Abstract
Schizophrenia is characterized by affective, cognitive, neuromorphological, and molecular abnormalities that may have a neurodevelopmental origin. MicroRNAs (miRNAs) are small noncoding RNA sequences critical to neurodevelopment and adult neuronal processes by coordinating the activity of multiple genes within biological networks. We examined the expression of 854 miRNAs in prefrontal cortical tissue from 100 control, schizophrenic, and bipolar subjects. The cyclic AMP-responsive element binding- and NMDA-regulated microRNA miR-132 was significantly down-regulated in both the schizophrenic discovery cohort and a second, independent set of schizophrenic subjects. Analysis of miR-132 target gene expression in schizophrenia gene-expression microarrays identified 26 genes up-regulated in schizophrenia subjects. Consistent with NMDA-mediated hypofunction observed in schizophrenic subjects, administration of an NMDA antagonist to adult mice results in miR-132 down-regulation in the prefrontal cortex. Furthermore, miR-132 expression in the murine prefrontal cortex exhibits significant developmental regulation and overlaps with critical neurodevelopmental processes during adolescence. Adult prefrontal expression of miR-132 can be down-regulated by pharmacologic inhibition of NMDA receptor signaling during a brief postnatal period. Several key genes, including DNMT3A, GATA2, and DPYSL3, are regulated by miR-132 and exhibited altered expression either during normal neurodevelopment or in tissue from adult schizophrenic subjects. Our data suggest miR-132 dysregulation and subsequent abnormal expression of miR-132 target genes contribute to the neurodevelopmental and neuromorphological pathologies present in schizophrenia.
Keywords: psychiatric disorders, epigenetics, methylation, gene networks
Schizophrenia is a chronic neuropsychiatric disorder characterized by affective and cognitive symptoms. Neuromorphological defects include decreased prefrontal and hippocampal volume, neocortical pyramidal neuron size, and dendritic spine density. Risk for schizophrenia is associated with neurodevelopmental disturbances during the prenatal and early postnatal period, and by additional pathogenesis associated with brain maturation during adolescence and early adulthood (1–3). Many candidate schizophrenia risk genes regulate neurodevelopmental functions (4, 5). In normal brain development, ∼50% of synapses produced are lost during adolescence by a process that is activity-dependent and requires coordinated activity of NMDA and AMPA receptors (6). Thus, dysregulation of synaptic pruning through NMDA/AMPA receptor-mediated mechanisms has the potential to disrupt cognitive development. Hypoactive NMDA signaling has been implicated in schizophrenia (7, 8). Notably, NMDA receptor (NMDAR) antagonists induce psychosis and cognitive impairment in normal human subjects (9), and produce both positive and negative schizophrenia-like behaviors in animal models (10, 11).
Widespread alterations in neurodevelopment and neurotransmitter signaling may arise from multiple causes. MicroRNAs (miRNAs) have the ability to coregulate multiple biological pathways and have recently been implicated in schizophrenia (12, 13). MiRNAs are small noncoding RNAs that regulate the stability and translation of up to 60% of protein-coding mRNAs (14). Dysregulation of a single miRNA can be sufficient to alter the gene-expression profile and developmental trajectory of cells (15, 16). MiRNAs contribute to regulation of many mechanisms in the nervous system, including regulation of neuronal migration and differentiation, synaptic plasticity, and adult neurogenesis (17, 18).
We hypothesized that dysregulation of miRNAs contributes to the pathogenesis of schizophrenia (19). To evaluate this possibility, we examined the expression of over 800 miRNAs in dorsolateral prefrontal cortex (DL-PFC) from a large sample of control, schizophrenic, and bipolar subjects. We found disease-specific dysregulation of several miRNAs, including miR-132, a cyclic AMP-responsive element binding (CREB)-regulated miRNA associated with NMDAR signaling (20). We confirmed dysregulation of miR-132 and several of its mRNA targets by quantitative real-time PCR (qPCR) in a second set of human subjects and controls. Comparison of miR-132 gene targets with relevant microarray datasets identified significant overlap with genes developmentally down-regulated in the PFC during adolescence in mice. We confirmed inverse expression changes in miR-132 and several targets during normal postnatal development of the PFC in mice, and further demonstrated that treatment with the NMDAR antagonist MK-801 during the early postnatal period results in adult down-regulation of miR-132 expression in the PFC. Finally, we show that miR-132 regulates schizophrenia- and development-associated genes including DNMT3A, GATA2, and DPYSL3. These results implicate miR-132 and its mRNA targets in the etiology and pathology of schizophrenia.
Results
MicroRNA Microarray Analysis of Control, Schizophrenia, and Bipolar DL-PFC Samples.
Microarray analysis of RNA samples derived from BA46 of control (CON, n = 34), bipolar (BPD, n = 31), and schizophrenic (SCZ, n = 35) subjects obtained from the Stanley Medical Research Institute (SMRI) Array Collection (Table S1) showed that 483 of 854 assayed miRNAs were expressed above threshold in DL-PFC tissue. ANCOVA analysis using diagnosis (CON, SCZ, BPD) and sex as main effects, and brain pH and age as covariates, followed by false-discovery rate (FDR) correction, identified only the major and minor products of one precursor hairpin, miR-132 and miR-132*, to be significantly altered in schizophrenia (Table 1 and Dataset S1). Samples from subjects with bipolar disorder exhibited significant dysregulation of 10 miRNAs.
Table 1.
MiRNAs with FDR < 0.05 in the SMRI samples
| MiRNA | SCZ FDR | BPD FDR | SCZ fold-change | BPD fold-change |
| hsa-miR-132 | 0.016 | 0.399 | 0.789 | 0.885 |
| hsa-miR-132* | 0.022 | 0.213 | 0.802 | 0.863 |
| hsa-let-7b | 0.074 | 0.194 | 1.172 | 1.147 |
| hsa-miR-383 | 0.742 | 0.015 | 1.077 | 1.369 |
| hsa-miR-32* | 0.611 | 0.016 | 1.169 | 1.604 |
| hsa-miR-490-5p | 0.825 | 0.022 | 1.076 | 1.421 |
| hsa-miR-196b | 0.707 | 0.024 | 1.174 | 1.707 |
| hsa-miR-513-5p | 0.691 | 0.046 | 1.223 | 1.847 |
| hsa-miR-876-3p | 0.747 | 0.046 | 1.126 | 1.505 |
| hsa-miR-449b | 0.837 | 0.046 | 1.118 | 1.623 |
| hsa-miR-297 | 0.921 | 0.046 | 1.050 | 1.390 |
| hsa-miR-188-5p | 0.981 | 0.046 | 1.022 | 1.403 |
| hsa-miR-187 | 0.824 | 0.046 | 1.078 | 1.361 |
| hsa-miR-15b | 0.684 | 0.051 | 1.070 | 1.207 |
Boldface indicates that the miRNA had a P < 0.05 in one dataset (e.g., miR-132 in SCZ) but not the other dataset (e.g., miR-132 in BPD).
There was no overlap between the significantly dysregulated miRNAs identified in schizophrenia and bipolar samples. However, evaluation of all probes with an uncorrected P value < 0.05 identified 10 miRNAs dysregulated in both disorders, more than twice what would be expected by chance (Dataset S2A). All 10 probes were altered in the same direction across disorders, suggesting some shared molecular pathology. These data support recent genome-wide association study studies suggesting schizophrenia and bipolar disorder share overlapping risk alleles (21); however, these studies have not identified the biological nature of the shared risk loci. To address this discrepancy, we identified miRNAs with the strongest association between disorders by ranking diagnosis-associated miRNA expression by fold-change and uncorrected P value. This analysis identified six dysregulated miRNAs (miR-132, miR-320, miR-135, miR-105, miR-449, and miR-17-5p) common to schizophrenia and bipolar disorder: 754 protein-coding genes are targeted by two or more of these miRNAs, more than would be expected by chance (P < 0.0001), suggesting that these codysregulated miRNAs target functionally overlapping molecular networks (SI Materials and Methods and Dataset S2B).
Association of miRNA Microarray Data with Patient Demographics.
In addition to the ANCOVA test described above, a linear regression model determined whether significant alterations in miRNA expression were associated with demographic details other than diagnosis (Dataset S3). Both miR-132 and miR-132* were correlated with age. However, all three diagnostic groups were age-matched, precluding age as a factor in diagnosis-associated changes in miR-132. Three miRNAs, including miR-132, -132*, and -105, correlated with the level of lifetime antipsychotic units. Additional miRNAs were significantly associated with psychosis (miR-32*, -181d, -330-5p, -206, and -133a) or suicide (miR-206), but none were affected by sex or brain pH, the two main demographic details that differed significantly between diagnoses. In contrast to previous reports (22), we found no disease-associated differences in either the total amount of small RNA or the percent of total small RNA represented by the ∼22-nt mature miRNA population (Fig. S1A). Differences in patient demographics, brain region, RNA extraction techniques, or method of determining the percentage of small RNAs may contribute to the differences with previous findings.
Antipsychotic Drug Treatment Does Not Affect miR-132 Expression in Vivo.
To evaluate the potential for antipsychotic drug exposure to contribute to down-regulation of miR-132 in schizophrenic subjects, rats were treated with therapeutically relevant doses of haloperidol (0.25 mg/kg), risperidone (5 mg/kg), or vehicle (1% acetic acid) for 21 d using osmotic minipumps to ensure constant central D2-receptor occupancy at exposures comparable to that measured in humans (23). RNA from the frontal cortex of treated animals showed no change in expression of miR-132 compared with vehicle-treated rats, suggesting that miR-132 down-regulation is a core feature of schizophrenia, rather than a by-product of drug treatment (Fig. S1C). These findings are also in agreement with previously published studies (12).
qPCR Validation of Microarray Results.
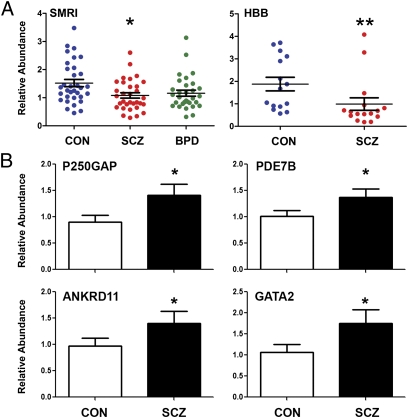
RNA from the SMRI samples and a second separate set of samples from control and schizophrenic subjects, obtained from the Harvard Brain Tissue Resource Center (HBB) (Table S2), was used to validate the miRNA microarray results by qPCR. qPCR analysis showed a significant decrease in miR-132 expression in both the SCZ and BPD samples from the SMRI collection, and in the schizophrenic patient samples from the HBB collection (Fig. 1A). Five additional miRNAs with nonsignificant but suggestive trends in the microarray were selected from the list in Dataset S2A for qPCR analysis (using the HBB samples); however, no significant changes were observed among these miRNAs, supporting the use of the strict FDR < P = 0.05 significance cutoff for microarray analysis.
Fig. 1.
Microarray analysis indicated that miR-132 expression was down-regulated in the PFC from schizophrenia subjects. (A) qPCR validation of miR-132 expression in the PFC from control (CON), schizophrenic (SCZ), and bipolar disorder (BPD) subjects from two separate populations (SMRI, HBB). Lines show mean + SEM, *P < 0.05, **P < 0.01. (B) Significant up-regulation of the expression of several putative miR-132 targets was validated by qPCR in the HBB samples. Group size for each sample set is presented in Tables S1 and S2; error bars = SEM.
Alteration of miR-132 Protein-Coding Targets in SCZ and BPD.
We identified 263 putative targets of miR-132 using the TargetScan prediction database. Ingenuity Pathway Analysis (IPA) software identified biological pathways associated with predicted miR-132 targets. Twenty-six pathways were significantly overrepresented, five of which were specific to, or enriched in, the nervous system (Dataset S4). The protein kinase A signaling pathway contained the largest number of significantly up-regulated miR-132 targets; also of note were pathways associated with synaptic long-term potentiation and long-term depression, neuronal CREB signaling, and DNA methylation. Importantly these pathways support both the glutamatergic and dopaminergic hypothesis of schizophrenia, and include several pathways, such as Reelin signaling, that have been definitively associated with schizophrenia (24).
The list of miR-132 targets was compared with publicly available gene-expression microarray data from the SMRI “A” and “C” array collections of DL-PFC tissue from control and schizophrenic subjects (www.stanleyresearch.org). Because miR-132 is down-regulated in the SCZ samples, only putative miR-132 targets significantly up-regulated (FDR-corrected P value < 0.05) in the SMRI dataset were considered. Twenty-six genes from the SCZ samples met these criteria (Dataset S5). A subset of seven genes, representing key biological pathways identified by IPA, were evaluated by qPCR. The known miR-132 target p250GAP (25), and four other genes, GATA2, FKBP2, ANKRD11, and PDE7B, were significantly up-regulated in samples from schizophrenic subjects (Fig. 1B and Fig. S1B). These results suggest that disease-associated miR-132 down-regulation may have a significant impact on the regulation of gene expression in the PFC of schizophrenic subjects.
Functional Role for miR-132 in Neurodevelopment.
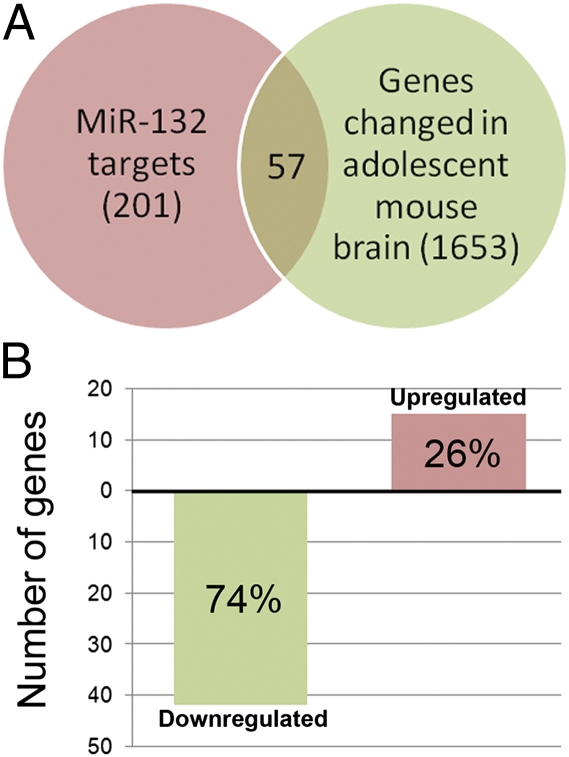
To identify potential functional roles for miR-132 in the brain, we queried the NextBio database of microarray studies (www.nextbio.com). From 99 datasets derived from cortical tissue, the study most enriched for predicted miR-132 targets was a microarray analysis of the changes in developing mouse PFC between postnatal weeks 2, 3, 4, 5, and 10 (26). The most significant overlap with miR-132 targets occurred between weeks 2 and 4 of development, equivalent to adolescence in humans and characterized by intensive NMDAR-dependent synaptic pruning. Of 201 miR-132 gene targets in the Nextbio database, 57 were significantly altered between 2 and 4 wk in the mouse PFC. The majority of transcripts were down-regulated, consistent with developmental up-regulation of miR-132 (P < 0.001) (Fig. 2 and Dataset S6). In addition to regulating schizophrenia-related biological pathways in adult brain, miR-132 may play a critical role in the neurodevelopmental changes that occur during adolescence and early adulthood.
Fig. 2.
The NextBio database was searched for microarray datasets that showed overlap with predicted miR-132 targets. (A) The most significant overlap was with genes that showed altered expression between postnatal weeks 2 and 4 in mouse PFC. (B) Of 201 predicted miR-132 targets, 57 showed a significant change in expression between postnatal weeks 2 and 4, with almost 75% of the targets down-regulated.
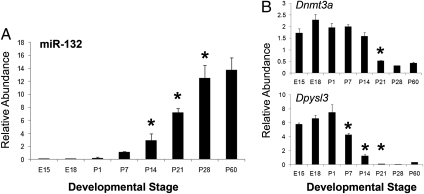
To test the hypothesis that miR-132 is developmentally regulated in the PFC, we evaluated expression of miR-132 in mRNA isolated from mouse PFC between embryonic day 12 (E12) through 2 mo of age (postnatal day 60, P60). MiR-132 expression was absent or low through P7, began to rise at P14, and increased almost fourfold between P14 and P28 (Fig. 3A). These results are similar to miR-132 expression patterns in the developing hippocampus (27). Three putative miR-132 target mRNAs identified as down-regulated in the developmental expression profile (26), Dnmt3a, Dypsl3, and Gmfb, were simultaneously examined. Two of the three putative targets, Dnmt3a and Dpyls3, showed developmental expression patterns with a significant negative correlation to miR-132 (Pearson's r = −0.94 and −0.84, respectively) (Fig. 3B). In contrast to the data from Semeralul et al. (26), Gmfb showed no age-related changes in expression. The full-length isoform of Dnmt3a, which predominates in the brain after birth, contains multiple 7-nt miR-132 binding sites in its 3′ UTR, and Dpysl3 contains a 8-nt miR-132 binding site in its 3′ UTR.
Fig. 3.
The expression of miR-132 (A) and two of its predicted protein-coding targets, Dnmt3a and Dpysl3 (B) in the PFC was measured from E12 through P60. MiR-132 expression was significantly up-regulated between postnatal weeks 2 and 4, but expression of Dnmt3a and Dpyls3 followed an opposite expression pattern. MiR-132 expression at P14 was significantly different from expression at P7, P21, P28, and P60 (P ≤ 0.01), Dnmt3a expression from P21 and later was significantly different from P14 and earlier (P ≤ 0.01), and Dpysl3 expression at P7 and P14 was significantly different from all other time points, and between those two time points (P ≤ 0.01). Error bars = SEM; *P < 0.05.
Validation of DNMT3A, GATA2, and DPYSL3 as miR-132 Targets.
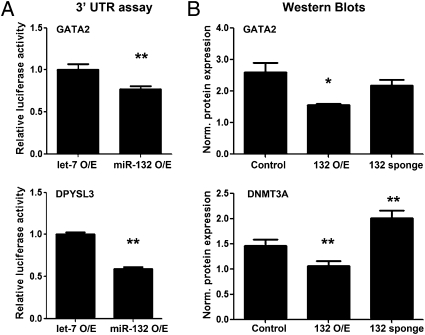
Consistent with qPCR analysis, overexpression of miR-132 in HEK293 cells significantly decreased reporter levels for the ∼1-kb 3′ UTR fragments of both Gata2 and Dpysl3 (Fig. 4A), although a Dnmt3a reporter was not affected. Evaluation of protein levels for native, full-length mRNA transcripts by Western blot showed overexpression of miR-132 in HEK-293 cells significantly decreased DNMT3A and GATA2 protein levels, and knockdown of miR-132 function resulted in significant increased DNMT3A but not GATA2 compared with cells transduced with a control vector (Fig. 4B and Fig. S1E) (28).
Fig. 4.
MiR-132 regulation of DNMT3A, GATA2, and DPYSL3. (A) Overexpression of miR-132 significantly down-regulated 3′ UTR luciferase reporter levels compared with the control vector. (B) Overexpression of miR-132 significantly reduced DNMT3A and GATA2 protein levels, and sponge-mediated knockdown of miR-132 function resulted in up-regulation of DNMT3A, but had no effect on GATA2. β-Actin, which was not affected by treatment, was used to normalize GATA2 and DNMT3A levels. O/E, overexpression; *P < 0.05, **P < 0.001.
The effect of miR-132 on DNMT3A protein levels but not reporter construct activity may be because of indirect regulation of DNMT3A by miR-132 through a yet-unidentified pathway, perhaps CREB-related, or because of technical constraints that prevented the entire DNMT3A 3′ UTR from being cloned into the reporter vector. The human DMNT3A 3′ UTR contains five possible seed matches for miR-132 in 6 kbps, three of which show evolutionary conservation. Factors outside the 3′ UTR seed region can also affect miRNA binding (29), and it is possible that the region of the DNMT3A UTR used in the reporter construct lacked nonseed sequences necessary for miR-132 binding.
In Vivo Regulation of miR-132 Expression by NMDA Signaling.
The proposed role of NMDAR-mediated hypofunction in schizophrenia (7) and previous evidence of NMDA regulation of miR-132 (20) led us to examine effects of the NMDAR antagonist MK-801 on miR-132 expression in adult and juvenile mice. Adult mice were injected once daily for 5 d with either saline or one of three doses of MK-801 (0.1, 0.3, or 0.6 mg/kg delivered intraperitoneally). MK-801 treatment resulted in a dose-dependent reduction in miR-132 expression compared with saline, indicating that 5-d inhibition of NMDAR signaling results in reduced expression of miR-132 in the prefrontal cortex (Fig. S2A).
To address consequences of neurodevelopmental disruption of NMDAR signaling on miR-132 expression, mouse pups were treated daily with a low dose (0.1 mg/kg) of MK-801 from P4 to P17. This treatment protocol has been shown to produce permanent behavioral and neurochemical changes that remain measureable throughout adulthood (10). Mice treated with MK-801 during postnatal development exhibited significant down-regulation of miR-132 in the PFC in adulthood (Fig. S2B). Thus, disruption of NMDAR signaling during postnatal development was associated with reduced adulthood expression of miR-132, as observed in schizophrenia.
To extend comparisons between mice treated with MK-801 during development and gene-expression patterns in schizophrenic patients, we tested mRNA expression of two miR-132 targets up-regulated in schizophrenia (p250GAP and Gata2) and two developmentally regulated miR-132 targets (Dnmt3a and Dpysl3) within the developmental MK-801 mouse model (Fig. S2 C and D). There was no effect of drug treatment on expression of any of the four genes at P14. In adult mice, p250GAP was significantly up-regulated in the PFC of developmentally treated MK-801 mice compared with controls, similar to the effect seen in the human HBB samples. There was a trend for Gata2 up-regulation in the MK-801 group, but it did not reach significance. Interestingly, both developmentally regulated genes showed sex-by-treatment effects: Dpysl3 was significantly up-regulated in MK-801–treated males compared with saline-treated males, but did not differ between female treatment groups, and Dnmt3a was significantly up-regulated in MK-801–treated females compared with saline-treated females, but did not differ between male treatment groups. This sex difference is consistent with the observed up-regulation of DNMT3A in adult female, but not male, schizophrenic patients (Fig. S2C and Dataset S5). Comparison of the P14 and P60 age groups showed significant up-regulation of Gata2 and down-regulation of Dpysl3 from P14 to P60. In contrast to the results presented in Fig. 3, Dnmt3a showed no difference between ages, a statistical effect driven by very high Dnmt3a expression in MK-801–treated adult females (Fig. S2D). Limited group size at P14 prevented further comparison of age × treatment × sex effects. Taken together, these results raise the possibility that the regulation of targets by miR-132 may be affected by both developmental stage and hormonal milieu, an aspect of miRNA activity that remains to be explored.
Alternate Animal Models of Disrupted NMDAR Signaling.
Global microRNA expression was also analyzed in two commonly used animal models of altered NMDAR signaling, the NR1 hypomorphic mouse (30) and nonhuman primates in withdrawal from chronic, low-dose phencyclidine (31). Surprisingly, neither model showed significant overlap with the results from the human SMRI samples. The data, and the implications of these results, are presented more thoroughly in the SI Materials and Methods (Dataset S7).
Discussion
We examined the expression of more than 800 miRNAs in DL-PFC tissue from a large cohort of control, schizophrenic, and bipolar subjects. Only one, miR-132, met rigorous statistical correction for diagnosis-associated expression. Of ∼200 bioinformatically predicted miR-132 targets, 26 were up-regulated in the PFC of schizophrenic subjects, including GATA2, PDE7B, ANKRD11, P250GAP, and FKBP2. We determined that expression of miR-132 in the PFC is regulated during development and by NMDAR signaling in the mouse. Finally, we demonstrated that several genes associated with neurodevelopment in mice and schizophrenia in humans, including DNMT3A, GATA2, and DPYSL3, are regulated by miR-132.
MicroRNA Dysregulation in Schizophrenia.
Early studies of a limited number (∼250) of miRNAs in cortical tissue from schizophrenia subjects showed little overlap. Perkins et al. identified 16 miRNAs with altered expression in schizophrenia (12), and Beveridge et al. found widespread alterations in miRNA processing and expression in samples from subjects diagnosed with schizophrenia (13, 22). No common miRNA was identified between reports, possibly because of the relatively small sample sizes, differences in brain regions, or differences in the method of RNA extraction. Our results show some overlap with those of Perkins et al., who found that miR-212, which is cotranscribed with miR-132, is down-regulated in the PFC (BA9) in schizophrenia subjects (12). Our data also overlap with recently published schizophrenia datasets, including data from Kim et al. (32) and Moreau et al. (33), who identified dysregulation of miR-132 and miR-212. MiR-181, which was identified as a schizophrenia-associated miRNA by Beveridge et al. (13), was significantly associated with the presence of psychotic symptoms in our dataset; notably, both datasets identified up-regulation of miR-181 in association with schizophrenia and psychotic symptoms. Furthermore, miR-449, which we identified as dysregulated in bipolar disorder, has been proposed as a blood biomarker for schizophrenia (34). Our data, combined with the existing literature, suggest that several miRNAs are reproducibly associated with schizophrenia and therefore represent candidate markers of the molecular pathology of schizophrenia.
Regulation of miR-132 Targets in the Adult Schizophrenic Brain.
MiR-132 has several known functions relevant to schizophrenia. First, miR-132 is enriched in the forebrain and is induced directly by activity-dependent CREB and ERK signaling (25). MiR-132 targets several genes associated with synaptic plasticity, including p250GAP and MECP2, and is predicted to regulate multiple members of neuronal activity-specific biological pathways, including long-term potentiation and long-term depression pathways and CREB signaling (20, 25, 28). We identified ∼25 genes predicted as miR-132 targets and significantly up-regulated in schizophrenia samples, including GATA2, PDE7B, and P250GAP. P250GAP regulates synaptic outgrowth (25), GATA2 is a transcription factor that regulates postmitotic neuronal differentiation (35), and PDE7B is a phosphodiesterase that has been genetically associated with schizophrenia by linkage mapping (36). These results indicate that miRNA and mRNA microarray data from human subjects can be combined to successfully identify biologically relevant miRNA targets and potentially therapeutic targets.
Regulation of miR-132 Expression by NMDAR Signaling and Neurodevelopmental Stage.
MiR-132 enhances activity-dependent synaptic plasticity through a positive feedback loop with the NMDAR: it is both induced by, and potentiates, NMDAR signaling (20, 27). We observed that pharmacologically inhibiting NMDA signaling in rodents, either chronically in the adult or during a critical postnatal developmental period, results in down-regulation of miR-132 expression and up-regulation of several miR-132 targets in the adult PFC. This process may set up a self-sustaining inhibitory cycle where reduced NMDAR activity in schizophrenia results in reduced miR-132 expression, leaving less miR-132 available to potentiate NMDA signaling. Surprisingly, NR1 hypomorphic mice do not show abnormalities in miR-132 expression (Dataset S7A), a finding that may be caused by the effects of strain background on miR-132 levels or the presence of the hypomorphic allele throughout the entirety of development. Additionally, NR1 hypomorphs lack a full complement of functional synaptic NMDARs; mice treated with NMDAR antagonists retain synaptic NMDARs in contact with internal cell signaling pathways (30). Nevertheless, this disparity requires further investigation.
MiR-132 expression exhibited a fourfold increase in the PFC between postnatal weeks 2 and 4, a period characterized by massive synaptic pruning, and is a strong candidate mechanism for producing neurodevelopmental susceptibility to schizophrenia requiring proper NMDAR signaling (6). Overlapping putative miR-132 target genes with developmentally regulated genes led to the confirmation of Dnmt3a and Dpysl3 as targets strongly down-regulated in the PFC during this period. Both genes contain miR-132 binding sites in their 3′ UTR, and show expression patterns that are the inverse of miR-132 expression. Dnmt3a is a de novo DNA methyltransferase in brain (37), and Dpysl3 is an NMDAR-regulated phosphoprotein involved in neurite outgrowth (38). Diminished miR-132 expression and resulting abnormal expression of Dpysl3 at this stage of development would be expected to negatively affect synaptic activity and outgrowth. These results suggest that miR-132 might play an important role in the pattern of gene expression in the PFC during adolescent neurodevelopment.
A second major process during late neurodevelopment is maturation of prefrontal GABAergic interneurons (39). GABAergic interneurons target excitatory pyramidal neurons, and the selective deletion of NR1 subunits in parvalbumin-positive neurons during postnatal development results in the appearance of schizophrenia-like behaviors in adulthood (40). Given the suggested role of miR-132 in potentiation of NMDAR signaling, abnormalities in miR-132 expression during this critical period may disrupt the molecular maturation of GABA interneurons. A core molecular characteristic of schizophrenia is reduced expression of the GABA-synthesizing enzyme GAD67, encoded by the GAD1 gene, and hypermethylation of schizophrenia risk genes, including GAD1 and REELIN, has been proposed as a cause of reduced GAD67 expression in schizophrenia (41). Notably, one of the miR-132 targets most strongly repressed during the period of peak miR-132 expression in adolescent mice is Dnmt3a, a DNA methyltransferase and confirmed miR-132 target. Reduced miR-132 levels in adolescents at risk for schizophrenia would be predicted to result in derepression of DNMT3A and subsequent hypermethylation of the genes targeted by DNMT3A. Our data do not prove a direct interaction between miR-132 and DNMT3A. However, we demonstrate that miR-132 regulates—directly or indirectly—RNA and protein levels of this important DNA methyltransferase. Furthermore, inhibition of NMDA signaling results in abnormal up-regulation of Dnmt3a expression in adulthood. The ultimate effect is still speculative, as the functions of the DNMT family members often overlap and may compensate for dysregulation of one family member.
Our results suggest that miR-132 down-regulation in the DL-PFC is a common molecular characteristic of schizophrenia and is associated with developmental and adult dysregulation of a number of miR-132 target genes, including p250GAP and DNMT3A. More broadly, other groups have reported that miRNA biogenesis is disrupted in patients with the 22q11 deletion, which causes schizophrenia-like symptoms (42), and a recent genome-wide association study report has linked miR-137 with schizophrenia (43). Although dysregulation of miR-137 has yet to be observed in schizophrenia, several of its potential biological functions overlap with those of miR-132, and the miR-132 target MECP2 regulates miR-137 expression (44). The ability of a single miRNA to coregulate the function of many genes within related signaling pathways suggests that dysregulation of miRNA expression has widespread biological consequences. Therefore, these recent studies linking miRNA dysregulation with schizophrenia have significant implications for both our understanding of the biological underpinnings of complex neuropsychiatric disorders and for development of novel, more effective therapeutics.
Materials and Methods
MiRNA microarray analysis was performed on 100 RNA samples from SMRI (Table S1). Subsequent qPCR analysis for miRNA and mRNA expression was performed on both SMRI and RNA from the HBB tissues. IPA and NextBio software were used for in silico analysis. The effect of miR-132 overexpression on three of the potential miR-132 targets identified above (Gata2, Dpyls3, Dnmt3a) was evaluated by 3′ UTR luciferase reporter assays, as previously described (28), or by Western blot analysis, using lentiviral constructs to transduce HEK293 cells with miR-132 overexpression or sponge cassettes. Animal experiments were performed on C57BL/6J mice from the Jackson Laboratory, or Sprague-Dawley rats from Taconic. Full experimental details are available in the SI Materials and Methods.
Supplementary Material
Acknowledgments
Postmortem brain tissue was donated by The Stanley Medical Research Institute and the Harvard Brain Tissue Resource Center. This work was funded by National Institute of Mental Health Grants 1R01MH08373301 (to C.W, and P.J.K.), F32MH084528 (to B.H.M), K99MH092321 (to B.H.M.), and 5R01MH057483 (to R.H.R).
Footnotes
Conflict of interest statement: R.J.K., T.A.L., S.D., and L.X. are employees of Pfizer Inc.; D.W. is the founder and owner of Ocean Ridge Biotechnology Inc.; M.S.L. is President and CSO of RxGen Inc.; and C.W. is a consultant for OPKO-CURNA.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1113793109/-/DCSupplemental.
References
- 1.Weinberger DR. Implications of normal brain development for the pathogenesis of schizophrenia. Arch Gen Psychiatry. 1987;44:660–669. doi: 10.1001/archpsyc.1987.01800190080012. [DOI] [PubMed] [Google Scholar]
- 2.Lewis DA, Levitt P. Schizophrenia as a disorder of neurodevelopment. Annu Rev Neurosci. 2002;25:409–432. doi: 10.1146/annurev.neuro.25.112701.142754. [DOI] [PubMed] [Google Scholar]
- 3.Rapoport JL, Addington AM, Frangou S, Psych MR. The neurodevelopmental model of schizophrenia: Update 2005. Mol Psychiatry. 2005;10:434–449. doi: 10.1038/sj.mp.4001642. [DOI] [PubMed] [Google Scholar]
- 4.Mei L, Xiong WC. Neuregulin 1 in neural development, synaptic plasticity and schizophrenia. Nat Rev Neurosci. 2008;9:437–452. doi: 10.1038/nrn2392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jaaro-Peled H, et al. Neurodevelopmental mechanisms of schizophrenia: Understanding disturbed postnatal brain maturation through neuregulin-1-ErbB4 and DISC1. Trends Neurosci. 2009;32:485–495. doi: 10.1016/j.tins.2009.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mirnics K, Middleton FA, Lewis DA, Levitt P. Analysis of complex brain disorders with gene expression microarrays: Schizophrenia as a disease of the synapse. Trends Neurosci. 2001;24:479–486. doi: 10.1016/s0166-2236(00)01862-2. [DOI] [PubMed] [Google Scholar]
- 7.Ross CA, Margolis RL, Reading SA, Pletnikov M, Coyle JT. Neurobiology of schizophrenia. Neuron. 2006;52:139–153. doi: 10.1016/j.neuron.2006.09.015. [DOI] [PubMed] [Google Scholar]
- 8.DeVito LM, et al. Serine racemase deletion disrupts memory for order and alters cortical dendritic morphology. Genes Brain Behav. 2011;10:210–222. doi: 10.1111/j.1601-183X.2010.00656.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Buchanan RW, et al. The Cognitive and Negative Symptoms in Schizophrenia Trial (CONSIST): The efficacy of glutamatergic agents for negative symptoms and cognitive impairments. Am J Psychiatry. 2007;164:1593–1602. doi: 10.1176/appi.ajp.2007.06081358. [DOI] [PubMed] [Google Scholar]
- 10.du Bois TM, Huang XF. Early brain development disruption from NMDA receptor hypofunction: Relevance to schizophrenia. Brain Res Brain Res Rev. 2007;53:260–270. doi: 10.1016/j.brainresrev.2006.09.001. [DOI] [PubMed] [Google Scholar]
- 11.Enomoto T, Noda Y, Nabeshima T. Phencyclidine and genetic animal models of schizophrenia developed in relation to the glutamate hypothesis. Methods Find Exp Clin Pharmacol. 2007;29:291–301. doi: 10.1358/mf.2007.29.4.1075358. [DOI] [PubMed] [Google Scholar]
- 12.Perkins DO, et al. microRNA expression in the prefrontal cortex of individuals with schizophrenia and schizoaffective disorder. Genome Biol. 2007;8:R27. doi: 10.1186/gb-2007-8-2-r27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Beveridge NJ, et al. Dysregulation of miRNA 181b in the temporal cortex in schizophrenia. Hum Mol Genet. 2008;17:1156–1168. doi: 10.1093/hmg/ddn005. [DOI] [PubMed] [Google Scholar]
- 14.Bartel DP. MicroRNAs: Target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lim LP, et al. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- 17.Kosik KS. The neuronal microRNA system. Nat Rev Neurosci. 2006;7:911–920. doi: 10.1038/nrn2037. [DOI] [PubMed] [Google Scholar]
- 18.Cheng LC, Pastrana E, Tavazoie M, Doetsch F. miR-124 regulates adult neurogenesis in the subventricular zone stem cell niche. Nat Neurosci. 2009;12:399–408. doi: 10.1038/nn.2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miller BH, Wahlestedt C. MicroRNA dysregulation in psychiatric disease. Brain Res. 2010;1338:89–99. doi: 10.1016/j.brainres.2010.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cheng HY, et al. microRNA modulation of circadian-clock period and entrainment. Neuron. 2007;54:813–829. doi: 10.1016/j.neuron.2007.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Purcell SM, et al. International Schizophrenia Consortium Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature. 2009;460:748–752. doi: 10.1038/nature08185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Beveridge NJ, Gardiner E, Carroll AP, Tooney PA, Cairns MJ. Schizophrenia is associated with an increase in cortical microRNA biogenesis. Mol Psychiatry. 2009;15:1176–1189. doi: 10.1038/mp.2009.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kapur S, VanderSpek SC, Brownlee BA, Nobrega JN. Antipsychotic dosing in preclinical models is often unrepresentative of the clinical condition: A suggested solution based on in vivo occupancy. J Pharmacol Exp Ther. 2003;305:625–631. doi: 10.1124/jpet.102.046987. [DOI] [PubMed] [Google Scholar]
- 24.Fatemi SH. Reelin mutations in mouse and man: From reeler mouse to schizophrenia, mood disorders, autism and lissencephaly. Mol Psychiatry. 2001;6:129–133. doi: 10.1038/sj.mp.4000129. [DOI] [PubMed] [Google Scholar]
- 25.Wayman GA, et al. An activity-regulated microRNA controls dendritic plasticity by down-regulating p250GAP. Proc Natl Acad Sci USA. 2008;105:9093–9098. doi: 10.1073/pnas.0803072105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Semeralul MO, et al. Microarray analysis of the developing cortex. J Neurobiol. 2006;66:1646–1658. doi: 10.1002/neu.20302. [DOI] [PubMed] [Google Scholar]
- 27.Nudelman AS, et al. Neuronal activity rapidly induces transcription of the CREB-regulated microRNA-132, in vivo. Hippocampus. 2010;20:492–498. doi: 10.1002/hipo.20646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Edbauer D, et al. Regulation of synaptic structure and function by FMRP-associated microRNAs miR-125b and miR-132. Neuron. 2010;65:373–384. doi: 10.1016/j.neuron.2010.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Grimson A, et al. MicroRNA targeting specificity in mammals: Determinants beyond seed pairing. Mol Cell. 2007;27:91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mohn AR, Gainetdinov RR, Caron MG, Koller BH. Mice with reduced NMDA receptor expression display behaviors related to schizophrenia. Cell. 1999;98:427–436. doi: 10.1016/s0092-8674(00)81972-8. [DOI] [PubMed] [Google Scholar]
- 31.Jentsch JD, Roth RH. The neuropsychopharmacology of phencyclidine: From NMDA receptor hypofunction to the dopamine hypothesis of schizophrenia. Neuropsychopharmacology. 1999;20:201–225. doi: 10.1016/S0893-133X(98)00060-8. [DOI] [PubMed] [Google Scholar]
- 32.Kim AH, et al. MicroRNA expression profiling in the prefrontal cortex of individuals affected with schizophrenia and bipolar disorders. Schizophr Res. 2010;124:183–191. doi: 10.1016/j.schres.2010.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moreau MP, Bruse SE, David-Rus R, Buyske S, Brzustowicz LM. Altered microRNA expression profiles in postmortem brain samples from individuals with schizophrenia and bipolar disorder. Biol Psychiatry. 2011;69:188–193. doi: 10.1016/j.biopsych.2010.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lai CY, et al. MicroRNA expression aberration as potential peripheral blood biomarkers for schizophrenia. PLoS ONE. 2011;6:e21635. doi: 10.1371/journal.pone.0021635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Alenina N, Bashammakh S, Bader M. Specification and differentiation of serotonergic neurons. Stem Cell Rev. 2006;2:5–10. doi: 10.1007/s12015-006-0002-2. [DOI] [PubMed] [Google Scholar]
- 36.Ikeda M, et al. Identification of novel candidate genes for treatment response to risperidone and susceptibility for schizophrenia: Integrated analysis among pharmacogenomics, mouse expression, and genetic case-control association approaches. Biol Psychiatry. 2010;67:263–269. doi: 10.1016/j.biopsych.2009.08.030. [DOI] [PubMed] [Google Scholar]
- 37.Feng J, et al. Dnmt1 and Dnmt3a maintain DNA methylation and regulate synaptic function in adult forebrain neurons. Nat Neurosci. 2010;13:423–430. doi: 10.1038/nn.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kowara R, Moraleja KL, Chakravarthy B. PLA(2) signaling is involved in calpain-mediated degradation of synaptic dihydropyrimidinase-like 3 protein in response to NMDA excitotoxicity. Neurosci Lett. 2008;430:197–202. doi: 10.1016/j.neulet.2007.10.036. [DOI] [PubMed] [Google Scholar]
- 39.Tseng KY, O'Donnell P. Dopamine modulation of prefrontal cortical interneurons changes during adolescence. Cereb Cortex. 2007;17:1235–1240. doi: 10.1093/cercor/bhl034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Belforte JE, et al. Postnatal NMDA receptor ablation in corticolimbic interneurons confers schizophrenia-like phenotypes. Nat Neurosci. 2010;13:76–83. doi: 10.1038/nn.2447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Guidotti A, et al. Decrease in reelin and glutamic acid decarboxylase67 (GAD67) expression in schizophrenia and bipolar disorder: A postmortem brain study. Arch Gen Psychiatry. 2000;57:1061–1069. doi: 10.1001/archpsyc.57.11.1061. [DOI] [PubMed] [Google Scholar]
- 42.Stark KL, et al. Altered brain microRNA biogenesis contributes to phenotypic deficits in a 22q11-deletion mouse model. Nat Genet. 2008;40:751–760. doi: 10.1038/ng.138. [DOI] [PubMed] [Google Scholar]
- 43.Ripke S, et al. Schizophrenia Psychiatric Genome-Wide Association Study (GWAS) Consortium Genome-wide association study identifies five new schizophrenia loci. Nat Genet. 2011;43:969–976. doi: 10.1038/ng.940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Szulwach KE, et al. Cross talk between microRNA and epigenetic regulation in adult neurogenesis. J Cell Biol. 2010;189:127–141. doi: 10.1083/jcb.200908151. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.