Abstract
The tyrosine kinase c-Src is activated in a large proportion of breast cancers, in which it is thought to play a key role in promoting the malignant phenotype. c-Src activity is also elevated in transgenic mouse models of breast cancer, including the widely used polyomavirus middle-T antigen (PyVmT) model, which provides an opportunity to study the importance of c-Src in mammary tumorigenesis. However, germline c-Src deletion in mammary epithelial and stromal compartments complicates the interpretation of in vivo tumorigenesis studies as a result of severe defects in mammary gland development. We have therefore engineered a mouse strain in which deletion of c-Src can be targeted to the mammary epithelium. We demonstrate that mammary epithelial disruption of c-Src impairs proliferation and tumor progression driven by PyVmT in vivo. Whereas related kinases substitute for c-Src in PyVmT signaling, c-Src ablation impairs cell cycle progression with decreased cyclin expression and elevated expression of cyclin-dependent kinase inhibitors. Our data indicate that c-Src has essential and unique functions in proliferation and tumor progression in this mouse model that may also be important in certain contexts in some human breast cancers.
Keywords: mammary tumor progression, transgenic models of breast cancer, tyrosine kinase signaling, tumor cell proliferation, conditional gene targeting
The tyrosine kinase c-Src is an important transducer of signals from receptor tyrosine kinases, integrins, and other receptors. By phosphorylating a wide range of substrates, activated c-Src controls many cellular processes including cytoskeletal remodeling, proliferation, and survival. Deregulation of this signaling can contribute to oncogenic transformation, and mutationally activated c-Src is well known as the first identified oncogene. Although such mutations are very rare in human cancer, clinical studies have shown elevated expression and activation of c-Src in human breast cancers (1) and other tumor types (2). In vitro data also support a crucial role for c-Src in human breast cancer cells. For example, genetic and biochemical analyses of ErbB2-overexpressing mammary tumor cells have demonstrated a direct interaction between the kinase domains of c-Src and ErbB2, leading to activated oncogenic signaling and disruption of epithelial polarity (3, 4).
Evidence for the importance of c-Src in mammary tumorigenesis also derives from transgenic mouse models of breast cancer. Mammary epithelial expression of constitutively activated c-Src in transgenic mice is weakly oncogenic, with induction of hyperplasias and scirrhous-type tumors after a long latency (5). However, endogenous c-Src is overexpressed and activated in mammary tumors from transgenic mice expressing polyomavirus middle-T antigen (PyVmT) or activated ErbB2 under the transcriptional control of the mouse mammary tumor virus (MMTV) promoter/enhancer (6, 7). Germline deletion of c-Src dramatically impairs mammary tumorigenesis in the MMTV-PyVmT tumor model (7). However, germline knockout of c-Src inhibits mammary ductal outgrowth (8), which could contribute to the observed impairment in tumorigenesis. Stromal c-Src deletion may also affect the MMTV-PyVmT/c-Src-null cross. For example, defects in cells of the macrophage/osteoclast lineage in the c-Src–null strain (9) may be important because loss of macrophage function blocks tumor progression in the MMTV-PyVmT model (10).
To clarify the role of mammary epithelial c-Src expression during in vivo tumorigenesis, we have used Cre-LoxP technology to develop a conditional knockout of c-Src. By using a mouse strain in which Cre recombinase is targeted specifically to the mammary epithelium (11), we have assessed the impact of mammary epithelial c-Src disruption on tumorigenesis in MMTV-PyVmT mice. Mammary epithelial c-Src ablation delayed tumor onset and initially decreased tumor burden. Significantly, tumors that developed in mice bearing conditional src alleles were derived from mammary epithelial cells that failed to express MMTV-Cre and therefore escaped Cre-mediated recombination. Premalignant lesions with c-Src deletion rarely progressed beyond the hyperplastic stage as a result of a failure to proliferate. Abolishing c-Src expression in cultured PyVmT tumor cells by using RNAi or Cre-LoxP strategies also impaired proliferation and restored epithelial cell–cell adhesion. c-Src was not required for PyVmT phosphorylation and associated signaling in these cells, but its silencing or deletion decreased cyclin expression and increased the expression and activity of cell cycle inhibitory proteins. These observations suggest that c-Src plays a critical role in oncogene-mediated transformation of the mammary epithelium by driving cell cycle progression.
Results
Conditional Deletion of c-Src in the Mouse Mammary Epithelium.
The conditional c-Src KO mouse was generated by using the strategy depicted in Fig. S1A and described in SI Materials and Methods. To target c-Src deletion specifically to mammary epithelial cells, mice with targeted src alleles (c-SrcL/L) were crossed with MMTV-Cre mice (11). By using PCR to detect the recombined allele, we confirmed that src deletion was specific to mammary glands in the presence of Cre recombinase (Fig. S1B). Ablation of c-Src protein in the mammary epithelium of c-SrcL/L/Cre mice was also confirmed by immunoblotting (Fig. S1C).
Germline deletion of c-Src severely delays mammary ductal outgrowth (8), although the requirements for c-Src in mammary epithelial cells and stromal cells are unclear. As developmental effects can confound tumorigenesis studies, we first assessed the impact of mammary epithelial c-Src deletion on the initial stages of mammary gland development. Whole-mount analysis of mammary glands from c-SrcL/L/Cre and WT/Cre (c-Src+/+/Cre) mice at 4, 6, 8, and 12 wk of age revealed no differences in branching and ductal outgrowth (Fig. S1D). To confirm functional expression of Cre, c-SrcL/L/Cre mice were crossed with mice bearing the GTRosa26 reporter allele (12). Staining whole-mounted mammary glands for β-gal activity confirmed Cre expression and hence c-Src deletion in the mature mammary epithelium (Fig. S1E). Therefore, c-Src expression in mammary epithelial cells is not required during the initial stages of mammary gland development.
Mammary Epithelial-Specific c-Src Ablation Impairs PyVmT-Driven Tumorigenesis.
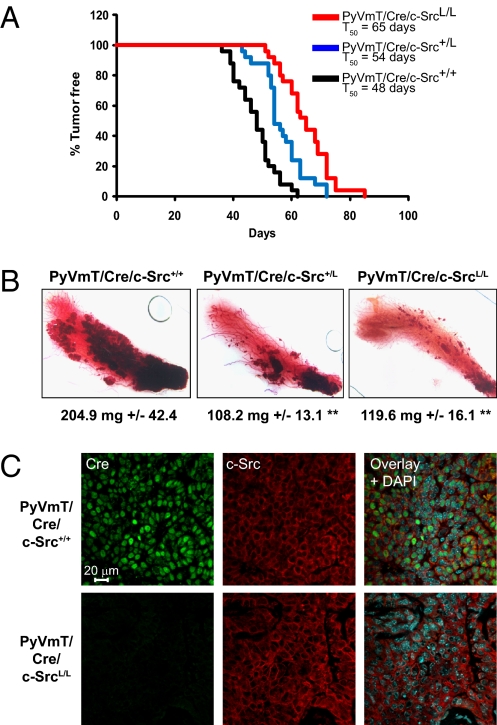
To study the role of mammary epithelial c-Src in tumorigenesis, the conditional c-Src strain was crossed with mice bearing MMTV-Cre and MMTV-PyVmT (13) transgenes, generating mice with WT src alleles (c-Src+/+/Cre/PyVmT) and one copy (c-Src+/L/Cre/PyVmT) or two copies of the conditional src allele (c-SrcL/L/Cre/PyVmT). Cohorts of 25 female animals were monitored for mammary tumor development by physical palpation. Conditional deletion of one or both src alleles modestly delayed tumor onset, with c-SrcL/L/Cre/PyVmT mice developing tumors 17 d later on average compared with controls (Fig. 1A). Conditional c-Src deletion also decreased tumor burden in 8-wk-old PyVmT mice as determined by whole-mount analyses of mammary glands (Fig. 1B).
Fig. 1.
Effect of mammary epithelial c-Src deletion on tumorigenesis in MMTV-PyVmT mice. (A) Tumor onset in PyVmT/Cre/c-Src+/+ (black), -c-Src+/L (blue), and -c-SrcL/L (red) mice. T50 is the age at which 50% of mice have palpable mammary tumors. (B) Representative whole mounts of inguinal mammary glands from 8-wk-old virgin PyVmT/Cre/c-Src+/+ (n = 7), -c-Src+/L (n = 5), and -c-SrcL/L (n = 7) mice. The average mass ± SEM is indicated below each image (**P < 0.01, unpaired Student t test vs. WT). (C) Expression of Cre and c-Src in sections of PyVmT/Cre/c-Src+/+ and PyVmT/Cre/c-SrcL/L tumors was determined by IF. Images are representative of at least five tumors.
In studies of conditional FAK or β1 integrin deletion in the MMTV-PyVmT model, tumors developed from mammary epithelial cells that did not express MMTV-Cre and therefore lacked recombination of the targeted alleles (14, 15). Consistent with this “escapee” phenomenon, mammary tumors from Cre/PyVmT mice bearing one or two floxed src alleles expressed levels of c-Src equivalent to controls (Fig. S1F), and Cre expression was detectable in c-Src+/+/Cre/PyVmT tumors but not in c-SrcL/L/Cre/PyVmT tumors by using immunofluorescence (IF; Fig. 1C). Tumors of both genotypes also expressed comparable levels of the related Src family kinases (SFKs) Fyn and c-Yes (Fig. S1G). Functional Cre expression was also undetectable in the lung metastases of SrcL/L/Cre/PyVmT mice by using the GTRosa26 reporter gene, but could be readily detected in the metastases of c-Src+/+/Cre/PyVmT mice (Fig. S2A). There were no differences in the incidence of lung metastases or average metastatic burden (Fig. S2B). These data indicate that tumors in SrcL/L/Cre/PyVmT mice developed from mammary epithelial cells that did not express Cre.
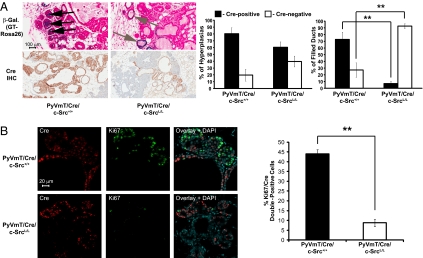
The MMTV-PyVmT model undergoes a well defined progression through distinct premalignant stages, with hyperplasia progressing to mammary intraepithelial neoplasia and then adenocarcinoma, mimicking human breast cancer progression (16). To study the role of c-Src in this process, we examined the formation and progression of premalignant mammary lesions by using IHC and the GTRosa26 Cre reporter. We identified a pronounced bias against Cre expression (and hence c-Src deletion) in premalignant lesions of mice with conditional src alleles, with only 2.8% of completely filled ducts expressing Cre, compared with more than 70% in WT controls (Fig. 2A). A bias against Cre expression in earlier, unfilled hyperplastic ducts was not statistically significant (Fig. 2A). These data suggest that c-Src deletion prevents PyVmT mammary tumor progression beyond the early hyperplastic stage. To determine whether this is due to impaired proliferation or increased cell death, we used IF to detect either Ki67 (a marker of proliferation) or cleaved caspase-3 (a marker of apoptosis) simultaneously with Cre. Although rare, we were able to identify a sufficient number of c-Src–null mammary lesions in 8-wk-old c-SrcL/L/Cre/PyVmT mice for quantitative analysis. These lesions contained significantly fewer Ki67/Cre double-positive cells than WT controls (Fig. 2B), and no significant differences in cleaved caspase-3/Cre double-positive cells were observed (Fig. S2C), suggesting that mammary epithelial c-Src deletion inhibits proliferation without inducing apoptosis. There were no evident differences in the localization of PyVmT in WT or c-Src–deficient hyperplastic cells (Fig. S2D). IF analysis confirmed the loss of c-Src protein in Cre-positive mammary epithelial cells of c-SrcL/L/Cre/PyVmT mice and also detected residual activated SFKs in mammary lesions lacking c-Src (Fig. S3 A and B). Fyn and c-Yes were expressed in WT and c-Src–deficient mammary lesions with no obvious differences (Fig. S3 C and D). Therefore, although related SFKs are expressed and activated in the absence of c-Src in vivo, they are unable to promote tumor progression.
Fig. 2.
c-Src deletion impairs MMTV-PyVmT mammary tumor progression by blocking proliferation. (A) Mammary glands from 8-wk-old PyVmT/Cre/c-Src+/+ (n = 4) and PyVmT/Cre/c-SrcL/L (n = 8) mice bearing the GTRosa26 Cre-reporter allele were stained for β-gal activity (Left, Upper) or mammary gland sections were stained with an anti-Cre antibody (Left, Lower). (Scale bars: 100 μm.) Bar graphs show the percentage of hyperplastic (Middle) and filled ducts (Right) with Cre IHC staining (error bars represent SEM; **P < 0.01, unpaired Student t test). (B) Left: IF using anti-Cre and -Ki67 antibodies on mammary gland sections from 8-wk-old PyVmT/Cre/c-Src+/+ (n = 5) and PyVmT/Cre/c-SrcL/L (n = 10) mice. (Scale bar: 20 μm.) Right: Quantification of Cre/Ki67 double-positive cells (error bars represent SEM; **P < 0.01, unpaired Student t test).
A recent study found that treatment of MMTV-PyVmT mice with a multikinase inhibitor targeting c-Src induced differentiation of mammary lesions and tumors, with altered expression of cytokeratins, ectopic expression of β-casein and increased E-cadherin localization to adherens junctions (17). H&E staining did not identify histological changes associated with differentiation in c-Src–deficient lesions, nor could we detect β-casein staining by IF (Fig. S4 A and B). All cells in WT and c-Src–deficient lesions were cytokeratin 8-positive, whereas cytokeratin 14 expression was sporadic, as previously reported (16) (Fig. S4 C and D). However, c-Src–deficient mammary lesions exhibited stronger membrane E-cadherin staining at regions of intercellular contact than WT controls (Fig. S4D). Therefore, although cell–cell adhesion may be affected by c-Src deletion in hyperplastic lesions, we find no further evidence for altered differentiation status.
Loss of c-Src Expression in PyVmT Mammary Tumor Cells Impairs Proliferation and Restores Cell–Cell Adhesion.
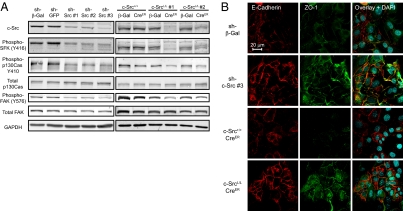
Because c-Src–null mammary tumors did not develop in transgenic mice, we used in vitro approaches to generate c-Src–deficient PyVmT tumor cells for further studies. Mammary tumor cells isolated from PyVmT/Src+/+ and PyVmT/c-SrcL/L mice were transduced with lentiviruses to express shRNAs targeting c-Src or retroviruses to express a CreER fusion protein. Lentiviruses encoding β-gal or GFP shRNAs or a retrovirus encoding β-gal were used as controls. Efficient silencing was achieved with three independent shRNAs targeting different regions of the c-Src mRNA (Fig. 3A). Two independent PyVmT/c-SrcL/L/CreER cell lines exhibited robust, tamoxifen-independent c-Src deletion, whereas no effect was observed in a PyVmT/Src+/+/CreER cell line (Fig. 3A and Fig. S5A). c-Src expression in PyVmT/c-SrcL/L/CreER cells recovered to control levels after several passages, indicating depletion of c-Src–null cells from the culture (Fig. S5B). Therefore, c-SrcL/L/CreER cell lines could not be continuously cultured and were used only at early passage. Consistent with loss of c-Src activity, tyrosine phosphorylation of FAK and p130Cas was reduced in c-Src–deficient cell lines (Fig. 3A). c-Src–deficient cells also formed colonies that were more epithelioid in appearance and exhibited a more localized pattern of E-cadherin and ZO-1 staining at sites of intercellular contact compared with controls (Fig. 3B). This suggests that the loss of c-Src in PyVmT tumor cells increases the formation of cell–cell junctions, an observation consistent with our in vivo findings and with published data on the role of c-Src in disrupting epithelial adhesion (18). As with c-Src–deficient mammary lesions in vivo, the pattern of PyVmT intracellular localization was not affected in c-Src–deficient cell lines (Fig. S5C).
Fig. 3.
c-Src silencing or ablation in PyVmT tumor cells decreases phosphorylation of Src targets and increases cell–cell adhesion. (A) Cell lines expressing c-Src shRNAs (Left) or CreER (Right) and relevant controls were lysed and immunoblotted for the indicated proteins. (B) IF staining was used to visualize the adherens junction marker E-cadherin and the tight junction marker ZO-1 in cell lines stably expressing c-Src shRNAs or CreER and relevant controls. (Scale bar: 20 μm.)
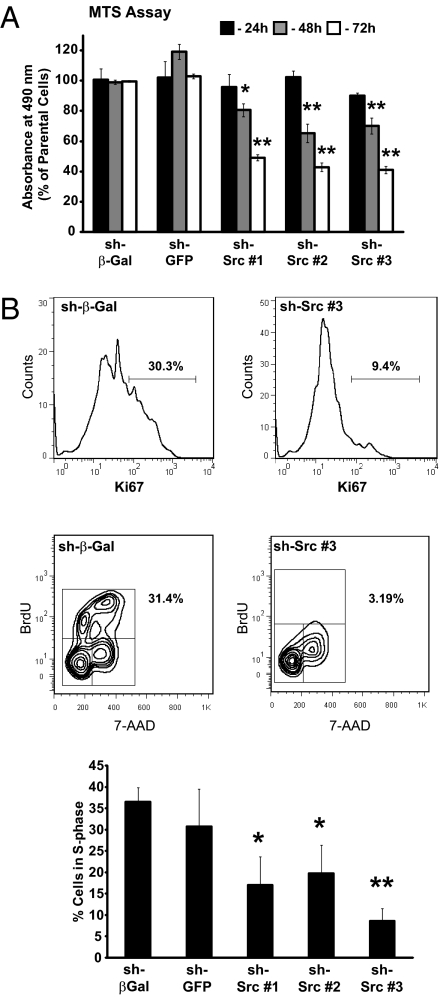
To study the requirement for c-Src in tumor cell proliferation in vitro, we used assays based on metabolism or nucleic acid content. c-Src–deficient cells exhibited a clear growth defect in both assays (Fig. 4A and Fig. S6 A and B). To characterize this phenotype further, cells were treated for 3 h with BrdU, stained with anti-Ki67 and anti-BrdU antibodies, and analyzed by flow cytometry to identify cells that had entered the cell cycle and progressed into S-phase. Deletion or silencing of c-Src reduced Ki67 expression and significantly decreased BrdU labeling (Fig. 4B and Fig. S6 C and D), suggesting that c-Src is required for cell cycle progression. Detection of cell surface annexin V staining by using flow cytometry revealed that c-Src silencing had no effect on apoptosis (Fig. S7A). However, annexin V staining was significantly increased by Cre-mediated c-Src deletion (Fig. S7B), suggesting that c-Src deletion in established PyVmT cells increases apoptosis. This may contribute to the loss of the c-Src–null population in these cell lines.
Fig. 4.
c-Src silencing in PyVmT tumor cells decreases proliferation and impairs cell cycle progression. (A) MTS proliferation assays (n = 3) were performed on c-Src–silenced or control cell lines. Data are normalized to values from parental cells (error bars represent SEM; *P < 0.05 and **P < 0.01, unpaired Student t test vs. sh-β-Gal control). (B) Upper: Flow cytometry analysis of Ki67 expression in c-Src–silenced cell lines and sh-β-Gal control. Data are representative of two independent experiments. The percentage of Ki67-positive cells is indicated. Lower: BrdU incorporation (y axis) and DNA content (7-AAD; x axis) in c-Src–silenced and control cell lines was determined by flow cytometry. The percentage of cells in S-phase (i.e., BrdU-positive) is indicated on the plots and in the bar graph (n = 3). Error bars represent SEM; *P < 0.05 and **P < 0.01, unpaired Student t test versus sh-β-Gal control.
To analyze further the ability of c-Src–deficient PyVmT cells to form mammary tumors in vivo, c-Src–deficient and control cells were injected into the mammary fat pad of immunocompromised mice. Tumor onset was significantly delayed in mice receiving c-Src–deficient cell lines (Fig. S8 A and B). However, when these tumors had formed, they grew at the same rate as in the controls. Tumors derived from c-Src–deficient cells had levels of c-Src expression equivalent to controls (Fig. S8C), indicating that these tumors developed from cells that did not undergo deletion or silencing of c-Src.
Silencing or Deletion of c-Src Inhibits Proliferation of PyVmT Tumor Cells by Altering the Expression of Cyclins and CDKIs.
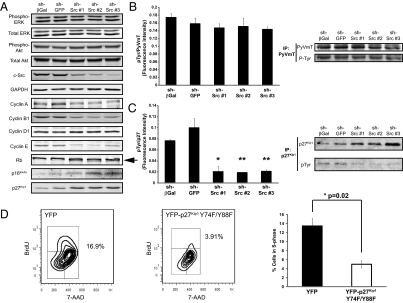
Tyrosine-phosphorylated PyVmT promotes proliferation by activating classical mitogenic signaling through the Ras-MAP kinase and PI3-kinase pathways (19, 20). As SFKs phosphorylate tyrosine residues on PyVmT, loss of PyVmT tyrosine phosphorylation may explain the proliferation defect in cells lacking c-Src. To test this hypothesis, we assessed the phosphorylation status of PyVmT, ERK, and Akt in c-Src–deficient and control cell lines. No differences in ERK or Akt phosphorylation were observed, suggesting that c-Src is dispensable for activation of the Ras-MAP kinase and PI3-kinase pathways in these cells (Fig. 5A and Fig. S9A). Quantitative analysis of PyVmT immunoprecipitates from control and c-Src–silenced cells revealed no differences in PyVmT tyrosine phosphorylation (Fig. 5B). We also observed similar levels of PyVmT tyrosine phosphorylation in cells with Cre-mediated c-Src deletion and controls (Fig. S9A). Interestingly, immunoblotting for Fyn and c-Yes revealed elevated expression of both kinases in c-Src–deficient cell lines (Fig. S9B). Immunoprecipitation experiments demonstrated enhanced activation of Fyn and c-Yes in c-Src–deficient cells, as well as an increase in their recruitment to PyVmT (Fig. S9B). This suggests that the requirement for c-Src in the proliferation of PyVmT tumor cells is not related to its ability to activate PyVmT by tyrosine phosphorylation, as other SFKs can substitute for c-Src in this capacity. Consistent with a continued dependence of PyVmT activation on SFK activity in the absence of c-Src, PyVmT tyrosine phosphorylation was reduced in both control and c-Src–deficient cells treated with SFK inhibitors (Fig. S9C).
Fig. 5.
c-Src silencing in PyVmT tumor cells does not affect PyVmT signaling but alters the expression of cell cycle regulators. (A) Lysates of c-Src–silenced or control PyVmT cell lines were immunoblotted for the indicated proteins. The arrow identifies the hypophosphorylated form of Rb. (B) PyVmT was immunoprecipitated and immunoblotting was performed with the indicated antibodies. The bar graph shows quantitative fluorescent immunoblotting (n = 3). Error bars represent SEM; no significant differences were observed (unpaired Student t test). (C) p27Kip1 was immunoprecipitated from cell lysates and immunoblotting was performed by using the indicated antibodies. The bar graph shows quantitative fluorescent immunoblotting (n = 3). Error bars represent SEM; *P < 0.05 and **P < 0.01, unpaired Student t test versus sh-β-Gal control. (D) BrdU incorporation (y axis) and DNA content (7-AAD; x axis) in YFP and YFP-p27Kip1 Y74F/Y88F-expressing cells was determined by flow cytometry. The percentage of cells in S-phase is indicated on the plots and in the bar graph (n = 3). Error bars are SEM; *P < 0.05, unpaired Student t test.
Because the cell cycle defect in c-Src–deficient PyVmT tumor cells cannot be explained by effects on the immediate events of PyVmT signaling, we examined the expression of key cell cycle regulators. The expression of A-, B-, and E-type cyclins was reduced in c-Src–deficient PyVmT cells, whereas cyclin D1 expression was unaffected (Fig. 5A and Fig. S9D). Strikingly, c-Src–deficient PyVmT cells expressed elevated levels of the cyclin-dependent kinase (CDK) inhibitor (CDKI) p27Kip1 and cells with stable c-Src silencing also expressed the CDKI p16Ink4a (Fig. 5A and Fig. S9D). Because c-Src can phosphorylate p27Kip1, reducing its CDK inhibitory function and promoting its degradation (21), we examined p27Kip1 tyrosine phosphorylation by immunoprecipitation and quantitative fluorescent immunoblotting. c-Src–silenced cells had significantly less tyrosine-phosphorylated p27Kip1 than controls (Fig. 5C). Consistent with a loss of CDK activity in c-Src–deficient cells, the faster-migrating, hypophosphorylated form of the CDK target retinoblastoma (Rb) was detectable in c-Src–silenced cells but not controls (Fig. 5A). To examine the role of p27Kip1 tyrosine phosphorylation in PyVmT tumor cells, we expressed a YFP-tagged mutant p27Kip1 with two major c-Src target sites mutated to phenylalanine (21) in primary cultures and analyzed cell cycle status by flow cytometry. Compared with the YFP-expressing control, cells transfected with p27Kip1 Y74F/Y88F incorporated significantly less BrdU (Fig. 5D) and expressed reduced levels of Ki67 (Fig. S9E), indicating a similar cell cycle block to that observed in c-Src–deficient PyVmT cells. These observations suggest that loss of c-Src expression in PyVmT mammary tumor cells interferes with proliferation by decreasing cyclin expression and up-regulating CDKIs, impairing CDK function and cell cycle progression.
Discussion
We have used a unique mouse model carrying a conditional src allele to identify a crucial role for c-Src in tumor progression in the MMTV-PyVmT model of breast cancer. Our approach has circumvented interpretational difficulties encountered in studies that used germline knockout of c-Src (7, 8). In particular, we have shown that mammary epithelial-specific c-Src disruption has no impact on mammary ductal outgrowth. However, loss of c-Src in the mammary epithelium delayed tumor onset and reduced tumor burden. Significantly, tumors and lung metastases in c-SrcL/L/Cre/PyVmT mice developed from an escapee population of mammary epithelial cells lacking Cre expression. In contrast to observations with conditional β1 integrin deletion (14), c-Src–deficient cells could form small hyperplastic lesions, although they exhibited a severe proliferative defect and very rarely progressed to form completely filled ducts. This suggests a critical role for c-Src in promoting the conversion of hyperplastic mammary epithelium to mammary intraepithelial neoplasia and subsequently adenocarcinoma. This is reminiscent of the phenotype caused by mammary epithelial deletion of FAK in the MMTV-PyVmT model (15). We also observed a reduced amount of tyrosine phosphorylated FAK in cultured PyVmT mammary tumor cells with loss of c-Src expression. These data are consistent with the role of FAK as an important downstream target of c-Src (22).
To examine the mechanism of c-Src function in PyVmT-driven proliferation, we used Cre-LoxP and RNAi approaches to generate c-Src–deficient cell lines. The requirement for c-Src in PyVmT-induced proliferation both in vivo and in vitro suggests that it plays a fundamental role in this process. Because c-Src can bind to PyVmT and phosphorylate key tyrosine residues required for signaling, we reasoned that a reduction in PyVmT tyrosine phosphorylation may be involved. Interestingly, we found no evidence that this was the case. One possible explanation is that other SFKs compensate in the early events of PyVmT signaling. In support of this idea, c-Src–deficient PyVmT cells express elevated levels of activated Fyn and, particularly, c-Yes. Our data suggest that PyVmT activation in c-Src–deficient cells becomes dependent on increased recruitment of these SFKs. This is consistent with the previous observation that c-Yes can account for a significant proportion of PyVmT-associated tyrosine kinase activity in c-Src–deficient mice in vivo, although it cannot fully compensate during mammary tumorigenesis (7).
To account for the proliferation defect in c-Src–deficient PyVmT tumor cells, we present evidence that loss of c-Src leads to reduced expression of cyclins A, B, and E and increased expression of CDKIs (p27Kip1 and p16Ink4a). By using quantitative biochemical analysis, we also demonstrate that p27Kip1 tyrosine phosphorylation is highly dependent on c-Src in PyVmT tumor cells. This may be crucial, as tyrosine phosphorylation of p27Kip1 is involved in the activation of p27Kip1–cyclin D1–Cdk4 complexes and releases inhibition of Cdk2 (21, 23), permitting phosphorylation and inactivation of Rb, synthesis of E-type cyclins, and progression into S-phase. Because the expression of A- and B-type cyclins occurs during late G2 and M phases, respectively (24), their depletion in c-Src–deficient cells may be a result of inefficient progression beyond the G1–S transition. This mechanism is supported by our finding that the Y74F/Y88F mutant of p27Kip1, which cannot be phosphorylated by c-Src (21), also blocks the proliferation of PyVmT cells. Loss of p27Kip1 phosphorylation may therefore at least partially cause the proliferation defect observed in c-Src–deficient tumor cells. Importantly, however, roles of other c-Src targets cannot be ruled out, presenting an intriguing area for further study. The expression of p16Ink4a in c-Src–deficient PyVmT cells is also interesting given the recent finding that a multikinase inhibitor targeting c-Src decreases the expression of the histone trimethylase EZH2 in PyVmT tumors (17). This epigenetic regulator mediates silencing of p16Ink4a expression (25) and also suppresses the formation of cell–cell junctions (26), which are restored in c-Src–deficient PyVmT cells both in vitro and in vivo.
Although PyVmT is a mouse-specific oncogene, the MMTV-PyVmT model does replicate important features of human breast cancer (13, 16). The data presented in this study therefore suggest that therapeutic targeting of c-Src may be of benefit in breast cancer. Notably, clinical trials of several SFK inhibitors have recently been conducted in patients with breast cancer. Although the results of these trials suggest that inhibitors of c-Src and related kinases are of modest clinical efficacy in the majority of late-stage breast cancers, the potential use of these drugs in patients with specific subtypes of breast cancer or in combination with other therapies remains a promising area of investigation (27).
Materials and Methods
Transgenic Mice.
The c-Src conditional knockout strategy is described in SI Materials and Methods. MMTV-PyVmT, MMTV-Cre, and GTRosa26 mice were described previously (11–13). Mammary tumor formation in MMTV-PyVmT mice was monitored by twice-weekly palpation. All experiments involving mice were conducted in accordance with McGill University animal care guidelines.
Mammary Gland Whole Mounts and β-Gal Assay.
Mammary glands (no. 4 inguinal) were fixed overnight in acetone. Glands were stained in hematoxylin for 24 h, destained in 70% ethanol/1% HCl, washed in 100% ethanol, dehydrated overnight in xylenes, and mounted using Permount (Fisher Scientific). For in situ β-gal assays, mammary glands were processed as described previously (14).
Histology and Immunostaining of Tissue Sections.
Tissues were fixed overnight in 10% neutral buffered formalin, paraffin-embedded, and sectioned at 4 μm. In situ β-gal staining of lung lobes and mammary glands was performed as described previously (14, 15). Pulmonary metastases were counted by microscopic analysis of H&E- or β-gal–stained sections. For IHC and IF, samples were processed as described previously (15) (SI Materials and Methods).
Primary Cell Culture, Transfection, and Viral Transduction.
Tumors at 8 wk after palpation were processed using a McIlwain tissue chopper (Mickle Laboratory Engineering), dissociated in collagenase B/Dispase II (Roche) for 2 h at 37 °C, washed with PBS solution plus 1 mM EDTA, and plated in DMEM supplemented with 2% FBS and Mammary Epithelial Growth Supplement (Invitrogen). CreER and β-gal in the retroviral vector pQxi were used for virus production in 293VSV cells [a gift of Alain Nepveu (Goodman Cancer Research Center, McGill University, Montreal, Quebec, Canada)]. and subsequent transductions as previously described (28). Lentiviral shRNAs targeting c-Src, β-gal, or GFP (Open Biosystems) were used according to the manufacturer's instructions. Transduced cell lines were selected and maintained in DMEM plus Mammary Epithelial Growth Supplement with 2 μg/mL puromycin (Sigma). The p27Kip1 Y74F/Y88F mutant was a gift of Joyce Slingerland (Sylvester Comprehensive Cancer Center, University of Miami, Miami, FL). Cells were transfected with pEYFP control or pEYFP-p27Kip1 Y47F/Y88F using NanoJuice (Merck) according to the manufacturer's instructions.
Proliferation Assays.
CyQuant (Invitrogen) and CellTiter Aqueous MTS (Promega) proliferation assays were performed according to the manufacturer's protocols using 1,000 (Cyquant) or 2,500 (CellTiter) cells per well in 96-well optical-bottom plates (Nunc).
Mammary Fat Pad Injection Assay.
Stable cell lines (2.5 × 105 cells in PBS solution) were injected into the no. 4 inguinal mammary fat pad of athymic nude (NCr) mice (Taconic). Mice were monitored twice weekly for tumor formation by palpation. Tumor growth was assessed by weekly caliper measurements.
Immunoprecipitation and Immunoblotting.
Immunoprecipitation and immunoblotting were performed as described previously (4), with fluorescent detection using the Odyssey imaging system (Li-COR Biosciences) according to the manufacturer's protocols. Immunoprecipitation was performed by using 5 μg antibody per milligram total protein. Primary antibodies were Akt1, Phospho-Akt S473, phospho-p130Cas, ERK, Phospho-ERK, and Phospho-Src from Cell Signaling; p130Cas, Cyclin B1, FAK, and Rb from BD Biosciences; cyclin A, Grb2, p16, p27, and c-Yes from Santa Cruz; Phospho-FAK Y576, Fyn, P-Tyr, c-Src, and Vinculin from Millipore; Cyclin D1 from Invitrogen; Cyclin E from Abcam and GAPDH from Novus.
IF.
IF staining of cells on glass coverslips was performed as described previously (4) and is detailed in SI Materials and Methods.
Flow Cytometry.
The FITC and APC BrdU Flow Kits, PE-Ki67 antibody, and PE-Annexin V Apoptosis Detection Kit (BD Biosciences) were used according to the manufacturer's instructions. Cells were analyzed on an LSR II flow cytometer (BD Biosciences).
Supplementary Material
Acknowledgments
We thank Bill Papavasiliou and Cynthia Lavoie for technical assistance, Ken McDonald and Mathieu Tremblay for assistance with flow cytometry analysis, and Janice Penney and Michelle Read for generation of transgenic mice. This study was supported by a fellowship from the US Department of Defense (to H.W.S.), a Canadian Institutes of Health Research Frederick Banting and Charles Best Canada Graduate scholarship (to R.V.M.), Canada Research Chair in molecular oncology (W.J.M.), a Canadian Breast Cancer Research Alliance grant, and National Institutes of Health Grant 5P01GA-099031-05.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. K.P. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1018861108/-/DCSupplemental.
References
- 1.Ottenhoff-Kalff AE, et al. Characterization of protein tyrosine kinases from human breast cancer: Involvement of the c-src oncogene product. Cancer Res. 1992;52:4773–4778. [PubMed] [Google Scholar]
- 2.Kim LC, Song L, Haura EB. Src kinases as therapeutic targets for cancer. Nat Rev Clin Oncol. 2009;6:587–595. doi: 10.1038/nrclinonc.2009.129. [DOI] [PubMed] [Google Scholar]
- 3.Kim H, et al. The c-Src tyrosine kinase associates with the catalytic domain of ErbB-2: Implications for ErbB-2 mediated signaling and transformation. Oncogene. 2005;24:7599–7607. doi: 10.1038/sj.onc.1208898. [DOI] [PubMed] [Google Scholar]
- 4.Marcotte R, Zhou L, Kim H, Roskelly CD, Muller WJ. c-Src associates with ErbB2 through an interaction between catalytic domains and confers enhanced transforming potential. Mol Cell Biol. 2009;29:5858–5871. doi: 10.1128/MCB.01731-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Webster MA, Cardiff RD, Muller WJ. Induction of mammary epithelial hyperplasias and mammary tumors in transgenic mice expressing a murine mammary tumor virus/activated c-src fusion gene. Proc Natl Acad Sci USA. 1995;92:7849–7853. doi: 10.1073/pnas.92.17.7849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Muthuswamy SK, Siegel PM, Dankort DL, Webster MA, Muller WJ. Mammary tumors expressing the neu proto-oncogene possess elevated c-Src tyrosine kinase activity. Mol Cell Biol. 1994;14:735–743. doi: 10.1128/mcb.14.1.735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guy CT, Muthuswamy SK, Cardiff RD, Soriano P, Muller WJ. Activation of the c-Src tyrosine kinase is required for the induction of mammary tumors in transgenic mice. Genes Dev. 1994;8:23–32. doi: 10.1101/gad.8.1.23. [DOI] [PubMed] [Google Scholar]
- 8.Kim H, Laing M, Muller W. c-Src-null mice exhibit defects in normal mammary gland development and ERalpha signaling. Oncogene. 2005;24:5629–5636. doi: 10.1038/sj.onc.1208718. [DOI] [PubMed] [Google Scholar]
- 9.Lowe C, et al. Osteopetrosis in Src-deficient mice is due to an autonomous defect of osteoclasts. Proc Natl Acad Sci USA. 1993;90:4485–4489. doi: 10.1073/pnas.90.10.4485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lin EY, Nguyen AV, Russell RG, Pollard JW. Colony-stimulating factor 1 promotes progression of mammary tumors to malignancy. J Exp Med. 2001;193:727–740. doi: 10.1084/jem.193.6.727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Andrechek ER, et al. Amplification of the neu/erbB-2 oncogene in a mouse model of mammary tumorigenesis. Proc Natl Acad Sci USA. 2000;97:3444–3449. doi: 10.1073/pnas.050408497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Soriano P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat Genet. 1999;21:70–71. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
- 13.Guy CT, Cardiff RD, Muller WJ. Induction of mammary tumors by expression of polyomavirus middle T oncogene: A transgenic mouse model for metastatic disease. Mol Cell Biol. 1992;12:954–961. doi: 10.1128/mcb.12.3.954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.White DE, et al. Targeted disruption of beta1-integrin in a transgenic mouse model of human breast cancer reveals an essential role in mammary tumor induction. Cancer Cell. 2004;6:159–170. doi: 10.1016/j.ccr.2004.06.025. [DOI] [PubMed] [Google Scholar]
- 15.Lahlou H, et al. Mammary epithelial-specific disruption of the focal adhesion kinase blocks mammary tumor progression. Proc Natl Acad Sci USA. 2007;104:20302–20307. doi: 10.1073/pnas.0710091104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maglione JE, et al. Transgenic Polyoma middle-T mice model premalignant mammary disease. Cancer Res. 2001;61:8298–8305. [PubMed] [Google Scholar]
- 17.Hebbard L, et al. Control of mammary tumor differentiation by SKI-606 (bosutinib) Oncogene. 2010;30:301–312. doi: 10.1038/onc.2010.412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Avizienyte E, et al. Src-induced de-regulation of E-cadherin in colon cancer cells requires integrin signalling. Nat Cell Biol. 2002;4:632–638. doi: 10.1038/ncb829. [DOI] [PubMed] [Google Scholar]
- 19.Schaffhausen BS, Roberts TM. Lessons from polyoma middle T antigen on signaling and transformation: A DNA tumor virus contribution to the war on cancer. Virology. 2009;384:304–316. doi: 10.1016/j.virol.2008.09.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Webster MA, et al. Requirement for both Shc and phosphatidylinositol 3′ kinase signaling pathways in polyomavirus middle T-mediated mammary tumorigenesis. Mol Cell Biol. 1998;18:2344–2359. doi: 10.1128/mcb.18.4.2344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chu I, et al. p27 phosphorylation by Src regulates inhibition of cyclin E-Cdk2. Cell. 2007;128:281–294. doi: 10.1016/j.cell.2006.11.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mitra SK, Schlaepfer DD. Integrin-regulated FAK-Src signaling in normal and cancer cells. Curr Opin Cell Biol. 2006;18:516–523. doi: 10.1016/j.ceb.2006.08.011. [DOI] [PubMed] [Google Scholar]
- 23.Larrea MD, et al. Phosphorylation of p27Kip1 regulates assembly and activation of cyclin D1-Cdk4. Mol Cell Biol. 2008;28:6462–6472. doi: 10.1128/MCB.02300-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Malumbres M, Barbacid M. Cell cycle, CDKs and cancer: A changing paradigm. Nat Rev Cancer. 2009;9:153–166. doi: 10.1038/nrc2602. [DOI] [PubMed] [Google Scholar]
- 25.Bracken AP, et al. The Polycomb group proteins bind throughout the INK4A-ARF locus and are disassociated in senescent cells. Genes Dev. 2007;21:525–530. doi: 10.1101/gad.415507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Herranz N, et al. Polycomb complex 2 is required for E-cadherin repression by the Snail1 transcription factor. Mol Cell Biol. 2008;28:4772–4781. doi: 10.1128/MCB.00323-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mayer EL, Krop IE. Advances in targeting SRC in the treatment of breast cancer and other solid malignancies. Clin Cancer Res. 2010;16:3526–3532. doi: 10.1158/1078-0432.CCR-09-1834. [DOI] [PubMed] [Google Scholar]
- 28.Ory DS, Neugeboren BA, Mulligan RC. A stable human-derived packaging cell line for production of high titer retrovirus/vesicular stomatitis virus G pseudotypes. Proc Natl Acad Sci USA. 1996;93:11400–11406. doi: 10.1073/pnas.93.21.11400. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.