation of Isl1, re-expression of Lhx6 and acquisition of a GABAergic phenotype. Based on these findings, we suggest that Lhx7 controls the generation of GABAergic and cholinergic interneuron lineages in the striatum by gating the cross inhibitory interaction between Lhx6 and Isl1 (model in Fig. 6). It should be noted that although the down-regulation of Lhx6 is crucial for cholinergic differentiation (8, 10, 12–14), it is not sufficient for instructing cholinergic identity in interneuron precursors because Lhx6-null animals do not show an increase in the number of cholinergic interneurons. Accordingly, our observation in this report that the residual cholinergic neurons in Lhx7 mutants retain Isl1 but show subtle changes in morphology and expression of mature cholinergic markers suggests that Lhx7 (perhaps in combination with other factors such as Isl1) has additional roles in cholinergic differentiation. Future studies could address in detail the molecular mechanisms by which Lhx7 contributes to the commitment and maintenance of the cholinergic lineage during maturation of the mammalian striatum and perhaps in adult animals.
The notion of a stable cellular identity has been challenged in different systems by studies using gene ablation or overexpression of defined sets of transcription factors to reprogram mature cellular phenotypes (35). Thus, deletion of Pax5 from B cells respecifies them into T cells, a process that requires cell division and the formation of an undifferentiated intermediate (36). In contrast, in vivo conversion of pancreatic exocrine cells into islet β cells (37) by the expression of key developmental factors does not involve reversal to a pluripotent stage or reentry into cell cycle. Similarly, the cholinergic to GABAergic switch we have observed in the Lhx7 conditional mutants is unlikely to require cell division as we found no reexpression of the cell cycle marker Ki67. Lhx7-depleted cholinergic neurons adopt a neuronal subtype identity (GABAergic) that represents an alternative postmitotic fate of the common precursor during physiologic specification. This is consistent with the idea that in the forebrain epigenetic possibilities are determined at the last cell division and subsequent changes in expression profile of postmitotic precursors (such the GABAergic vs. cholinergic choice by striatal interneuron precursors) are reversible without the need for extensive chromatin remodelling. It has been suggested that cross-antagonism between transcription factors that generate two cell fates could be a regulatory mechanism that allows physiological transitions between these fates (35). Maintaining the flexibility to switch between two types of striatal interneurons by Lhx7-controlled expression of Isl1 and Lhx6 could fine tune the necessary balance of striatal interneuron subtypes during development. By retaining phenotypic plasticity beyond the initial specification, forebrain neurons could contribute to the ability of the CNS to cope with dynamic changes imposed by developmental requirements, learning, or disease.
Materials and Methods
Animals.
Generation of the Lhx7fl allele is described in SI Materials and Methods. All animal studies were carried out under a United Kingdom Home Office Project License in a Home Office-designated facility.
In Situ Hybridization Histochemistry and Immunohistochemistry.
In situ hybridization (ISH) on sections from embryonic and adult brain were carried out as described in SI Materials and Methods (38). The ChAT riboprobe was generated from a 2.2-kb rat cDNA (gift from C. Goridis, Institut de Biologie de l'Ecole normale supérieure, Paris, France).
Quantifications.
For all quantifications, the result are expressed as means ± SD, and statistical analysis of significance was performed using a two-tailed Student t test (Prism 5; GraphPad). The level of significance is indicated by *P < 0.5, **P < 0.05, or ***P < 0.005 in the figures.
Measurement of Nuclear Size and Fluorescence Intensity.
To evaluate the nuclear size of β-gal+ cells and quantify the expression of β-gal, ChAT, and Lhx6 in these cells a series of confocal images were analyzed with ImageJ with user-designed macros to establish a signal/noise threshold, select particles, and create areas of local background for each particle. The size of the particles was measured on the β-gal channel, and intensity of fluorescence, subtracted by the local background, was calculated for each particle on the three channels corresponding to β-gal, ChAT, and Lhx6. The resulting data were analyzed using Igor Pro (Wavemetrics).
In Vitro Focal Electroporation and Organotypic Slice Culture.
Organotypic slice culture and focal electroporation were performed as previously described (39). For details, see SI Materials and Methods.
Supplementary Material
Acknowledgments
This work was funded by the Medical Research Council.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1109251109/-/DCSupplemental.
References
- 1.Flames N, et al. Delineation of multiple subpallial progenitor domains by the combinatorial expression of transcriptional codes. J Neurosci. 2007;27:9682–9695. doi: 10.1523/JNEUROSCI.2750-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Guillemot F. Cell fate specification in the mammalian telencephalon. Prog Neurobiol. 2007;83:37–52. doi: 10.1016/j.pneurobio.2007.02.009. [DOI] [PubMed] [Google Scholar]
- 3.Shirasaki R, Pfaff SL. Transcriptional codes and the control of neuronal identity. Annu Rev Neurosci. 2002;25:251–281. doi: 10.1146/annurev.neuro.25.112701.142916. [DOI] [PubMed] [Google Scholar]
- 4.Zhou FM, Wilson CJ, Dani JA. Cholinergic interneuron characteristics and nicotinic properties in the striatum. J Neurobiol. 2002;53:590–605. doi: 10.1002/neu.10150. [DOI] [PubMed] [Google Scholar]
- 5.Bolam JP, Hanley JJ, Booth PA, Bevan MD. Synaptic organisation of the basal ganglia. J Anat. 2000;196:527–542. doi: 10.1046/j.1469-7580.2000.19640527.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Marin O, Anderson SA, Rubenstein JL. Origin and molecular specification of striatal interneurons. J Neurosci. 2000;20:6063–6076. doi: 10.1523/JNEUROSCI.20-16-06063.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nóbrega-Pereira S, et al. Postmitotic Nkx2-1 controls the migration of telencephalic interneurons by direct repression of guidance receptors. Neuron. 2008;59:733–745. doi: 10.1016/j.neuron.2008.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fragkouli A, van Wijk NV, Lopes R, Kessaris N, Pachnis V. LIM homeodomain transcription factor-dependent specification of bipotential MGE progenitors into cholinergic and GABAergic striatal interneurons. Development. 2009;136:3841–3851. doi: 10.1242/dev.038083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Flames N, Marín O. Developmental mechanisms underlying the generation of cortical interneuron diversity. Neuron. 2005;46:377–381. doi: 10.1016/j.neuron.2005.04.020. [DOI] [PubMed] [Google Scholar]
- 10.Liodis P, et al. Lhx6 activity is required for the normal migration and specification of cortical interneuron subtypes. J Neurosci. 2007;27:3078–3089. doi: 10.1523/JNEUROSCI.3055-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Elshatory Y, Gan L. The LIM-homeobox gene Islet-1 is required for the development of restricted forebrain cholinergic neurons. J Neurosci. 2008;28:3291–3297. doi: 10.1523/JNEUROSCI.5730-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fragkouli A, et al. Loss of forebrain cholinergic neurons and impairment in spatial learning and memory in LHX7-deficient mice. Eur J Neurosci. 2005;21:2923–2938. doi: 10.1111/j.1460-9568.2005.04141.x. [DOI] [PubMed] [Google Scholar]
- 13.Mori T, et al. The LIM homeobox gene, L3/Lhx8, is necessary for proper development of basal forebrain cholinergic neurons. Eur J Neurosci. 2004;19:3129–3141. doi: 10.1111/j.0953-816X.2004.03415.x. [DOI] [PubMed] [Google Scholar]
- 14.Zhao Y, et al. The LIM-homeobox gene Lhx8 is required for the development of many cholinergic neurons in the mouse forebrain. Proc Natl Acad Sci USA. 2003;100:9005–9010. doi: 10.1073/pnas.1537759100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lewandoski M, Meyers EN, Martin GR. Analysis of Fgf8 gene function in vertebrate development. Cold Spring Harb Symp Quant Biol. 1997;62:159–168. [PubMed] [Google Scholar]
- 16.Gong S, et al. Targeting Cre recombinase to specific neuron populations with bacterial artificial chromosome constructs. J Neurosci. 2007;27:9817–9823. doi: 10.1523/JNEUROSCI.2707-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Srinivas S, et al. Cre reporter strains produced by targeted insertion of EYFP and ECFP into the ROSA26 locus. BMC Dev Biol. 2001;1:4. doi: 10.1186/1471-213X-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Phelps PE, Brady DR, Vaughn JE. The generation and differentiation of cholinergic neurons in rat caudate-putamen. Brain Res Dev Brain Res. 1989;46:47–60. doi: 10.1016/0165-3806(89)90142-9. [DOI] [PubMed] [Google Scholar]
- 19.Arvidsson U, Riedl M, Elde R, Meister B. Vesicular acetylcholine transporter (VAChT) protein: A novel and unique marker for cholinergic neurons in the central and peripheral nervous systems. J Comp Neurol. 1997;378:454–467. [PubMed] [Google Scholar]
- 20.Sobreviela T, et al. TrkA-immunoreactive profiles in the central nervous system: Colocalization with neurons containing p75 nerve growth factor receptor, choline acetyltransferase, and serotonin. J Comp Neurol. 1994;350:587–611. doi: 10.1002/cne.903500407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kawaguchi Y. Neostriatal cell subtypes and their functional roles. Neurosci Res. 1997;27:1–8. doi: 10.1016/s0168-0102(96)01134-0. [DOI] [PubMed] [Google Scholar]
- 22.Matamales M, et al. Striatal medium-sized spiny neurons: Identification by nuclear staining and study of neuronal subpopulations in BAC transgenic mice. PLoS ONE. 2009;4:e4770. doi: 10.1371/journal.pone.0004770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hobert O. Regulatory logic of neuronal diversity: Terminal selector genes and selector motifs. Proc Natl Acad Sci USA. 2008;105:20067–20071. doi: 10.1073/pnas.0806070105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cheng L, et al. Tlx3 and Tlx1 are post-mitotic selector genes determining glutamatergic over GABAergic cell fates. Nat Neurosci. 2004;7:510–517. doi: 10.1038/nn1221. [DOI] [PubMed] [Google Scholar]
- 25.Nakatani T, Minaki Y, Kumai M, Ono Y. Helt determines GABAergic over glutamatergic neuronal fate by repressing Ngn genes in the developing mesencephalon. Development. 2007;134:2783–2793. doi: 10.1242/dev.02870. [DOI] [PubMed] [Google Scholar]
- 26.Sieber MA, et al. Lbx1 acts as a selector gene in the fate determination of somatosensory and viscerosensory relay neurons in the hindbrain. J Neurosci. 2007;27:4902–4909. doi: 10.1523/JNEUROSCI.0717-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Azim E, Jabaudon D, Fame RM, Macklis JD. SOX6 controls dorsal progenitor identity and interneuron diversity during neocortical development. Nat Neurosci. 2009;12:1238–1247. doi: 10.1038/nn.2387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Batista-Brito R, et al. The cell-intrinsic requirement of Sox6 for cortical interneuron development. Neuron. 2009;63:466–481. doi: 10.1016/j.neuron.2009.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bolam JP, Wainer BH, Smith AD. Characterization of cholinergic neurons in the rat neostriatum. A combination of choline acetyltransferase immunocytochemistry, Golgi-impregnation and electron microscopy. Neuroscience. 1984;12:711–718. doi: 10.1016/0306-4522(84)90165-9. [DOI] [PubMed] [Google Scholar]
- 30.Haas CA, et al. Axotomy-induced c-JUN expression in young medial septal neurons is regulated by nerve growth factor. Neuroscience. 1998;87:831–844. doi: 10.1016/s0306-4522(98)00188-2. [DOI] [PubMed] [Google Scholar]
- 31.Lazo OM, Mauna JC, Pissani CA, Inestrosa NC, Bronfman FC. Axotomy-induced neurotrophic withdrawal causes the loss of phenotypic differentiation and downregulation of NGF signalling, but not death of septal cholinergic neurons. Mol Neurodegener. 2010;5:5. doi: 10.1186/1750-1326-5-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Altar CA, et al. Recovery of cholinergic phenotype in the injured rat neostriatum: Roles for endogenous and exogenous nerve growth factor. J Neurochem. 1992;59:2167–2177. doi: 10.1111/j.1471-4159.1992.tb10108.x. [DOI] [PubMed] [Google Scholar]
- 33.Gage FH, et al. NGF receptor reexpression and NGF-mediated cholinergic neuronal hypertrophy in the damaged adult neostriatum. Neuron. 1989;2:1177–1184. doi: 10.1016/0896-6273(89)90184-0. [DOI] [PubMed] [Google Scholar]
- 34.Förander P, Söderström S, Humpel C, Strömberg I. Chronic infusion of nerve growth factor into rat striatum increases cholinergic markers and inhibits striatal neuronal discharge rate. Eur J Neurosci. 1996;8:1822–1832. doi: 10.1111/j.1460-9568.1996.tb01326.x. [DOI] [PubMed] [Google Scholar]
- 35.Graf T, Enver T. Forcing cells to change lineages. Nature. 2009;462:587–594. doi: 10.1038/nature08533. [DOI] [PubMed] [Google Scholar]
- 36.Cobaleda C, Jochum W, Busslinger M. Conversion of mature B cells into T cells by dedifferentiation to uncommitted progenitors. Nature. 2007;449:473–477. doi: 10.1038/nature06159. [DOI] [PubMed] [Google Scholar]
- 37.Zhou Q, Brown J, Kanarek A, Rajagopal J, Melton DA. In vivo reprogramming of adult pancreatic exocrine cells to beta-cells. Nature. 2008;455:627–632. doi: 10.1038/nature07314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schaeren-Wiemers N, Gerfin-Moser A. A single protocol to detect transcripts of various types and expression levels in neural tissue and cultured cells: In situ hybridization using digoxigenin-labelled cRNA probes. Histochemistry. 1993;100:431–440. doi: 10.1007/BF00267823. [DOI] [PubMed] [Google Scholar]
- 39.Heng JI, et al. Neurogenin 2 controls cortical neuron migration through regulation of Rnd2. Nature. 2008;455:114–118. doi: 10.1038/nature07198. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.