Abstract
MicroRNAs (miRNAs) are small RNAs that play a regulatory role in numerous and diverse eukaryotic cellular processes. Virus-encoded miRNAs have garnered much interest, although the functions of most remain to be deciphered. To date, readily detectable, evolutionarily conserved natural miRNAs have only been identified from viruses with DNA genomes. Combined with the fact that most miRNAs are generated from endonucleolytic cleavage of longer transcripts, this finding has led to a common conception that naturally occurring RNA viruses will not encode miRNAs to avoid unproductive cleavage of their genomes or mRNAs. Here we demonstrate that the bovine leukemia virus (BLV), a retrovirus with an RNA genome, encodes a conserved cluster of miRNAs that are transcribed by RNA polymerase III (pol III). Thus, the BLV miRNAs avoid the conundrum of genome/mRNA cleavage because only the subgenomic pol III transcripts are efficiently processed into miRNAs. BLV infection is strongly associated with B-cell tumors in cattle. Because most cells in BLV-associated tumors express little viral mRNAs or proteins, exactly how BLV contributes to tumorigenesis has remained a decades-long unsolved mystery. One BLV miRNA, BLV-miR-B4, shares partial sequence identity and shared common targets with the host miRNA, miR-29. As miR-29 overexpression is associated with B-cell neoplasms that resemble BLV-associated tumors, our findings suggest a possible mechanism contributing to BLV-induced tumorigenesis.
Keywords: mir-29a, mir-943, chronic lymphocytic leukemia, noncoding RNA
MicroRNAs (miRNAs) are small ∼22-nucleotide regulatory RNAs encoded by most Eukaryotes and some viruses (reviewed in refs. 1–3). To date, almost all viral miRNAs described derive from viruses with DNA genomes. There are several reports that one RNA virus (HIV, a retrovirus) might encode miRNAs, but the relevance of these reports remains controversial (4). Independent laboratories have failed to verify their existence (5, 6). Furthermore, the putative HIV miRNAs suffer from one or more of the following characteristics: they are not evolutionarily conserved among different strains, they are expressed at extremely low levels, and they lack any well-defined function (4, 5, 7). Thus, a widely accepted example of any naturally-occurring RNA virus-encoded miRNA is lacking, though alternative viewpoints do exist (8).
Infection with bovine leukemia virus (BLV) is associated with naturally arising B-cell tumors in cattle, and experimentally induced tumors in sheep (reviewed in ref. 9). A complete understanding of how BLV induces tumorigenesis remains enigmatic, particularly as most of these tumor cells are positive for an integrated BLV proviral genome but lack abundant expression of BLV-encoded RNA polymerase II (pol II) transcripts or proteins (9–11). Using an algorithm designed to identify RNA polymerase III (pol III)-encoded miRNAs as our starting point, we identify a cluster of five miRNAs expressed from the BLV genome. Unlike most host miRNAs, these miRNAs are not processed by the endonuclease Drosha, and this allows the viral pol II genomic and mRNA transcripts to escape cleavage. One of these miRNAs, BLV-miR-B4, shares perfect identity with the target-specifying portion (seed region, nucleotides 2–8) of the host miRNA, miR-29. We demonstrate that both BLV-miR-B4 and bovine miR-29a target two transcripts previously associated with miR-29–induced B-cell tumorigenesis in mice (12, 13). The implications of these results with regards to BLV-induced tumorigenesis are discussed.
Results
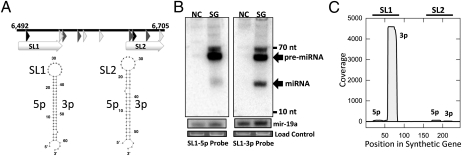
The vast majority of known miRNAs are transcribed by pol II. The one clear exception to this comes from a virus—the murine herpesvirus 68 (MHV68)—which encodes numerous miRNAs derived from pol III-transcribed tRNA-like precursor structures (6). We reasoned it is likely that other viruses might also use pol III to generate miRNAs, and therefore developed an algorithm to predict such miRNAs (Fig. S1). The algorithm takes into account common features of the RNA pol III class II promoter and terminator sequences (14) combined with structural-based ab initio precursor miRNA (pre-miRNA) prediction. We designed the promoter-prediction portion of the algorithm based on the sequences of tRNA, Alu elements, 7SL, vaRNAs, and MHV68 tRNA-like miRNAs. As numerous families of DNA viruses have already been extensively probed for miRNA expression (3), and retroviruses are the only RNA viruses with a DNA intermediate in their replication cycle, we chose to focus our initial efforts on the Retroviridae. We applied the algorithm to all retroviral reference genome sequences in the National Center for Biotechnology Information (NCBI) genome collection. Interestingly, we observed predicted pol III miRNA genes for a number of Spumaviruses and only a single member of the Orthoretrovirinae, BLV, a delta retrovirus (Table S1). We synthesized predicted genes for candidate miRNAs from macaque simian foamy virus and BLV (Fig. 1A and Fig. S2A), including the putative pol III promoter and terminator, into a vector lacking a mammalian promoter. We then transfected HEK293T cells, harvested RNA, and screened for miRNA expression via Northern blot analysis. This analysis failed to identify any miRNAs from the macaque simian foamy virus (Fig. S2B); however, we readily detected a strong signal deriving from a discrete band for the BLV miRNA candidate (Fig. 1B). To better map this candidate miRNA, we conducted next-generation sequencing on small RNAs from our transfection experiments. This analysis again failed to identify any miRNAs from the macaque simian foamy virus (Fig. S2C), but produced reads consistent with miRNA production from the BLV candidate that mapped to the predicted stem-loop structures (Fig. 1C). Thus, these results provided one line of evidence that BLV encodes a miRNA.
Fig. 1.
A combined computational and synthetic approach identifies a candidate BLV-encoded miRNA. (A) Schematic of the organization of the predicted and synthesized BLV miRNA gene region. The predicted A Box-like, B Box-like, and terminator-like sequences are represented by black, gray, and white triangles respectively. Predicted stem-loop structures (SL1 and SL2) are indicated with white arrows, and Mfold RNA secondary-structure predictions for the predicted miRNA stem-loop sequences are presented below. The genomic coordinates refer to BLV reference sequence NC_001414. (B) Northern blot analysis of 293T cells transfected with the synthetic BLV miRNA gene construct (SG lanes) or negative control empty vector (NC lanes). As a load control and to provide a rough comparison of levels to a host miRNA, blots were stripped and reprobed for host mir-19a. An additional load control is ethidium bromide-stained low molecular weight RNA. (C) Small RNA profiling of 293T cells transfected with the synthetic BLV miRNA gene construct plotted as previously described (44). The vertical axis depicts small RNA read coverage observed; the horizontal axis shows the relative position within the synthetic gene sequence. Stem-loops SL1 and SL2 correspond to miRNAs BLV miR-B4 and B5 in subsequent analysis.
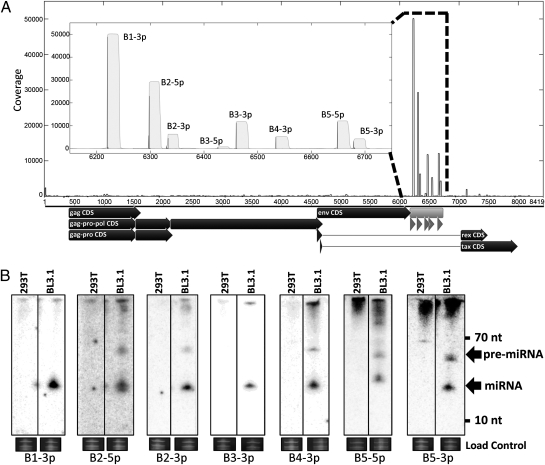
To investigate whether the predicted miRNA is expressed in the context of BLV-infected cells, we conducted high-throughput small RNA profiling on the BL3.1 cell line, a bovine B-cell line that is persistently infected and shedding low levels of BLV (15). We not only confirmed the expression of our computationally predicted candidate miRNAs (sequences corresponding to SL1 and SL2 are now referred to as miRs-B4 and B5 for consistency with viral miRNA nomenclature), but also identified abundant sequence reads that mapped additional putative BLV miRNAs from five total hairpin precursor structures (Fig. 2A, Fig. S3, and Table S2). Importantly, some of the BLV candidate miRNAs are more abundant than most host miRNAs, supporting their likely biological relevance (Table S3). All of these candidate miRNAs map to a small portion of the BLV genome that is not known to code for protein expression.
Fig. 2.
Small RNA profiling and Northern blots identify BLV-encoded miRNAs expressed in BL3.1 cells. (A) Small RNAs sequenced from BL3.1 cells were mapped to the BLV genome. (Inset) Detail of the newly identified miRNA-encoding region with black impulses representing start counts and gray filled area representing coverage. Below the x axis of the main graph, annotated coding sequences are displayed as black arrows. The miRNA-encoding region is indicated in light gray and individual miRNAs are represented by dark gray arrows. B4 and B5 correspond to the synthetic sequences SL1 and SL2 of Fig. 1, respectively. (B) Northern blot analysis confirms candidate miRNAs. RNA harvested from the persistently infected BL3.1 cell line or the negative control 293T cells was probed with oligonucleotides specific to the abundantly mapped sequences. The load control is ethidium bromide-stained low molecular weight RNA.
Next, we assayed two different cell lines persistently infected with BLV for expression of these candidate miRNAs. Total RNA was harvested from BL3.1 and the fetal lamb kidney (FLK) cell line chronically infected with BLV (FLK-BLV) and Northern blot analysis was conducted. This analysis confirmed the existence of the abundantly mapped small RNAs for all five candidate miRNAs in the BL3.1 cell line; additionally, we were able to detect the star strands (less abundant processed derivatives) from two of the five precursor structures (Fig. 2B). Furthermore, we were able to detect the majority of these small RNAs in the FLK-BLV cell line (Fig. S4). The fact that we clearly map and detect small RNA derivatives from predicted hairpin structures that resemble known pre-miRNAs is consistent with these candidates being bona fide miRNAs.
We examined the evolutionary conservation of the viral-derived miRNAs by aligning all NCBI-deposited full-length BLV genomic sequences along with the sequence of the miRNA-encoding region that we cloned from the BL3.1 cell line. Interestingly, the identified miRNAs appear to be well-conserved across isolates and the seed sequences of the predominant arms for four of the five miRNAs are identical across all examined isolates (Fig. S5A). We hypothesized that all of the BLV miRNAs are derived via pol III transcription and reasoned that the putative pol III promoter and terminator elements should be conserved across the viral isolates. Therefore, we searched for promoter-like and terminator-like sequences across the alignment (Fig. S5B). Indeed, the majority of these elements are conserved. Notably, predicted and conserved A Box sequences are present in every pre-miRNA stem loop and at least partially overlap the 5p arm of each. The fact that both the miRNA sequences and their associated predicted promoter and terminator sequences are well-conserved across all deposited BLV isolates is consistent with one or more conserved functional roles for the newly identified BLV miRNAs.
After examination of the miRNA sequences, we noted that the original computationally predicted miRNA, BLV-miR-B4-3p, shares nucleotide identity with the host miRNA family miR-29 in nucleotide positions 1–8 (Fig. S5). Importantly, this region encompasses the so-called “seed” region of the miRNA, which is of great importance in specifying complementary direct miRNA:mRNA target associations (16). Although the majority of virus-encoded miRNAs do not share any sequence identity with host miRNAs (3), there are a few important examples that do (17–19). Because our findings suggest that BLV-miR-B4 functions as a likely analog to the host miRNA miR-29a, we focused our efforts on better understanding this miRNA.
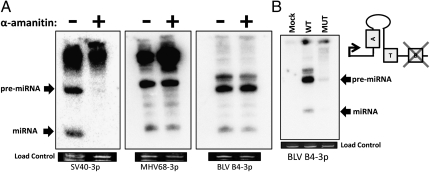
We first explored our initial predictions of whether BLV-miR-B4 is transcribed by pol III. We used a previously published strategy (20): treatment of HEK293T cells with the appropriate concentration of α-amanitin is sufficient to block pol II-mediated transcription but not pol III. We transfected cells with the plasmid expressing BLV-miR-B4 from its endogenous promoter in the presence or absence of α-amanitin. As shown in Fig. 3A, α-amanitin inhibits both the pre-miRNA and mature miRNA of the pol II- transcribed SV40 miRNA. As expected, expression of the control pol III-derived MHV68 miR-M1-7 miRNA was resistant to α-amanitin treatment. Importantly, the BLV transcripts were unaffected by α-amanitin treatment, thus supporting our initial pol III prediction (Fig. 3A). As an independent test, we generated a version of the BLV-miR-B4 expression vector, whereby three nucleotides in the predicted B Box were mutated. As expected, the B Box mutant showed severe inhibition when transfected samples were assayed by Northern blot analysis (Fig. 3B). Thus, we conclude that as predicted from our algorithm, BLV-miR-B4 is transcribed by pol III.
Fig. 3.
BLV miRNA B4 is transcribed by RNA Pol III. (A) Northern blot analysis of SV40-S1, MHV68-M1-7, and BLV-B4 miRNA expression vectors transfected into HEK293T cells with or without treatment of the RNA pol II inhibitor α-amanitin. The load control is ethidium bromide-stained low molecular weight RNA. (B) Northern blot analysis of RNA from 293T cells transfected with the wild-type or B Box mutant BLV-B4 miRNA expression vectors. The load control is ethidium bromide-stained low molecular weight RNA. Schematic indicates organization of the BLV miR-B4 promoter region and location of the B Box promoter element mutation.
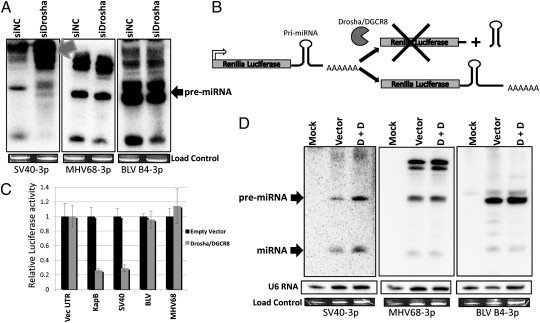
Drosha is the endonuclease responsible for excising most pre-miRNA structures from pol II transcripts (21). Previously, Bogerd et al. demonstrated that biogenesis of the MHV68 pre-miRNAs is independent of Drosha processing (20). Because, like MHV68, the BLV primary miRNA transcripts are transcribed by pol III, we hypothesized that biogenesis of the BLV miRNAs would be independent of Drosha. We cotransfected HEK293T cells with previously characterized siRNAs against Drosha (22) and vectors expressing either the BLV miR-B4 miRNA or Drosha-dependent or -independent control miRNAs (Fig. 4A). Knockdown of Drosha resulted in a marked decrease in the SV40 pre-miRNA with a concurrent increase in slower mobility bands that represent the primary uncleaved primary miRNA (pri-miRNA) transcripts. These results were expected because the SV40 pri-miRNA is transcribed by pol II and processed by Drosha. In contrast, knocking down Drosha had little effect on the control MHV68 or the BLV pri-miRNA and pre-miRNA transcripts (Fig. 4A). We note that cotransfection of the Drosha siRNA also results in a slight decrease in the final Dicer product miRNA expressed from both the MHV68 and BLV vectors (Fig. 4A). Despite this finding, unlike the SV40 construct, we never observed any increase in the pri-miRNA transcripts for MHV68 or BLV, thus ruling out a role for Drosha in processing these transcripts. We speculate that this phenomenon could result from unequal transfection or a previously unknown mechanism of cross-talk between Drosha and Dicer levels. Irrespective of this, the SV40 transcripts clearly behave differently than BLV transcripts in terms of Drosha sensitivity, and these results are internally controlled because the assay measures inverse trends in the pri-miRNA and pre-miRNAs from the same RNA samples. However, transfection of either MHV68-M1-7 (a control Dicer-dependent miRNA) or BLV-miR-B4 expression vectors in a Dicer hypomorphic cell line results in an increase of the ratio of pre-miRNA:miRNA, which is consistent with BLV miRNA biogenesis being Dicer-dependent (Fig. S6B). Thus, we conclude that the biogenesis of BLV-miR-B4 is Dicer-dependent but independent of Drosha processing.
Fig. 4.
BLV encoded miRNAs are not processed by Drosha. (A) Northern blot analysis of the SV40-S1, MHV68-M1-7, and BLV-B4 miRNA expression vectors cotransfected into HEK293T cells with either Drosha siRNA (siDrosha) or a negative control (siNC) siRNA. Gray arrow indicates accumulated unprocessed miRNA precursors. The load control is ethidium bromide-stained low molecular weight RNA. (B) Schematic of luciferase-based Drosha cleavage assay. Drosha-cleavable stem-loop structures located in the 3′UTR of the luciferase reporter result in endonucleolytic cleavage of pre-mRNA transcripts and reduced luciferase activity. (C) Renilla luciferase constructs with control pri-miRNAs or the entire BLV miRNA cluster-encoding region cloned into the 3′ UTR region were either cotransfected with empty vector or Drosha and DGCR8-expression vectors. The control Renilla reporter containing the vector-derived 3′UTR is labeled “Vec UTR.” The relative luciferase activity ratio of Renilla to firefly is graphed and normalized to the Renilla luciferase vector 3′ UTR. Error bars indicate SD of three replicates. (D) Northern blot analysis of RNA from HEK293 cells cotransfected with SV40-S1, MHV68-M1-7, or BLV-B4 miRNA expression vectors and empty vector (Vector) or Drosha and DGCR8 expression vectors (D + D). The load control is ethidium bromide-stained low molecular weight RNA. Northern blot analysis of U6 snRNA is shown as an additional load control.
It is a common notion that RNA virus genomes will not contain pre-miRNA structures to avoid endonuclease-mediated cleavage of the genome, antigenome, and mRNAs. We hypothesized that BLV uses subgenomic pol III transcripts to circumvent the need for endonucleolytic cleavage of its pol II transcripts. Although we demonstrated that the BLV miRNAs are derived from pol III transcripts via a Drosha-independent mechanism, it was still formally possible that the BLV pre-miRNA hairpin structures are susceptible to Drosha cleavage in the context of pol II transcripts. To test the susceptibility of pol II transcripts containing the BLV pre-miRNAs, we took advantage of an assay that we had previously developed (23). Drosha and its binding partner DGCR8 (together referred to as “Microprocessor”) are cotransfected with luciferase constructs containing putative Drosha substrates within the 3′ UTR of the derived transcripts (Fig. 4B). We cloned the complete BLV miRNA-encoding region (positions 6133–6739) containing all five hairpins into the 3′ UTR region of a Renilla luciferase expression vector (Fig. S6A). This context is analogous to some viral mRNA transcripts (e.g., Gag-Pol or Env) that contain the miRNA cluster within their 3′ UTR. If Drosha is able to cleave the pri-miRNAs in the pol II context, then luciferase levels should decrease. If however, the BLV pre-miRNA hairpins are resistant to Drosha cleavage, then luciferase activity should be unaffected. The luciferase reporter vector was cotransfected with either control parental vector or Drosha/DGCR8 expression vectors. As shown in Fig. 4C, when Drosha/DGCR8 is cotransfected with the SV40 and KapB Renilla control reporters [both give rise to well-characterized Drosha-dependent viral pre-miRNAs (23, 24)], the luciferase activity significantly decreases. However, no decrease was observed for the reporter constructs containing the control Drosha-independent MHV68-M1-7 pre-miRNA. Importantly, the luciferase levels from the reporter containing the entire BLV miRNA cassette did not decrease upon cotransfection of Drosha/DGCR8 (Fig. 4C). This result demonstrates that Drosha does not cleave RNA polymerase II transcripts containing the BLV pre-miRNAs. To further confirm these results via an independent assay, we conducted Northern blot analysis on cells transfected with the various luciferase reporters in the presence or absence of exogenous Microprocessor (Fig. 4D). Increased Drosha activity led to readily detectable increased pre-miRNA and miRNA bands from the SV40 pre-miRNA, consistent with the SV40 pre-miRNA being a product of Drosha cleavage. However, no increase was observed for the control MHV68 pre-miRNA or the BLV pre-miRNA (Fig. 4D). Combined, these results show that the BLV miRNAs are not efficiently processed from pol II transcripts.
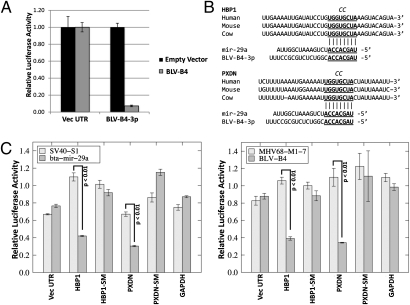
To determine whether BLV-miR-B4 is biologically functional, RNA-induced silencing complex (RISC) activity was measured via luciferase assay. To allow sensitive detection, two perfectly complementary target sites were cloned into the Renilla luciferase vector 3′ UTR. When the luciferase vector with the BLV-miR-B4-3p miRNA target sites was cotransfected with an empty expression vector, the Renilla luciferase activity did not significantly change (Fig. 5A). However, when the BLV miRNA expression vector was cotransfected with the Renilla target construct, the Renilla luciferase activity was significantly reduced relative to the firefly luciferase (Fig. 5A). These results indicate that the BLV-miR-B4 miRNA is active within the RISC complex.
Fig. 5.
BLV-B4-3p is a functional analog of host miRNA mir-29. (A) RISC reporter assay for BLV miR-B4. Cells were cotransfected with empty miRNA expression vector (empty vector) or the BLV miRNA expression vector (BLV-B4) and reporters containing the vector-derived (Vec UTR) or BLV miRNA complementary regions (BLV-B4-3p) in the 3′ UTR region. The luciferase activity of Renilla-normalized firefly activity is shown. Error bars indicate SD of three independent replicates. (B) Alignment of human and cow predicted mir-29 binding sites in the 3′UTRs of tumor suppressor genes HBP1 and PXDN based on previously identified bona fide murine miR-29a targets (12, 13). The conserved seed complementary region is underlined and in bold. The two nucleotide substitution in the seed mutant (SM) reporters is indicated in italics at the top of each alignment. (C) miR-29a and BLV miR-B4 share common mRNA targets. (Left) Bovine mir-29a or SV40 miRNA (SV40-S1) pol II expression vectors were cotransfected into HEK293T cells with the indicated Renilla reporter and control firefly luciferase vectors. (Right) BLV miR-B4 or MHV68 miR-M1-7 pol III expression vectors were cotransfected into HEK293T cells with the indicated Renilla and control firefly luciferase vectors. The abbreviation “SM” indicates a 2-nt swap seed mutant 3′UTR. For all treatments, relative luciferase levels are normalized to transfection with empty miRNA expression vector. Error bars indicate SD of three replicates and P values were calculated using Student’s t-test.
The fact that BLV-miR-B4 shares a seed sequence with miR-29a strongly suggests that these miRNAs will have some overlapping targets. Independent studies have demonstrated that overexpression of miR-29a in lymphocytes is sufficient to drive hyperproliferative disorders (12, 13). Interestingly, Pekarsky et al. (25) demonstrate that miR-29a is overexpressed in a subset of the mild form of human chronic lymphocytic leukemia (CLL) and artificial overexpression of miR-29a induces a CLL-like phenotype in mice (13, 25). As it has been well documented that naturally and experimentally arising BLV-associated tumors also resemble the indolent form of CLL (9, 26, 27), we wanted to test if BLV-miR-B4 would regulate host transcripts previously implicated as miR-29a targets in murine tumor-inducing studies (12, 13). As targets, we chose HMG-box transcription factor 1 (HBP1) from Han et al. (12) and peroxidasin homolog (PXDN) from Santanam et al. (13), both of which are tumor suppressors associated with down-regulation in B-cell tumors deriving from exogenous expression of miR-29a. We first determined if the miR-29a binding sites from these genes are conserved with the bovine genes. Indeed, the seed complement regions in the 3′ UTRs are perfectly conserved between the human, mouse, and bovine genomes (Fig. 5B). Next, we generated chimeric luciferase reporters containing the entire bovine 3′ UTR from these genes. We cotransfected miRNA expression vectors with either of these reporters or controls containing the entire GAPDH 3′ UTR sequence or vector 3′ UTR (both lack seed complements to miR-29) or HBP1 or PXDN 3′UTRs with two nucleotide mutations in the predicted seed complementary regions. As expected, expression of the bovine miR-29a negatively regulated both reporters but not the negative controls (vector 3′ UTR, GAPDH 3′ UTR, nor either of the seed mutant 3′ UTRs) (Fig. 5C). Expression of irrelevant pol II- or pol III-derived control miRNAs (from SV40 and MHV68, respectively) had little effect on any of these reporters (Fig. 5C). Thus, we demonstrate that the transcript regulation first uncovered for murine miR-29a is evolutionarily conserved with the bovine transcripts. Most importantly, BLV-miR-B4 also specifically negatively regulates the bovine HBP1 and PXDN reporters (Fig. 5C). These data demonstrate that BLV-miR-B4 shares some mRNA targets in common with miR-29a and therefore can serve as a functional analog.
Discussion
There are now over 250 known virus-encoded miRNAs and the vast majority are derived from pol II transcripts encoded by DNA viruses (3). A particular advantage for viral utilization of miRNAs is their presumed lack of immunogenicity. Although an in-depth functional understanding is lacking for most viral miRNAs, the field is progressing rapidly, with ongoing discovery of both host and viral targets. Although limited studies exist, a preliminary model suggests that DNA viruses will use miRNAs for varied activities, including: regulation of latent/persistent infection, evasion of the innate and adaptive immune responses, and promotion of cell viability (3).
No naturally occurring RNA viral miRNAs are widely accepted [albeit several laboratory-generated RNA viruses do support miRNA or miRNA-like expression (28–30)]. Because of the potential problem of having endonuclease-sensitive sites within the genome, it has been speculated that natural RNA virus genomes do not contain miRNAs (8, 31). Because retroviruses have a DNA replication intermediate in their replication cycle and maintain long-term persistent infections, miRNAs could offer advantages to this class of RNA viruses similar to the roles that miRNAs play in DNA viruses. The key challenge for retroviruses would be to either tolerate some level of cis cleavage within the genome during pre-miRNA processing [as is the case for some laboratory retroviral expression vectors (32)], or to evolve genomic regions that are somehow resistant to cleavage in the context of the full genome. Here, we provide evidence that BLV overcomes this obstacle by encoding for pre-miRNA structures that are only competent to be processed into miRNAs when generated from subgenomic pol III-derived transcripts.
We designed an algorithm that identifies RNA pol III-transcribed miRNAs using both predicted gene structure and RNA secondary structural motifs. Using this bioinformatics approach, we identified BLV-miR-B4. It is worth noting that although several of the BLV miRNAs described here were missed by our initial in silico screen, all possess the hallmarks of pol III promoter elements, including the A and B Boxes and the poly T terminator. Our algorithm was originally designed to recognize pol III, class II promoters with a linear ordering of A Box, B Box, and terminator sequence. However, not all of the BLV miRNAs are predicted to follow this ordering as some predicted terminator sequences precede predicted B Box sequences (Fig. S5A). Future endeavors will include modifications of our predictive algorithms to accommodate these observations, as well as searching for promoter elements common to the other classes of pol III promoters. Therefore, we speculate it is likely that other retroviruses exist that use pol III promoters but were missed by our algorithm. Thus, future miRNA discovery efforts should not be limited to the DNA viruses.
What is the advantage to BLV of using pol III promoters? We hypothesize that a major advantage for BLV is the ability to encode subgenomic small transcripts that can only be processed into miRNAs in this context. Because the longer pol II genomic and mRNA transcripts are resistant to endonuclease processing, this mechanism provides an effective means for generating high levels of the derivative miRNAs without negatively affecting genome or mRNA integrity. Thus, BLV miRNA biogenesis resembles some man-made retroviral shRNA vectors that use pol III transcripts (33, 34). These vectors are effective at generating biologically active levels of small RNAs and are readily grown to high titers in a laboratory setting. Additionally, using pol III might offer some advantages in the context of lymphocytes. Although it is generally accepted that pol III promoters are less amenable to fine-tune regulation provided by pol II transcription factors, it is notable that the only other known pol III miRNAs derive from MHV68. Like BLV, MHV68 forms long-term persistent infections in B lymphocytes. Thus, the possibility that similar B-cell signaling events regulate both MHV68 and BLV microRNA production awaits future study.
BLV is associated with B-cell tumors in naturally infected cattle and experimentally infected sheep. The tumors are somewhat atypical, being of B-cell origin, but are positive for the CD5 marker (9, 35, 36). It has been noted that cells from BLV-associated tumors in cattle do not have increased proliferation but rather have reduced turnover of CD5+ B-cells, thus contributing to the tumorigenic state (9). This phenotype bears a striking resemblance to the indolent form of CLL, which is also a CD5+ B-cell tumor (27). In fact, it has even been proposed that BLV-induced tumors in sheep and cattle could serve as a valuable model to better understand human CLL (26). A mechanistic understanding of how BLV promotes tumorigenesis remains enigmatic. Essentially all tumor cells are positive for the viral genome; however, very little, if any, viral mRNAs and proteins are detected in most cells (9–11). Our results demonstrating that one of the BLV miRNAs is a functional analog of miR-29a could provide a contributing mechanism for BLV-mediated tumorigenesis. In a screen of human CLL tumors, miR-29a was overexpressed (25). Importantly, in mouse models, exogenous expression of miR-29a is sufficient to induce different classes of B-cell tumors (12, 13). When miR-29a is exogenously driven in mice via the Emu promoter, the tumors derived strongly resemble CLL (13). Taken together, our data support the hypothesis that BLV-miR-B4 provides a growth and survival advantage to aid in maintaining infection in vivo. In addition, BLV-miR-B2-5p also shares seed identity with host miRNA, miR-943. Although little is known regarding the function of miR-943, it is notable that miR-943 is induced when the tumor suppressor p53 activity is blocked, and is found at elevated levels in the stem population of primary human mammalian epithelial cells (37). This finding implies a possible oncogenic role for miR-943. Finally, because necessarily all BLV-derived miRNAs share partial complementarity (and in some cases this includes perfect seed complementarity) to the BLV pol II transcripts, it is possible that the pol III-derived BLV miRNAs play a role in binding to and enforcing silencing of BLV pol II transcripts, thus contributing to the low BLV protein expression observed in BLV-associated tumors (9–11). Future studies will directly address the role of all five BLV miRNAs in in vivo oncogenesis and the autoregulation of viral protein expression.
In summary, we demonstrate that an RNA virus expresses abundant, evolutionarily conserved miRNAs, including at least one that functions as an analog to a host oncogenic miRNA. These findings open up a role for noncoding RNAs in retroviral-associated tumorigenesis, and suggest the possibility that other retroviruses exist that use noncoding RNAs in their infectious cycles.
Materials and Methods
Small RNA Northern blot analysis was performed as described (38). Specific probes are listed in Table S3. Small RNA libraries for SOLiD sequencing were prepared as described previously (39). RNA secondary structure predictions were generated using Mfold Web server (40, 41) or the RNAfold Web server (42) as indicated. Conservation of HBP1 and PXDN mir-29 target sites was assessed using Targetscan (43). RNA polymerase III dependence assay was performed essentially as described (20). Drosha knockdown was performed as described (44).
Full detailed methods are provided in SI Materials and Methods.
Supplementary Material
Acknowledgments
The authors thank Drs. Louis Mansky (University of Minnesota), Scott Tibbetts (University of Florida), and Gisela Heidecker (National Cancer Institute, Frederick, MD) for reagents; Dr. Stephen Trent for use of equipment; and Drs. L. Mansky and Jackie Dudley and the members of the C.S.S. laboratory for comments regarding this manuscript. This work was supported by Grant R01AI077746 from the National Institutes of Health, Grant RP110098 from the Cancer Prevention and Research Institute of Texas, and a University of Texas at Austin Institute for Cellular and Molecular Biology fellowship.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
See Commentary on page 2695.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1116107109/-/DCSupplemental.
References
- 1.Boss IW, Renne R. Viral miRNAs: Tools for immune evasion. Curr Opin Microbiol. 2010;13:540–545. doi: 10.1016/j.mib.2010.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cullen BR. Viruses and microRNAs: RISCy interactions with serious consequences. Genes Dev. 2011;25:1881–1894. doi: 10.1101/gad.17352611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Grundhoff A, Sullivan CS. Virus-encoded microRNAs. Virology. 2011;411:325–343. doi: 10.1016/j.virol.2011.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chiang K, Rice AP. Mini ways to stop a virus: MicroRNAs and HIV-1 replication. Future Virol. 2011;6:209–221. doi: 10.2217/fvl.10.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lin J, Cullen BR. Analysis of the interaction of primate retroviruses with the human RNA interference machinery. J Virol. 2007;81:12218–12226. doi: 10.1128/JVI.01390-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pfeffer S, et al. Identification of microRNAs of the herpesvirus family. Nat Methods. 2005;2:269–276. doi: 10.1038/nmeth746. [DOI] [PubMed] [Google Scholar]
- 7.Sun G, Rossi JJ. MicroRNAs and their potential involvement in HIV infection. Trends Pharmacol Sci. 2011;32:675–681. doi: 10.1016/j.tips.2011.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Houzet L, Jeang KT. MicroRNAs and human retroviruses. Biochim Biophys Acta. 2011;1809:686–693. doi: 10.1016/j.bbagrm.2011.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gillet N, et al. Mechanisms of leukemogenesis induced by bovine leukemia virus: Prospects for novel anti-retroviral therapies in human. Retrovirology. 2007;4:18. doi: 10.1186/1742-4690-4-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gaynor EM, Mirsky ML, Lewin HA. Use of flow cytometry and RT-PCR for detecting gene expression by single cells. Biotechniques. 1996;21:286–291. doi: 10.2144/96212rr02. [DOI] [PubMed] [Google Scholar]
- 11.Kettmann R, Cleuter Y, Gregoire D, Burny A. Role of the 3′ long open reading frame region of bovine leukemia virus in the maintenance of cell transformation. J Virol. 1985;54:899–901. doi: 10.1128/jvi.54.3.899-901.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Han YC, et al. microRNA-29a induces aberrant self-renewal capacity in hematopoietic progenitors, biased myeloid development, and acute myeloid leukemia. J Exp Med. 2010;207:475–489. doi: 10.1084/jem.20090831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Santanam U, et al. Chronic lymphocytic leukemia modeled in mouse by targeted miR-29 expression. Proc Natl Acad Sci USA. 2010;107:12210–12215. doi: 10.1073/pnas.1007186107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Orioli A, Pascali C, Pagano A, Teichmann M, Dieci G. RNA polymerase III transcription control elements: Themes and variations. Gene. 2011;493:185–194. doi: 10.1016/j.gene.2011.06.015. [DOI] [PubMed] [Google Scholar]
- 15.Harms JS, Splitter GA. Impairment of MHC class I transcription in a mutant bovine B cell line. Immunogenetics. 1992;35:1–8. doi: 10.1007/BF00216620. [DOI] [PubMed] [Google Scholar]
- 16.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gottwein E, et al. A viral microRNA functions as an orthologue of cellular miR-155. Nature. 2007;450:1096–1099. doi: 10.1038/nature05992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Skalsky RL, et al. Kaposi's sarcoma-associated herpesvirus encodes an ortholog of miR-155. J Virol. 2007;81:12836–12845. doi: 10.1128/JVI.01804-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhao Y, et al. Critical role of the virus-encoded microRNA-155 ortholog in the induction of Marek's disease lymphomas. PLoS Pathog. 2011;7:e1001305. doi: 10.1371/journal.ppat.1001305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bogerd HP, et al. A mammalian herpesvirus uses noncanonical expression and processing mechanisms to generate viral MicroRNAs. Mol Cell. 2010;37:135–142. doi: 10.1016/j.molcel.2009.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 22.Han J, et al. Posttranscriptional crossregulation between Drosha and DGCR8. Cell. 2009;136:75–84. doi: 10.1016/j.cell.2008.10.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lin YT, Sullivan CS. Expanding the role of Drosha to the regulation of viral gene expression. Proc Natl Acad Sci USA. 2011;108:11229–11234. doi: 10.1073/pnas.1105799108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sullivan CS, Grundhoff AT, Tevethia S, Pipas JM, Ganem D. SV40-encoded microRNAs regulate viral gene expression and reduce susceptibility to cytotoxic T cells. Nature. 2005;435:682–686. doi: 10.1038/nature03576. [DOI] [PubMed] [Google Scholar]
- 25.Pekarsky Y, et al. Tcl1 expression in chronic lymphocytic leukemia is regulated by miR-29 and miR-181. Cancer Res. 2006;66:11590–11593. doi: 10.1158/0008-5472.CAN-06-3613. [DOI] [PubMed] [Google Scholar]
- 26.Debacq C, et al. Reduced cell turnover in bovine leukemia virus-infected, persistently lymphocytotic cattle. J Virol. 2003;77:13073–13083. doi: 10.1128/JVI.77.24.13073-13083.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Defoiche J, et al. Reduction of B cell turnover in chronic lymphocytic leukaemia. Br J Haematol. 2008;143:240–247. doi: 10.1111/j.1365-2141.2008.07348.x. [DOI] [PubMed] [Google Scholar]
- 28.Rouha H, Thurner C, Mandl CW. Functional microRNA generated from a cytoplasmic RNA virus. Nucleic Acids Res. 2010;38:8328–8337. doi: 10.1093/nar/gkq681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shapiro JS, Varble A, Pham AM, Tenoever BR. Noncanonical cytoplasmic processing of viral microRNAs. RNA. 2010;16:2068–2074. doi: 10.1261/rna.2303610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Varble A, et al. Engineered RNA viral synthesis of microRNAs. Proc Natl Acad Sci USA. 2010;107:11519–11524. doi: 10.1073/pnas.1003115107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cullen BR. Five questions about viruses and microRNAs. PLoS Pathog. 2010;6:e1000787. doi: 10.1371/journal.ppat.1000787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Scott GK, et al. Coordinate suppression of ERBB2 and ERBB3 by enforced expression of micro-RNA miR-125a or miR-125b. J Biol Chem. 2007;282:1479–1486. doi: 10.1074/jbc.M609383200. [DOI] [PubMed] [Google Scholar]
- 33.Brummelkamp TR, Bernards R, Agami R. Stable suppression of tumorigenicity by virus-mediated RNA interference. Cancer Cell. 2002;2:243–247. doi: 10.1016/s1535-6108(02)00122-8. [DOI] [PubMed] [Google Scholar]
- 34.Rubinson DA, et al. A lentivirus-based system to functionally silence genes in primary mammalian cells, stem cells and transgenic mice by RNA interference. Nat Genet. 2003;33:401–406. doi: 10.1038/ng1117. [DOI] [PubMed] [Google Scholar]
- 35.Letesson JJ, Mager A, Mammerickx M, Burny A, Depelchin A. B cells from bovine leukemia virus- (BLV) infected sheep with hematological disorders express the CD5 T cell marker. Leukemia. 1990;4:377–379. [PubMed] [Google Scholar]
- 36.Matheise JP, Delcommenne M, Mager A, Didembourg CH, Letesson JJ. CD5+ B cells from bovine leukemia virus infected cows are activated cycling cells responsive to interleukin 2. Leukemia. 1992;6:304–309. [PubMed] [Google Scholar]
- 37.Chang CJ, et al. p53 regulates epithelial-mesenchymal transition and stem cell properties through modulating miRNAs. Nat Cell Biol. 2011;13:317–323. doi: 10.1038/ncb2173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McClure LV, Lin YT, Sullivan CS. Detection of viral microRNAs by Northern blot analysis. Methods Mol Biol. 2011;721:153–171. doi: 10.1007/978-1-61779-037-9_9. [DOI] [PubMed] [Google Scholar]
- 39.Chen CJ, Kincaid RP, Seo GJ, Bennett MD, Sullivan CS. Insights into Polyomaviridae microRNA function derived from study of the bandicoot papillomatosis carcinomatosis viruses. J Virol. 2011;85:4487–4500. doi: 10.1128/JVI.02557-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mathews DH, Sabina J, Zuker M, Turner DH. Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J Mol Biol. 1999;288:911–940. doi: 10.1006/jmbi.1999.2700. [DOI] [PubMed] [Google Scholar]
- 41.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hofacker IL, Fontana W, Stadler PF, Bonhoeffer M, Schuster P. Fast folding and comparison of RNA secondary structures. Monatsh Chem. 1994;125:167–188. [Google Scholar]
- 43.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 44.Lin YT, et al. Small RNA profiling reveals antisense transcription throughout the KSHV genome and novel small RNAs. RNA. 2010;16:1540–1558. doi: 10.1261/rna.1967910. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.